Abstract
Total extracted DNA from two heavily polychlorobiphenyl-contaminated soils was analyzed with respect to biphenyl dioxygenase sequences and activities. This was done by PCR amplification and cloning of a DNA segment encoding the active site of the enzyme. The translated sequences obtained fell into three similarity clusters (I to III). Sequence identities were high within but moderate or low between the clusters. Members of clusters I and II showed high sequence similarities with well-known biphenyl dioxygenases. Cluster III showed low (43%) sequence identity with a biphenyl dioxygenase from Rhodococcus jostii RHA1. Amplicons from the three clusters were used to reconstitute and express complete biphenyl dioxygenase operons. In most cases, the resulting hybrid dioxygenases were detected in cell extracts of the recombinant hosts. At least 83% of these enzymes were catalytically active. Several amino acid exchanges were identified that critically affected activity. Chlorobiphenyl turnover by the enzymes containing the prototype sequences of clusters I and II was characterized with 10 congeners that were major, minor, or not constituents of the contaminated soils. No direct correlations were observed between on-site concentrations and rates of productive dioxygenations of these chlorobiphenyls. The prototype enzymes displayed markedly different substrate and product ranges. The cluster II dioxygenase possessed a broader substrate spectrum toward the assayed congeners, whereas the cluster I enzyme was superior in the attack of ortho-chlorinated aromatic rings. These results demonstrate the feasibility of the applied approach to functionally characterize dioxygenase activities of soil metagenomes via amplification of incomplete genes.
INTRODUCTION
Environmental pollution by polychlorobiphenyls (PCBs) poses a specific problem for bioremediation, as it typically consists of industrial mixtures of dozens of different congeners. Even if it is broad in substrate range, no single pathway is able to metabolize all the PCBs in such mixtures. Thus, the recruitment of novel biocatalysts that may support their removal is of considerable interest. A key enzyme in the aerobic catabolism of PCBs is biphenyl dioxygenase (BphA), which carries out the initial attack on the inert aromatic nucleus. It belongs to class II of aryl-hydroxylating dioxygenases (ARHDOs) that typically hydroxylate substituted benzenes, like toluenes and biphenyls (7). This enzyme represents a catabolic bottleneck, as its substrate range is typically narrower than that of subsequent pathway enzymes (9, 13, 43). Moreover, its regiospecificity is a crucial parameter, as it codetermines whether the initial dioxygenation products become dead-end metabolites or can be further transformed.
A number of enzyme-engineering projects have been carried out to obtain BphAs with altered or broadened substrate ranges (6, 14, 19, 43). Another approach is the detection and isolation of naturally occurring but so far inaccessible enzymatic activities by metagenomic methods (24, 31). Such techniques seem promising for discovering novel biocatalysts, as only a tiny fraction of existing microorganisms are apparently culturable under laboratory conditions (3). Metagenomic approaches have been applied to characterize the diversity of dioxygenase sequences at a number of PCB-polluted sites (1, 10, 17). Sequences with high, as well as with low, similarities to those known from culturable organisms have been detected, depending on the examined site and probably also on the PCR primers used. So far, however, these investigations have been limited to the determination of (incomplete) gene sequences. Therefore, it has remained unclear how many detected sequences belonged to active enzymes. Moreover, it was impossible to deduce detailed substrate and product specificities from the translated DNA sequences.
Previously, we developed a sequence-based strategy that permits the characterization of enzymatic properties of BphA and other class II ARHDO activities, whose genes are only fragmentarily amplified (9, 18). It is applicable to DNA from any source, including metagenomes. In this approach, a “donor” segment that encodes the catalytic center is amplified. This is fused with sequences of a “recipient” bphA gene cluster that is efficiently expressed in an appropriate host. It was confirmed that the substrate ranges of the resulting hybrid dioxygenases are dependent on the nature of the donor segment (9, 18).
Here, we report the use of this system for a first characterization of dioxygenase activities encoded by the metagenomes of two soil samples from a heavily contaminated site near the city of Wittenberg, Germany. This site has previously been characterized with respect to its PCB profile and bacterial community structure (25, 26).
MATERIALS AND METHODS
Chemicals.
Chlorobiphenyl (CB) congeners (99% purity) were obtained from Lancaster Synthesis (White Lund, Morecombe, England), Promochem (Wesel, Germany), or Restek (Sulzbach, Germany).
Isolation of DNA from soil samples.
Soil sampling and storage have been described previously (25, 26). DNA was isolated using the FastDNA Spin Kit for Soil from Qbiogene Bio 101 (MP Biomedicals, Heidelberg, Germany) according to the protocol of the supplier. Briefly, up to 500 mg of soil was mixed with 978 μl of sodium phosphate buffer and 122 μl of MT buffer, and cells were disrupted for 30 s in a Qbiogene FastPrep instrument (MP Biomedicals, Heidelberg, Germany) at a rate of 5.5 m/s. After centrifugation (12,000 × g; 30 s), the supernatant was mixed with 250 μl of protein precipitation solution (PPS). The precipitated proteins were removed by centrifugation (12,000 × g; 5 min). The supernatant was mixed with 1 ml of binding matrix suspension. After settling of the matrix, 500 μl of the supernatant was discarded, and the resuspended matrix was placed in two subsequent batches onto a SpinFilter and centrifuged (12,000 × g; 1 min). After addition of 500 μl of SEWS-M (salt/ethanol wash solution) to the filter and centrifugation (12,000 × g; 1 min), it was air dried, and the DNA was eluted with 50 μl of DES (ultrapure water). After another centrifugation (12,000 × g; 1 min) of the filter, the eluate was collected; supplemented with 0.1 vol of 10 mM Tris-HCl, pH 8; and stored at −20°C.
