Abstract
For improvement of tolerance to oxidative stress in Bifidobacterium longum 105-A, we introduced the Bacillus subtilis catalase gene (katE) into it. The transformant showed catalase activity (39 U/mg crude protein) in the intracellular fraction, which increased survival by ∼100-fold after a 1-h exposure to 4.4 mM H2O2, decreased de novo H2O2 accumulation, and increased survival in aerated cultures by 105-fold at 24 h. The protection level was better than that conferred by exogenously added catalase.
TEXT
As probiotics, some species of Bifidobacterium are widely used in various therapeutics (11, 19) and food products (9, 10, 27). To be effective, bifidobacteria must remain viable until reaching the intestinal tract. However, as bifidobacteria are obligate anaerobic bacteria, their sensitivity to O2 limits their survival and use in industrial applications (14, 24).
Superoxide anion (O2−) is a biologically quite toxic component (7). It produced by a one-electron reduction of dioxygen, which occurs widely in nature under aerobic conditions. The aerobic organisms express superoxide dismutase, which decomposes O2− and produces H2O2 (3, 7). In the presence of ferrous iron(II) or some other transition metals, H2O2 is converted to an OH radical by the Fenton reaction, which shows the highest toxicity among active oxygen (2, 3, 6). Thus, H2O2 decomposition is one of the most important processes in cell survival under aerobic conditions. Bifidobacteria also have an oxidase function which enables them to use O2 as an electron acceptor to reduce to H2O and H2O2 under aerobic conditions (5, 16, 21).
Eukaryotes have catalase and glutathione peroxidase (GPX) as an H2O2-decomposing system to survive under aerobic conditions (1, 3, 13). Little is known about GPX activity in prokaryotes, and GPX gene and glutathione synthesis pathways have not been detected in obligate anaerobes, including bifidobacteria (13, 20, 23). Catalase is an enzyme commonly found in aerobes and facultative anaerobes but is absent in almost all obligate anaerobes, including bifidobacteria (1, 3). NADH peroxidase was found in bifidobacteria, and some types of peroxidases were predicted, including thiol peroxidase, alkyl hydroperoxide reductase, and peptide methionine sulfoxide reductase. However, these peroxidases are unable to decompose the H2O2 produced by bifidobacteria under aerobic conditions (23, 26).
Catalases are classified in 2 groups, depending on the type of metal cofactors: heme-dependent catalase and manganese-dependent catalase (3, 7). B. subtilis KatE (heme-dependent catalase) is a well-known catalase that is used to improve the viability of some species of bacteria via heterologous expression (4, 18, 22). In this study, to reduce the toxicity from active oxygen, we therefore investigated the effects of expressing B. subtilis heme-dependent catalase on the oxidative stress resistance of Bifidobacterium longum 105-A. For comparison, the effects of exogenously added catalase on B. longum were also tested.
To express katE of B. subtilis, we constructed an expression vector, pBCAT001, derived from plasmid pKKT427 (28), by inserting the B. subtilis KatE gene within the promoter and terminator of hup of B. longum and then introduced it into B. longum 105-A and Escherichia coli UM255 (by a method described in the supplemental material; also see Table S1 and Fig. S1 in the supplemental material). To determine the activity of KatE, hemin (10 μM) (Sigma-Aldrich, St. Louis, MO) was added to the medium because bifidobacteria do not synthesize heme (23) (see Table S2 in the supplemental material). We analyzed the crude extracts from E. coli UM255(pKKT427, pBCAT001) and B. longum 105-A(pKKT427, pBCAT001) by SDS-PAGE. The 77-kDa band corresponding to B. subtilis KatE was clearly identified in E. coli UM255(pBCAT001) but was not identifiable in B. longum 105-A(pBCAT001) because its expression level was lower than 1/13 of that in E. coli UM255 (see Fig. S2 in the supplemental material). We considered this weak expression was caused by the low plasmid copy number (≈10 copies per cell), transcription, and translation strengths (12, 25). The catalase activity was examined by detecting bubble (O2) formation upon the addition of 30% H2O2 to the cell pellet. Catalase activity was only detected in the cell fraction—39 U/mg crude protein in B. longum 105-A(pBCAT001) versus less than 0.1 U/mg in B. longum 105-A(pKKT427)—under anaerobic conditions (optical density at 660 nm [OD660] of 1.0), whereas the extracellular catalase activity was less than 0.4%.