PCR with DNA from soil samples.
Reactions were carried out in PCR buffer containing 1.5 mM MgCl2 (Qiagen, Hilden, Germany), with about 50 ng of template DNA, 0.5 μM primers HDO2AF and HDO2AR (18), 0.25 mM deoxynucleoside triphosphates (dNTPs), 0.2 mg/ml bovine serum albumin (BSA), 1.6 μl of dimethyl sulfoxide (DMSO), and 1 unit of recombinant Taq DNA polymerase (Fermentas, St. Leon-Rot, Germany) in a total volume of 20 μl. The thermocycler program was as follows: 1 cycle of 30 s at 94°C; 30 cycles of 30 s at 60°C, followed by 90 s at 72°C, with an increment of 3 s per cycle; and 1 cycle of 600 s at 72°C.
Molecular-cloning techniques.
Restriction, ligation, dephosphorylation, agarose gel electrophoresis, and bacterial transformations were carried out following standard protocols (30).
Plasmid construction.
pAIA6099 is a derivative of pAIA6100 (18) that harbors a deletion of 12 codons within the MluI/AflII fragment of bphA1, which inactivates the gene. It was constructed as follows. Two segments of bphA1LB400 were PCR amplified, using pAIA111 (18) as a template and BPH1917 (CGCTCCAGGCACGCGTGGC [MluI recognition sequence underlined]) and BPH-2420MUT (GGGTGCCAGATCCGGAAGATCGTCATATGCTGGCCGACC [NdeI recognition sequence underlined]) or BPH2454MUT (ATGACGATCTTCCGGATCTGGCACCCTCGAGGTCCCAATG [XhoI recognition sequence underlined]) and BPH-2711 (AATCAGGGTGACCGGTCTGC [AgeI recognition sequence underlined]), respectively, as primers. Both products were fused in an overlap extension PCR with BPH1917 and BPH-2711 as primers. The amplicon was cleaved with MluI and AgeI and used to replace the corresponding fragment in pAIA50 (43). This introduced the deletion, as well as NdeI and XhoI sites, and yielded pAIA500. A part of its inactivated bphA1 gene was PCR amplified with primers HDO2AF and HDO2AR (18). The latter introduced an AflII site. The product was cleaved with MluI and AflII and used to replace the MluI/AflII fragment of pAIA6100.
Cloning of PCR products in TOPO vectors.
Taq DNA polymerase-generated PCR products were inserted into the T-overhang topoisomerase vector pCR-XL-TOPO (Invitrogen, Karlsruhe, Germany) according to the protocol of the supplier. Escherichia coli strain Top10 (Invitrogen, Karlsruhe, Germany) was transformed with the ligation reaction mixtures. Plasmid preparations from transformants were analyzed by restriction and agarose gel electrophoresis.
Subcloning of PCR products in pAIA6099.
The MluI/AflII fragments of the inserts of the TOPO clones were excised with these enzymes and were ligated into MluI/AflII-cleaved and dephosphorylated pAIA6099. E. coli strain Top10 or XL10-Gold (Stratagene, Amsterdam, The Netherlands) was transformed with the ligation reaction mixtures. Plasmid preparations from transformants were analyzed by restriction and agarose gel electrophoresis. Correct plasmids were used to transform E. coli BL21(DE3)(pLysS). The resulting clones were analyzed for correct plasmid size.
Preparation of resting cells.
Preparation of resting cells was carried out as previously described (33) with some modifications. Cells of E. coli BL21(DE3)(pLysS) harboring the respective plasmid were grown in LB medium at 30°C. At an optical density at 600 nm (OD600) of about 1.0, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 0.4 mM, and the incubation was continued for another 60 min. Cells were harvested, washed with 1 vol of 50 mM sodium phosphate buffer (pH 7.5), and resuspended in the same buffer to give the concentrations specified below.
Determination of specific ARHDO activity with biphenyl.
Biphenyl was placed into an Erlenmeyer flask at a final nominal concentration of 125 μM, and the solvent was evaporated. Five milliliters of resting cells (see above) was added to a final OD600 of 1, and the flask was shaken at room temperature at 120 rpm. At appropriate times, 640-μl samples were withdrawn, mixed with 160 μl of 5 N NaOH, and centrifuged for 3 min at 12,000 × g. The UV/visible light (Vis) spectra of the supernatants were recorded, using the resting-cell medium as a baseline. The absorption at 600 nm was set to zero. The formation rates of extradiol or meta-cleavage products (MCPs), expressed in milli-absorbance units (mAbs)/min, were determined from the linear parts of the resulting plots. Concentrations of wild-type (WT) and variant BphA1 subunits were determined by evaluation of digitalized images of SDS gels of cell extracts stained with Sypro-Ruby (5), using the AIDA 4.15 software (Raytest, Straubenhardt, Germany) and bovine serum albumin as a standard. The extracts were prepared with the Relay 96 Protein Screen (Invitrogen) according to the manufacturer's instructions.
Determination of CB catabolism by prototype hybrid dioxygenases.
Resting-cell suspensions containing 0.5% (wt/vol) glucose and E. coli BL21(DE3)(pLysS) harboring pAIA6B15 or pAIA6C18 at an OD600 of 2 were shaken in Erlenmeyer flasks at 30°C with nominal concentrations of single CBs of 125 μM. At various times, aliquots were withdrawn and centrifuged for 5 min at 12,000 × g. The UV/Vis spectra of the supernatants were recorded, and the rates of MCP formation were determined as described above.