We investigated the effect of KatE on the short-term H2O2 tolerance of B. longum 105-A. The survival rates of B. longum 105-A(pKKT427 pBCAT001) were determined by incubating cultures for 1 h in MRS medium with 4.4 mM H2O2 at 37°C. The survival rates of B. longum 105-A(pBCAT001) during the exponential and stationary phases were significantly increased by 120- and 103-fold, respectively, compared to that of B. longum 105-A(pKKT427) (Table 1). The data in Table 1 demonstrate that exponential-phase cells were more sensitive to H2O2 than stationary-phase cells.
Table 1.
Numbers of CFU/ml and survival rates after H2O2 exposure for 1 h
| Growth phase (OD660) | Strain | CFU/ml after exposure toa: |
Survival rateb | Fold survival rate increasec | |
|---|---|---|---|---|---|
| 0 mM H2O2 | 4.4 mM H2O2 | ||||
| Exponential (0.6) | B. longum 105-A(pKKT427) | (7.00 ± 0.34) × 108 | (1.20 ± 0.20) × 105 | 0.00017 | |
| B. longum 105-A(pBCAT001) | (8.00 ± 0.16) × 108 | (1.64 ± 0.26) × 107 | 0.021 | 120 | |
| Stationary (1.0) | B. longum 105-A(pKKT427) | (1.68 ± 0.22) × 109 | (2.10 ± 0.40) × 106 | 0.0013 | |
| B. longum 105-A(pBCAT001) | (1.56 ± 0.34) × 109 | (2.00 ± 0.14) × 108 | 0.13 | 103 | |
Data are the means ± standard deviations from three independent experiments.
Survival rates were determined by comparing the colony counts for exposure to 0 versus 4.4 mM H2O2 for 1 h at 37°C.
Survival rate increases were compared for B. longum 105-A(pBCAT001) and B. longum 105-A(pKKT427).
We also investigated the physiology of B. longum 105-A under aerobic conditions. B. longum 105-A(pKKT427, pBCAT001) reached a maximum growth rate at 12 h after inoculation under anaerobic conditions. The growth of B. longum 105-A(pBCAT001) was partially inhibited, and that of B. longum 105-A(pKKT427) was nearly stopped under aerobic culture conditions (Fig. 1A). To measure growth rates, cells were cultured and then plate counted. Although most of the B. longum 105-A(pKKT427) specimens survived 12 h of aerobic culture, cell growth began to sharply decrease and became almost unculturable after 24 h in aerobic culture. However, B. longum 105-A(pBCAT001) exhibited a high rate of survival (1 × 107 CFU/ml) at 24 h and only became unculturable after 48 h under aerobic conditions (Fig. 1B). The results demonstrate that the presence of KatE protected B. longum 105-A from aerobic culture-induced death.