Sequence determination and analysis.
DNA sequencing was carried out as previously described (4). DNA and protein sequence alignments and calculations of dendrograms were performed with Clustal W2 (15, 21) at the EBI website (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The dendrograms were displayed with the iTOL tool (22, 23; http://itol.embl.de/). Sequence database searches were done with the blastn and blastp programs (2) at the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Nucleotide sequence accession numbers.
Newly determined sequences have been deposited in the GenBank/EMBL/DDBJ database under accession numbers FR877587 to FR877632 and HE577113 to HE577117.
RESULTS AND DISCUSSION
Amplification of ARHDO gene segments from metagenomic DNA.
DNA was isolated from an uncontaminated and from two increasingly PCB-contaminated soil samples (A, B, and C) from a moorland in the vicinity of the city of Wittenberg, Germany (25). The polluted samples contained average PCB concentrations of approximately 1 g/kg or 10 g/kg, respectively. Using this DNA as a potential template, PCR amplifications targeting segments that encode the substrate range-determining cores of the alpha subunits of class II ARHDOs, here collectively termed BphAs, were attempted, using the previously established consensus primers HDO2AF and HDO2AR (18), which amplify fragments of about 720 bp.
It is clear that such an approach will probably not detect PCB-attacking dioxygenase sequences not belonging to class II. The fraction of all available class II sequences that will be amplified can only be estimated. It has been pointed out that theoretical considerations, as well as experimental results, suggest that the oligonucleotides used are able to amplify more than 80% of the cores of known sequences encoding alpha subunits of class II ARHDOs (18).
For high-efficiency and restriction-independent cloning, the amplicons were inserted into a T-overhang TOPO cloning vector. With soil A, only trace amounts of PCR products were observed, which were not further processed. The heavily contaminated soils B and C, however, yielded significant amounts of amplicons of the expected length, suggesting that bphA sequences are enriched and thus are likely to be functionally relevant in these soils.
Sequences of the alpha subunit core gene segments.
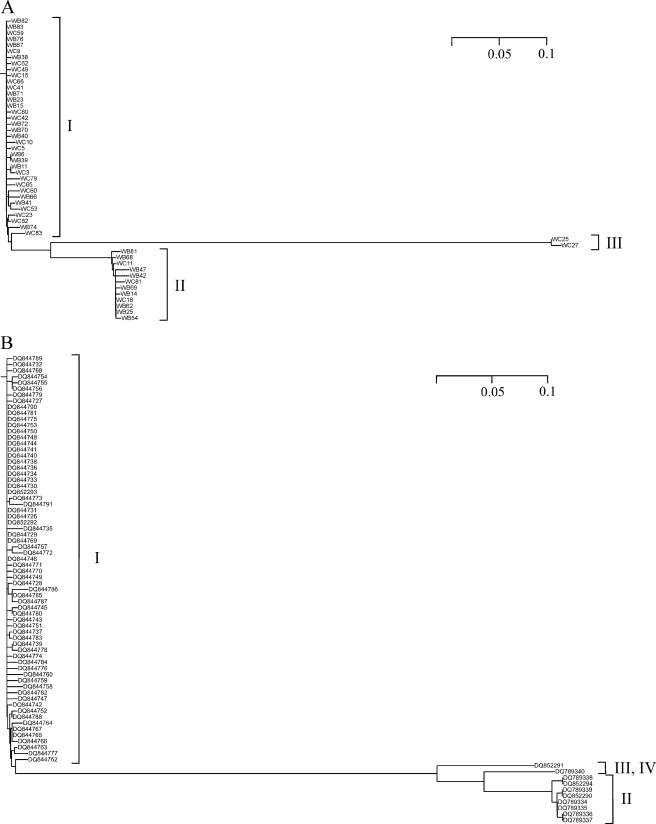
A total of 51 TOPO clones, 26 from soil B and 25 from soil C, were sequenced. Translation of the sequences showed that one contained a frameshift and four contained nonsense mutations. DNA and protein sequence alignments revealed that the sequences formed three similarity clusters, named I to III (Fig. 1A). They comprised 37, 12, and 2 sequences, respectively. Nucleotide and amino acid sequence identities between these clusters were 85 to 88%, 38 to 42%, and 37 to 38%, respectively (Table 1). Within a given cluster, nucleotide and amino acid sequences were 97 to 100% identical (Table 1). Of course, the possibility that minor sequence differences were due to PCR errors cannot be ruled out. However, the detection of similar microheterogeneities and of comparable frequencies of nonsense and frameshift mutations in metagenomic studies involving not PCR, but direct cloning of environmental DNA (36, 38), suggests that all or a major fraction of the apparent sequence diversity was of natural origin. Unequivocal consensus sequences could be determined for clusters I and II, as the bias toward one specific nucleotide or amino acid, respectively, was always very strong. For cluster I, the consensus sequence itself was found in 7 of 37 clones at the nucleotide level and in 10 of 37 clones at the amino acid level. For cluster II, the respective values were 1 and 3 for a total of 12 clones. Clones WB15 of cluster I and WC18 of cluster II, which contained the consensus sequences, are referred to below as prototypes. Cluster I sequences were predominant in both soils, and especially in soil C (Table 2). In contrast, the percentage of cluster II sequences was higher in soil B. Cluster III sequences were found only in soil C. This might indicate that enzymes or organisms, respectively (see below), belonging to clusters I and III can more readily cope with the more heavily contaminated site.
Fig 1.
Dendrograms of metagenomic BphA1 (A) and DitA1 (B) sequences from the Wittenberg site. The dendrograms were derived from sequence alignments. Similarity clusters are indicated by brackets and are designated by roman numerals. The scale bars give distances in amino acid substitutions per site. DitA1 sequences (40) are identified by database accession numbers.