Fig 1.
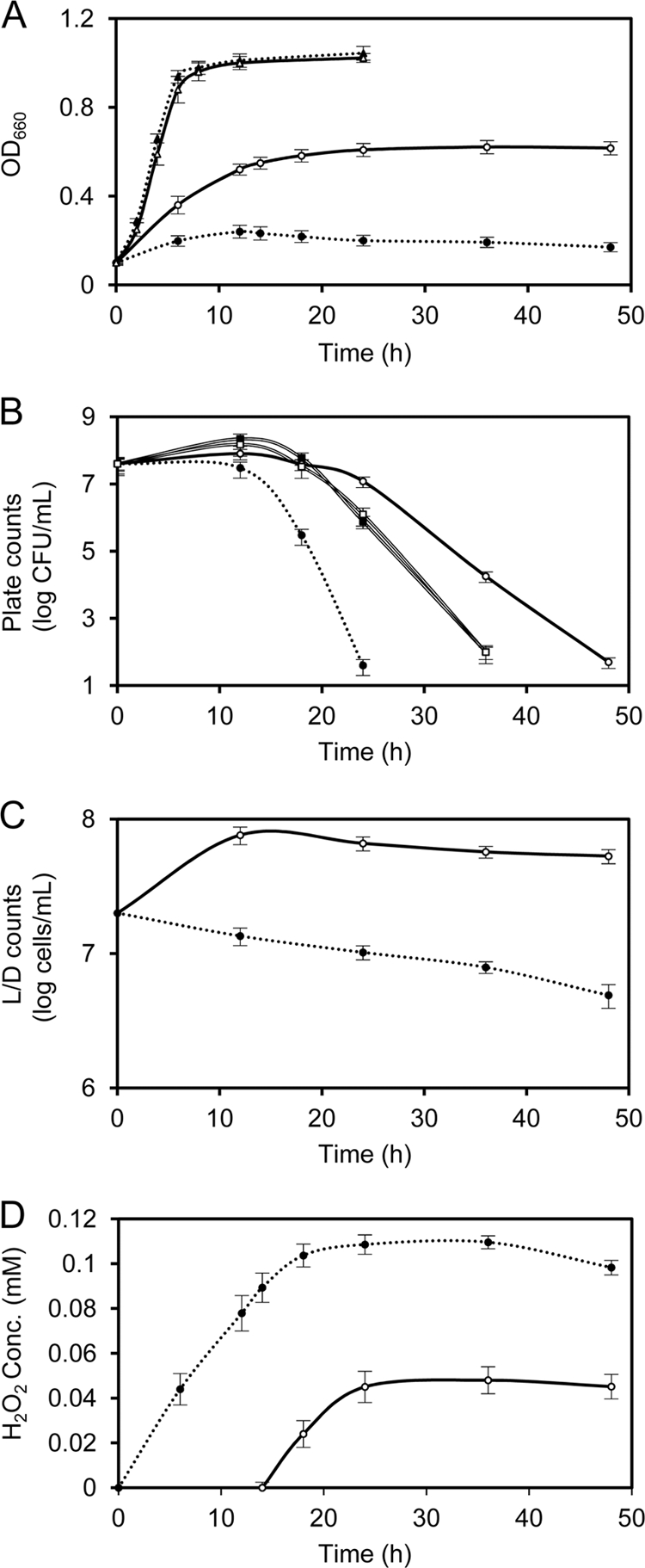
Cultured B. longum 105-A(pKKT427 or pBCAT001) under aerobic or anaerobic conditions. (A) Growth (OD660) of the cultured B. longum 105-A strain. (B) Counts of B. longum 105-A cultured under aerobic conditions. (C) LIVE/DEAD (L/D) assay of B. longum 105-A cultured under aerobic conditions. (D) H2O2 accumulation by B. longum 105-A cultured under aerobic conditions. Dotted line, B. longum 105-A(pKKT427); solid line, B. longum 105-A(pBCAT001); solid circles, B. longum 105-A(pKKT427) cultured under aerobic conditions; open circles, B. longum 105-A(pBCAT001) cultured under aerobic conditions; solid triangles, B. longum 105-A(pKKT427) cultured under anaerobic conditions; open triangles, B. longum 105-A(pBCAT001) cultured under anaerobic conditions; solid squares, B. longum 105-A(pKKT427) cultured using exogenously added catalase (3,000 U/ml medium) under aerobic conditions; open squares, B. longum 105-A(pKKT427) cultured using exogenously added catalase (100 U/ml medium) under aerobic conditions. The results presented correspond to the averages of three different assays. Error bars correspond to the standard errors of the mean values.
The concomitant generation of H2O2 was also measured (Fig. 1D). The accumulation of H2O2 in B. longum 105-A(pKKT427) increased for 18 h and peaked at 0.1 mM. H2O2 was scavenged by the genetically expressed catalase during the exponential phase, and it did not begin to accumulate in the medium of B. longum 105-A(pBCAT001) until the stationary phase. At this time, the cells became unculturable (Fig. 1B). Interestingly, the decrease in the growth of B. longum 105-A(pKKT427) was faster than that of B. longum 105-A(pBCAT001). This might be because the concentration of H2O2 in B. longum 105-A(pKKT427) was 2.4-fold higher than that of B. longum 105-A(pBCAT001) in aerated cultures. B. longum 105-A(pBCAT001) survived longer due to the increased period of time in which H2O2 had not accumulated. This difference in growth rates suggests that H2O2 was primarily responsible for B. longum 105-A becoming unculturable under aerobic conditions.
Lahtinen et al. reported that B. longum lost culturability quickly during storage, but the cells still maintained intact membranes (17). H2O2 is known to damage DNA and protein (1, 3, 7, 8); however, it is unknown whether H2O2 can easily damage the B. longum membrane. Therefore, we investigated whether it was possible that B. longum 105-A cells lost their culturability but maintained an intact membrane. These experiments were conducted using the LIVE/DEAD BacLight bacterial viability kit (L/D; Invitrogen). After 24 h in aerobic culture, B. longum 105-A containing pKKT427 remained relatively stable, and 1 × 107 “viable” cells/ml were maintained (Fig. 1C); however, the survival decreased to 1 × 101 to 1 × 102 CFU/ml (Fig. 1B). Based on this information, we were only able to make the decision that the cells had intact membranes, but it is still unknown whether the cells were dead. To confirm whether cells maintain viability, further studies are needed, such as examining the synthesis of DNA, RNA, and protein.
Some studies reported that adding exogenous catalase to the liquid medium improved aerobic growth of bifidobacteria (5, 15). Because H2O2 readily diffuses across cell membranes but exogenously added catalase cannot penetrate cell membranes (1, 2), we therefore compared the culturable B. longum 105-A strain protected by catalase expression with the culturable B. longum 105-A strain protected by the addition of exogenous catalase. Although the counts of B. longum 105-A(pKKT427) recovered when cultured under aerobic conditions with exogenously added catalase from bovine liver (100 and 3,000 U/ml medium) (C1345-1G; Sigma), the counts of B. longum 105-A(pKKT427) were similar regardless of the concentration of added catalase (Fig. 1). Interestingly, the counts of B. longum 105-A protected by addition of exogenous catalase were nearly identical to those of B. longum 105-A(pBCAT001) when aerobically cultured for 18 h, although the concentrations of exogenously added catalase were much higher than the levels of expressed catalase. B. longum 105-A that was protected by exogenously added catalase was unculturable after 36 h in aerated culture; however, B. longum 105-A(pBCAT001) did not become unculturable until 48 h in aerated culture. These results indicate that B. longum maintained intact cell membranes, whereas induced exogenously added catalase eliminated extracellular H2O2 but was unable to eliminate intracellular H2O2.
In conclusion, we have successfully expressed B. subtilis KatE in B. longum 105-A. The KatE-expressed transformant was able to grow and survive under aerobic conditions. This finding revealed that H2O2 accumulation is a primary factor of growth inhibition in bifidobacteria. The addition of catalase to the medium also protects bifidobacteria from oxidative stress; however, this effect was weaker than that of heterologously expressed catalase. To our knowledge, this is the first report on protecting bifidobacteria from oxidative stress by heterologous expression of catalase.
Supplementary Material
ACKNOWLEDGMENTS
This research was partially supported by the Ministry of Education, Culture, Sports, Science, and Technology in Japan [Grant-in Aid for Scientific Research (C) 20510189] and the C19 Kiyomi Yoshizaki research grant.
Footnotes
Published ahead of print 3 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Beckman JS, et al. 1988. Superoxide dismutase and catalase conjugated to polyethylene glycol increases endothelial enzyme activity and oxidant resistance. J. Biol. Chem. 263:6884–6892 [PubMed] [Google Scholar]
- 2. Behl C, Davis JB, Lesley R, Schubert D. 1994. Hydrogen peroxide mediates amyloid beta protein toxicity. Cell 77:817–827 [DOI] [PubMed] [Google Scholar]
- 3. Brioukhanov AL, Netrusov AI. 2004. Catalase and superoxide dismutase: distribution, properties, and physiological role in cells of strict anaerobes. Biochemistry (Mosc.) 69:949–962 [DOI] [PubMed] [Google Scholar]
- 4. Chen L, Keramati L, Helmann JD. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. U. S. A. 92:8190–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. de Vries W, Stouthamer AH. 1969. Factors determining the degree of anaerobiosis of Bifidobacterium strains. Arch. Mikrobiol. 65:275–287 [DOI] [PubMed] [Google Scholar]
- 6. Fenton HJH. 1894. Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 65:899–910 [Google Scholar]