Table 1.
Similarities of amino acid sequences encoded by the Wittenberg clones
| Sequence cluster | No. of clones sequenced | Amino acid sequence identity (%) |
|||
|---|---|---|---|---|---|
| Within the cluster | With cluster II | With cluster III | With the most similar sequence in the databasea | ||
| I | 37 | 97–100 | 85–88 | 38–42 | 96b |
| II | 12 | 97–100 | 37–38 | 100/93c | |
| III | 2 | 99 | 56/43d | ||
If the most similar sequence belongs to a putative enzyme, a second value is given, which refers to the most similar sequence of a nonputative enzyme.
Biphenyl dioxygenase alpha subunit from B. xenovorans LB400 (NCBI Protein Database accession no. ABE37059).
Table 2.
Distribution of DNA sequences of soils B and C between sequence clusters
| Sequence cluster | Sequences obtained from soil: |
|||
|---|---|---|---|---|
| B |
C |
|||
| No. | % | No. | % | |
| I | 17 | 65 | 20 | 80 |
| II | 9 | 35 | 3 | 12 |
| III | 0 | 0 | 2 | 8 |
| Sum | 26 | 100 | 25 | 100 |
A database search revealed that the sequence most closely related to the translated sequences of cluster I was that of the BphA alpha subunit (BphA1) of Burkholderia xenovorans strain LB400, showing 96% amino acid sequence identity with the WB15 prototype (Table 1). Of the dioxygenases experimentally shown to possess catalytic activity, BphA1 of Pseudomonas pseudoalcaligenes KF707, showed the highest degree of amino acid sequence identity (93%) with the sequences of cluster II (Table 1). Interestingly, the nucleotide sequence of the WC18 prototype was 100% identical with the sequences of putative bphA1 gene fragments from two strains isolated from the Wittenberg site, Burkholderia sp. strains WBF3 and WBF4 (Table 1). The two sequences of cluster III were most similar (56%) to a putative ring-hydroxylating dioxygenase alpha subunit from Burkholderia ambifaria IOP40-10 (Table 1). Of the dioxygenases experimentally shown to be catalytically active, BphA1 of Rhodococcus jostii RHA1 possessed the highest (43%) amino acid sequence identity (Table 1). Thus, the enzymes of clusters I and II belong to known ARHDO subfamilies, whereas the dioxygenases of cluster III appear to be part of a novel subfamily.
A number of previous studies also characterized PCB-polluted soils by using PCRs that target parts of the ARHDO class II alpha subunit gene. Capodicasa et al. (10) investigated bioreactors containing soils contaminated with 0.89 g PCB/kg. They found highly similar sequences that, in translated form, showed 92 to 99% amino acid identity to known sequences, mostly from cultivated organisms, like strain LB400 or KF707 or Pseudomonas sp. strain Cam-1. Aguirre de Cárcer et al. (1) examined soil with a PCB contamination of 0.18 g per kg of dry material. They discovered large sequence diversities before, as well as after, the introduction of willow trees for rhizomediation. Sequencing of 28 clones revealed that the translated sequences all showed similarities to characterized ARHDO sequences. However, they showed great heterogeneity among themselves, displaying between 10% and 100% amino acid sequence identity. Iwai et al. (17) investigated PCB-contaminated soil with the comparatively low concentration of 0.015 g/kg. They subjected PCR products directly to pyrosequencing, obtaining about 2,600 sequences of 175 or 200 nucleotides, depending on the primer used. In their analysis, the authors obtained 40 sequence clusters that contained newly determined, as well as database, sequences and 25 clusters that contained only novel sequences, indicating a wide variety of primary structures. In summary, these studies, including the present one, identified abundances of either highly similar or fairly diverse alpha subunit segment sequences. Diversity appeared to decrease with increasing PCB contaminations of the soil samples.
Witzig et al. (40) investigated the sequence diversity of alpha subunit segments of diterpenoid dioxygenase (DitA), which also belongs to the ARHDO family. This work is of interest here, because it also examined PCB-contaminated soil from the Wittenberg site. The authors determined 77 sequences of PCR products encoding the very same region of the alpha subunit as our amplicons. Their template DNAs originated from bacterial isolates, as well as from the metagenome. The latter sequences were obtained either after cloning or after electrophoretic separation of the amplicons. Their results resemble ours in several respects. For DitA1, as well as for BphA1, the large majority of sequences fall into two similarity clusters, I and II in Fig. 1. The relative sizes of the clusters are comparable. While the intercluster distances differ, intracluster sequence identities are similarly high at 96% or above. The assumption that both minor clusters represent the same group of organisms is corroborated by the finding that the bphA1 sequences of the Wittenberg isolates WBF3 and WBF4 are completely identical to those of clones WB25, WB62, and WC18, belonging to BphA1 cluster II, and that the ditA1 sequences of the latter strains (100% identity; accession no. DQ789336 and DQ789337) belong to DitA1 cluster II. As mentioned above, our PCR amplifications suggest an enrichment of bphA1 genes in the polluted soils. Thus, it appears likely that the ability to utilize certain CBs selected for certain bph operons and, as long as horizontal gene transfer plays no major role, thereby for certain taxa and that this selection is also reflected by the ditA1 genes. This interpretation agrees with the results of Witzig et al. (40) for isolates from the Wittenberg site, which suggests that the observed clustering of ditA1 sequences is paralleled by taxonomic clustering, as determined by gyrB sequencing. The widespread occurrence of dit genes in CB and aromatic hydrocarbon degraders in general may originate from the utilization of ubiquitous resin acids prior to introduction of the pollutants (40).