- 7. Fridovich I. 1998. Oxygen toxicity: a radical explanation. J. Exp. Biol. 201:1203–1209 [DOI] [PubMed] [Google Scholar]
- 8. Fridovich I. 1995. Superoxide radical and superoxide dismutases. Annu. Rev. Biochem. 64:97–112 [DOI] [PubMed] [Google Scholar]
- 9. Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. 2001. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am. J. Clin. Nutr. 74:833–839 [DOI] [PubMed] [Google Scholar]
- 10. Guyonnet D, et al. 2007. Effect of a fermented milk containing Bifidobacterium animalis DN-173 010 on the health-related quality of life and symptoms in irritable bowel syndrome in adults in primary care: a multicentre, randomized, double-blind, controlled trial. Aliment. Pharmacol. Ther. 26:475–486 [DOI] [PubMed] [Google Scholar]
- 11. Hamaji Y, et al. 2007. Strong enhancement of recombinant cytosine deaminase activity in Bifidobacterium longum for tumor-targeting enzyme/prodrug therapy. Biosci. Biotechnol. Biochem. 71:874–883 [DOI] [PubMed] [Google Scholar]
- 12. He J., Sakaguchi K., Suzuki T. 2012. Determination of the ribosome-binding sequence and spacer length between binding site and initiation codon for efficient protein expression in Bifidobacterium longum 105-A. J. Biosci. Bioeng., 113:442–444 [DOI] [PubMed] [Google Scholar]
- 13. Herbette S, Roeckel-Drevet P, Drevet JR. 2007. Seleno-independent glutathione peroxidases—more than simple antioxidant scavengers. FEBS J. 274:2163–2180 [DOI] [PubMed] [Google Scholar]
- 14. Jayamanne VS, Adams MR. 2006. Determination of survival, identity and stress resistance of probiotic bifidobacteria in bio-yoghurts. Lett. Appl. Microbiol. 42:189–194 [DOI] [PubMed] [Google Scholar]
- 15. Kawasaki S, Mimura T, Satoh T, Takeda K, Niimura Y. 2006. Response of the microaerophilic Bifidobacterium species, B. boum and B. thermophilum, to oxygen. Appl. Environ. Microbiol. 72:6854–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawasaki S, Satoh T, Todoroki M, Niimura Y. 2009. b-Type dihydroorotate dehydrogenase is purified as a H2O2-forming NADH oxidase from Bifidobacterium bifidum. Appl. Environ. Microbiol. 75:629–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lahtinen SJ, Gueimonde M, Ouwehand AC, Reinikainen JP, Salminen SJ. 2005. Probiotic bacteria may become dormant during storage. Appl. Environ. Microbiol. 71:1662–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loewen PC, Switala J. 1987. Multiple catalases in Bacillus subtilis. J. Bacteriol. 169:3601–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Long RT, et al. 2010. Bifidobacterium as an oral delivery carrier of oxyntomodulin for obesity therapy: inhibitory effects on food intake and body weight in overweight mice. Int. J. Obes. 34:712–719 [DOI] [PubMed] [Google Scholar]
- 20. Mills GC. 1957. Hemoglobin catabolism. I. Glutathione peroxidase, an erythrocyte enzyme which protects hemoglobin from oxidative breakdown. J. Biol. Chem. 229:189–197 [PubMed] [Google Scholar]
- 21. Poupard JA, Husain I, Norris RF. 1973. Biology of the bifidobacteria. Bacteriol. Rev. 37:136–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rochat T, et al. 2005. High-level resistance to oxidative stress in Lactococcus lactis conferred by Bacillus subtilis catalase KatE. Microbiology 151:3011–3018 [DOI] [PubMed] [Google Scholar]
- 23. Schell MA, et al. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shah NP, Ding WK, Fallourd MJ, Leyer G. 2010. Improving the stability of probiotic bacteria in model fruit juices using vitamins and antioxidants. J. Food Sci. 75:M278–M282 [DOI] [PubMed] [Google Scholar]
- 25. Takeuchi A, Matsumura H, Kano Y. 2002. Cloning and expression in Escherichia coli of a gene, hup, encoding the histone-like protein HU of Bifidobacterium longum. Biosci. Biotechnol. Biochem. 66:598–603 [DOI] [PubMed] [Google Scholar]
- 26. Talwalkar A, Kailasapathy K. 2004. The role of oxygen in the viability of probiotic bacteria with reference to L. acidophilus and Bifidobacterium spp. Curr. Issues Intest. Microbiol. 5:1–8 [PubMed] [Google Scholar]
- 27. Xiao JZ, et al. 2003. Effects of milk products fermented by Bifidobacterium longum on blood lipids in rats and healthy adult male volunteers. J. Dairy Sci. 86:2452–2461 [DOI] [PubMed] [Google Scholar]
- 28. Yasui K, et al. 2009. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res. 37:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