Reconstitution of complete BphA gene clusters and determination of gene expression.
In order to assess whether the environmental DNA fragments harbor the potential to encode parts of active dioxygenases and to establish sequence-function and sequence-specificity correlations, 21 of the different alpha subunit core sequences were used to reconstitute complete bphA gene clusters. This was done by supplementing them with the missing flanking sequences of the alpha subunit gene, as well as with the other three genes required for BphA systems. These encode the beta subunit, a ferredoxin, and a ferredoxin reductase. In principle, this was done as previously described, by exchanging the respective core segment of the cloned bphALB400 gene cluster for an amplified core fragment (18). The procedure was modified in that a newly constructed plasmid, pAIA6099, harboring an inactive bphA1 core segment that lacks 36 bp (for details, see Materials and Methods) was used for the replacement. This recipient plasmid also harbored genes bphBC from strain LB400, encoding the two subsequent catabolic pathway enzymes. Their presence enabled verification of whether the products of the initial dioxygenation were further metabolized. Many of the resulting MCPs possess characteristic electronic spectra which not only facilitate assessment of dioxygenase activity, but also permit, to some extent, assignment of the regiospecificity of the initial dioxygenation (see below).
The primary transformants of this reconstitutive subcloning were checked for genetic correctness, and the expression strain E. coli BL21(DE3)(pLysS) was transformed by the respective plasmids. The resulting clones, 14 of them belonging to cluster I, 6 to cluster II, and 1 to cluster III, were used for further analysis.
The cellular concentrations of dissolved alpha and beta subunits were determined by quantitation of SDS-PAGE band intensities. In most, but not all, cases, the concentrations of hybrid alpha subunits were similar to that of the parental BphA1LB400 (data not shown). The WT beta subunits were generally found in some excess compared to the concentrations of the alpha subunits. Three hybrid subunits were not detected. In accordance with this observation, no catalytic activity was observed (see below).
Catalytic activities of the hybrid enzymes and their correlation with amino acid substitutions.
The catalytic activities of the hybrid enzymes were quantitated with resting cells and biphenyl as a substrate (Table 3). It has been shown with this and a wide range of other substrates that the activities of BphB and BphC in these strains are not rate limiting (41, 43; C. Standfuß-Gabisch and B. Hofer, unpublished results). Of 14 hybrid BphAs belonging to cluster I, 10 were active, 3 were inactive, and in one case, no alpha subunit was found. Of 6 BphAs of cluster II, 5 were active; again, in one case, a large subunit was not detected. Also the alpha subunit of cluster III hybrid WC27 was not observed. In accordance with this, no activity was detected, either with biphenyl or with a range of other aromatic compounds, such as toluene, isopentylbenzene, diphenylmethane, and dibenzofuran. A possible reason for the absence of some hybrid subunits among the dissolved proteins is incompatibility between recipient and donor protein segments with respect to proper folding, leading to rapid proteolysis and/or precipitation of the hybrid. Of all 18 detected hybrids, 83% were definitely active, whereas 7% showed no activity under assay conditions. As hybrid formation, even with core segments from active ARHDOs, does not in all cases yield active enzymes, the percentage of “active” donor segments may actually be higher.
Table 3.
Activities of hybrid dioxygenases with biphenyl as a substrate and correlation with amino acid substitutions in BphA1a
| Clone name | Sequence cluster | Sp act [(pmol/min)/mg BphA1]b,c | Amino acid substitution relative to cluster prototyped | Comparisone of and comments on amino acid substitutions |
|---|---|---|---|---|
| WB6 | I | 21.6 | D279G | G and N also found at this position |
| WB11 | I | 30.0 | K291E | K invariant |
| WB15 | I | 41.9 | NAh | |
| WB40 | I | 49.1 | L309P | V also found at this position |
| WB41 | I | 59.9 | S283P | SBSRi; I, T, M, and L also found at this position |
| H343R | H invariant | |||
| WB70 | I | NADf | S274P | T and A also found at this position; probable steric clash between P274 and G273 |
| WB72 | I | 25.9 | T356A | V, W, and I also found at this position |
| WB74 | I | NAD, NPDg | I247V | L and M also found at this position |
| I341T | T, S, and A also found at this position | |||
| I375V | L and V also found at this position | |||
| WC5 | I | 75.9 | L304H | K and R also found at this position |
| WC10 | I | NAD | Q322R | SBSR; E also found at this position |
| A354T | S and P also found at this position | |||
| WC15 | I | 80.7 | F265L | Y also found at this position |
| WC23 | I | 0.904 | I375L | L and V also found at this position |
| N377S | T also found at this position; neighbor of SBSR F378 | |||
| WC42 | I | 94.9 | I339V | V also found at this position |
| WC65 | I | NAD | G271R | G invariant |
| Y370H | F also found at this position | |||
| WB14 | II | 14.2 | V352 M | V invariant |
| WB42 | II | 654 | E250G | G, D, and N also found at this position |
| V358A | V invariant | |||
| D361N | D invariant | |||
| WB47 | II | NAD, NPD | Y232C | Y invariant; neighbor of SBSR M231 and of Fe ligand H233 |
| T329P | T invariant | |||
| A360T | A invariant | |||
| WB68 | II | 639 | M247V | I and L also found at this position |
| WB69 | II | 744 | T260A | K and S also found at this position |
| WC18 | II | 500 | NA | |
| WC27 | III | NAD, NPD | NA |
Prototype lines are in boldface.
Standard deviations were ±30%.
Specific activity was calculated using a molar extinction coefficient of 33,200 for the MCP (35).
Amino acid numbering from BphA1LB400.
With 23 alpha subunit sequences of active benzene-type (class II) ARHDOs. Their NCBI protein database accession numbers are CAA56346, AAB07750, BAA06868, AAP74038, AAA26005, ADI95397, CAA06970, Q07944, AAC43632, AAC46390, ABE37059, AAB88813, Q52028, AAK14781, 1WQL_A, AAD12763, AAB36666, AAC03436, CAB99196, AAC44526, CAA08985, BAJ72245, and BAC01052. We changed the BAC01052 sequence in positions 351 to 359 to VWAFVVVDA because in this segment the database sequence obviously switched to the wrong reading frame.
NAD, no activity detected.
NPD, no protein (BphA1) detected.
NA, not applicable.
SBSR, substrate binding site residue; i.e., the amino acid is located within a distance of 6 Å from the biphenyl molecule in the BphALB400 structure 2XRX (20).
When amino acid deviations relative to the prototypes resulted in significant changes of catalytic activity, decreases rather than increases were observed, a behavior expected for the introduction of random amino acid substitutions into the prototype sequence. Below, amino acid deviations that strongly affected activity (Table 3) are discussed with respect to structure-function relationships. In cluster I, this was the case for the apparently inactive hybrids WB70, WC10, and WC65, as well as for hybrid WC23, which displayed 40-fold-decreased activity.
BphAWB70h (h denotes hybrid) harbors only a single substitution relative to its WB15 prototype, Ser274Pro (amino acid numbering from BphA1LB400). In order to examine which other residues are tolerated at this position in related ARHDOs, we neglected sequence data without a positive correlation with enzymatic activity and restricted our sequence alignment to 23 alpha subunit sequences of active class II ARHDOs. This showed that Thr and Ala also occur at this position. In the crystal structure of the closely related LB400 enzyme (20) (Protein Data Bank [PDB] ID 2XRX), Ser274 has no direct contact with the biphenyl substrate. Its replacement by Ala or Thr does not appear to lead to steric clashes. A Pro residue, however, would shift Gly273. This displacement could, via Met324, well be transmitted to His323, resulting in steric interference with the substrate.
Similarly, the two replacements in hybrid WC10, Gln322Arg and Ala354Thr, should not directly affect interactions with the substrate. Ser354, as well as Pro354, but not Thr354, was found in our compilation of active enzymes. In the LB400 structure, none of these 3 changes appears to cause steric interference. The only other residue found in position 322 is Glu, which is sterically similar to Gln. Although BphALB400 seems to be able to accommodate Glu, as well as Arg, at this position, it seems likely that the Gln322Arg exchange is mainly responsible for the inactivity of WC10, as it probably affects the positions of its direct main-chain neighbors, Gly321 and His323, which, according to the LB400 structure, make van der Waals contacts with the substrate.
BphAWB65h also harbors two changes, Gly271Arg and Tyr370His. In active enzymes, Phe, but not His, occurs at position 370. Gly271, however, is invariant. The crystal structure of the LB400 dioxygenase clearly shows that both residues are remote from the substrate-binding site and that His370, but not the large Arg271 side chain, can be accommodated without major rearrangements of the protein structure. This suggests that the latter exchange probably triggers structural changes that result in inactivity.
Hybrid WC23 contains two replacements in close proximity, Ile375Leu and Asn377Ser. Leu375 and Thr377, but not Ser377, are found in active enzymes. The smaller Ser would generate a cavity within the fold, unless this was prevented by shifts of Ser itself and of adjacent residues. This may well affect the active site, for example, the substrate-lining Phe378, which has been shown to be critical for dioxygenation (42).
Only one of the six cluster II hybrids, WB14, showed a remarkable (about 35-fold) reduction in activity. It contains only a single change, Val352Met. The LB400 structure indicates that this Val is quite remote from the active site and that a Met residue could be accommodated at position 352. Thus, a mechanism that triggers the observed drastic decrease in activity is not obvious. A crucial role of this Val residue is in agreement with its invariance in the compilation of active enzymes. We note, however, that changes of other invariant residues, such as Lys291, His343, Val358, and Asp361, did not lead to drastic losses of activity.
Assay of BphA prototypes for productive dioxygenation of selected CBs.
The strains producing the prototype enzymes BphAWB15h and BphAWC18h were assayed for dioxygenation of 10 CBs that were major, minor, or not constituents of the PCB mixture found at the Wittenberg site. It had previously been shown that the BphB and BphC enzymes, which were also synthesized by the recombinant strains, were able to convert ortho,meta-dioxygenated products of all of these CBs into MCPs (33, 34, 43). Initial dioxygenations that formed this type of further degradable catabolites were termed “productive.” The finding that BphC of strain LB400 is unable to convert meta,para-dihydroxylated biphenyl (11a) indicates that productive dioxygenations in the pathway examined here are directed to ortho and meta carbons. The 10 congeners used were di- or trisubstituted and possessed no unchlorinated ring. They contained all three types of monochlorinated rings. Six of them (2,2′-, 2,4′-, 4,4′, 3,4,2′-, 2,4,3′-, and 3,4,4′-CB) were present at the contaminated site in different amounts (Table 4), while the other four (3,3′-, 3,5,2′-, 2,3,3′-, and 3,5,4′-CB) were not detected (25).
Table 4.
Productive dioxygenation of various CBs by prototype hybrid BphAs
| CB chlorinated carbons | CB contamination of sitea | CB aqueous solubility (μM) | Rate of product formation (mAbsmaxe/h) |
|||
|---|---|---|---|---|---|---|
| BphAWB15h |
BphAWC18h |
|||||
| Mean | SD | Mean | SD | |||
| 2,2′ | + | 1.91b | 1,370 | 320 | 87 | 51 |
| 2,4′ | ++ | 3.47b | 1,170 | 270 | 626 | 223 |
| 3,4,2′ | +++ | 0.616b | 507 | 11 | 121 | 16 |
| 3,5,2′ | − | 0.501b | 133 | 41 | 46 | 10 |
| 3,3′ | − | 0.354b | NPOd | NPO | 37 | 13 |
| 2,3,3′ | − | 0.426c | 31 | 5 | 54 | 7 |
| 2,4,3′ | + | 0.776b | 59 | 9 | 46 | 11 |
| 4,4′ | +++ | 0.426b | NPO | NPO | 7 | 1 |
| 3,4,4′ | +++ | 0.301c | NPO | NPO | NPO | NPO |
| 3,5,4′ | − | 0.194c | NPO | NPO | NPO | NPO |
Gross classification of CB contaminations, based on gas chromatography data (25). −, not a constituent; +, minor constituent; ++, medium constituent; +++, major constituent of the Wittenberg site.
Experimental (28).
Calculated (39).
NPO, no product observed.
mAbsmax, milli-absorbance units at absorption maximum.
An overview of the results is shown in Table 4, where congeners are listed according to their type of monochlorinated ring. With a single exception (see below), productive dioxygenation was always directed toward the monochlorinated ring. The position of the chlorine at this ring largely determined the rate of dioxygenation. There was no obvious correlation with the aqueous solubilities of the congeners (Table 4).
Some simple rules can be deduced from the experimental data for correlations between the absorption maxima and substituent patterns of chlorinated MCPs (Table 5), which allow some assignments of the sites of the initial dioxygenations. (i) Substitutions at carbon 5, 4, or ortho (for numbering, see footnote b of Table 5) shift absorption maxima from values above 430 nm to increasingly lower values. (ii) In cases of multiple substitutions, these effects dominate in the order ortho > 4 > 5. Values based on these rules are given for the different MCPs in the “Expected” column of Table 5. In the subsequent two columns, they are compared with other experimental data. As can be seen, the observed values agree in many, but not all, cases.
Table 5.
Absorption maximaa of MCPs formed via dioxygenation of CBs by prototype hybrid BphAs and tentative assignments of initially oxidized carbons
| CB chlorinated carbons | CB potentially oxidized carbons | Resulting MCP |
|||||
|---|---|---|---|---|---|---|---|
| Chlorinated carbonsb | Expected λmax (nm)c,d | Previous experimental data |
BphAWB15h λmax (nm) | BphAWC18h λmax (nm) | |||
| λmax (nm) | Oxidized carbons | ||||||
| 2,2′ | 2,3 | 8 | ∼393 | 392c | 2,3h | 393 | 393 |
| 5,6 | 5,8 | ∼393 | 395e | 5,6e | 393 | 393 | |
| 2,4′ | 2,3 | 10 | >430 | 438c | 2,3h | 433 (399) | |
| 5,6 | 5,10 | ∼402 | 400 | ||||
| 2′,3′ | 3,8 | ∼393 | |||||
| 3,4,2′ | 2,3 | 3,8 | ∼393 | ||||
| 5,6 | 3,4,8 | ∼393 | |||||
| 2′,3′ | 9,10 | >430 | 440 (401)f | 2′,3′h | 435 (401) | ||
| 5′,6′ | 5,9,10 | ∼402 | 401 | ||||
| 3,5,2′ | 2,3 | 4,8 | ∼393 | ||||
| 2′,3′ | 9,11 | >430 | 435 (402)f | 2′,3′h | 435 (400) | ||
| 5′,6′ | 5,9,11 | ∼402 | 400 | ||||
| 3,3′ | 2,3 | 9 | >430 | NPOi | |||
| 5,6 | 4,9 | ∼410 | 430 (410)c | 5,6e,h | 395 | ||
| 425 (395)e | |||||||
| 2,3,3′ | 5,6 | 4,5,9 | ∼402 | ||||
| 2′,3′ | 8,9 | ∼393 | 385 ± 3 | 388j | |||
| 5′,6′ | 4,8,9 | ∼393 | 400 (370)c | 5′,6′h | 385 ± 3 | 388j | |
| 2,4,3′ | 2,3 | 3,9 | >430 | 433f | 2,3f | 420 (438) | |
| 5,6 | 3,5,9 | ∼402 | 437f | 5,6h | |||
| 2′,3′ | 8,10 | ∼393 | 388j | ||||
| 5′,6′ | 4,8,10 | ∼393 | 388j | ||||
| 4,4′ | 2,3 | 3,10 | >430 | 432e,g | 2,3e,h | NPO | 433 |
λmax. All values are ±2 nm unless otherwise indicated. The numbers in parentheses indicate final values in case of a shift of the absorption maximum during incubations.
Carbon numbering in MCP is as shown here: 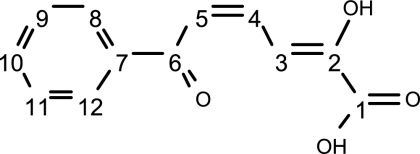 .
.
Data from reference 33.
Data from reference 32.
Data from reference 9.
Standfuß-Gabisch and Hofer, unpublished.
Data from reference 43.
Data from reference 34.
NPO, no product observed.
Absorption unstable.
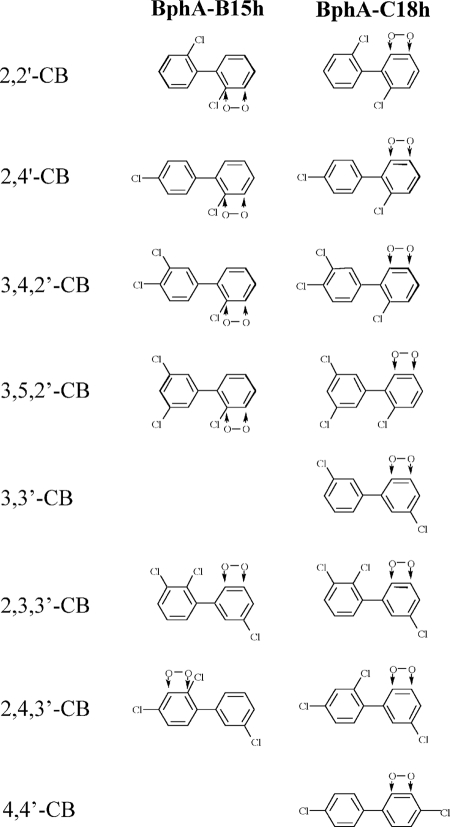
CBs with an ortho-monochlorinated ring were most readily turned over by both enzymes (Table 4). There was a fundamental difference, however, regarding the site of attack, as reflected (with the exception of 2,2′-CB) by the different absorption maxima of the resulting MCPs (Table 5). These indicate, as shown in Fig. 2, that BphAWC18h dioxygenated 2,4′-, 3,4,2′-, and 3,5,2′-CB at unchlorinated carbons (positions 5,6 or 5′,6′, respectively), whereas BphAWB15h attacked them at the semichlorinated side of the ring (positions 2,3 or 2′,3′, respectively). It appears very likely that the same scheme also applies to 2,2′-CB, where the resulting MCPs were neither expected nor found to possess a significant difference in their absorption maxima. Generally, the rates of attack of the ortho-monochlorinated ring were higher with BphAWB15h than with BphAWC18h. The two of these four CBs that were found in higher concentrations at the Wittenberg site (2,4′- and 3,4,2′-CB) were the best substrates for the latter enzyme. Such a correlation was less clear for BphAWB15h, as 2,2′-CB was also an excellent substrate for this dioxygenase.
Fig 2.
Regiospecificity of CB dioxygenation by cluster I and cluster II prototype hybrid enzymes. The sites of productive attack were deduced as shown in Table 5 and discussed in the text.
CBs with a meta-monochlorinated ring were much “slower” substrates with both enzymes (Table 4). Independent of the substitution pattern of the nonoxidized ring, BphAWC18h turned all three congeners over at similar rates. 3,3′-CB was dioxygenated at the unchlorinated side (Table 5). In an analogy with this, we assigned the same regiospecificity to the dioxygenations of 2,3,3′- and 2,4,3′-CB (Fig. 2), which also agrees with the finding that an attack involving meta-dechlorination has very rarely been observed (37). In contrast to BphAWC18h, the WB15 enzyme attacked the meta-monochlorinated ring only in 2,3,3′-CB. It also slowly dioxygenated 2,4,3′-CB; here, however, as deduced from Table 5, the attack was not directed against the monochlorinated ring, but against carbons 2 and 3 (Fig. 2), in agreement with the described preference of this enzyme for chlorinated ortho carbons. Only BphAWB15h showed a somewhat faster productive dioxygenation of 2,4,3′-CB, which is the only one of these three congeners that was found at the contaminated site.
For CBs possessing a para-chlorinated ring, almost no turnover was observed with both enzymes (Table 4). Only BphAWC18h slowly dioxygenated 4,4′-CB, which, like 3,4,4′-CB, belongs to the CBs that are predominant at the Wittenberg site.
In our assays, BphAWC18h showed a broader CB range than BphAWB15h. On the other hand, the latter enzyme clearly was a better catalyst for the dioxygenation of CBs with ortho-chlorinated rings. A correlation between the concentrations of the selected CBs at the polluted site and their rates of productive dioxygenation by the two prototype enzymes was not generally apparent. The expectation of such a correlation may well be based on an oversimplified view. Rapid dioxygenation of a given CB must not necessarily result in an evolutionary advantage. Indeed, it may result in a disadvantage, if accumulating catabolites exert toxic effects (8, 11, 12, 16, 27, 29). Moreover, the evolution of catalytic activity in the presence of substrate mixtures will necessarily lead to compromises, so that any given enzyme will only be able to efficiently transform a fraction of substrates. Thus, evolution toward the efficient utilization of a substrate other than those applied in our assays may obscure correlations with the congeners used here.
Concluding remarks.
The present work demonstrated the feasibility of the applied approach in retrieving not only ARHDO sequence information from metagenomic DNA but also experimental data on enzymatic properties, such as activity, substrate, and product ranges. In this context, it will be of interest to investigate in detail how much the sequence diversity within a given sequence cluster affects the substrate spectrum. Moreover, it will be intriguing to obtain active enzymes from the donor segments of similarity cluster III and, generally speaking, of other classes of ARHDOs by constructing alternative recipient gene clusters based on genes from strains such as RHA1 and polycyclic aromatic hydrocarbon (PAH) degraders.
Footnotes
Published ahead of print 10 February 2012
REFERENCES
- 1. Aguirre de Cárcer D, Martín M, Karlson U, Rivilla R. 2007. Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl-polluted soil after introduction of willow trees for rhizoremediation. Appl. Environ. Microbiol. 73:6224–6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Amann RI, Ludwig W, Schleifer K-H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartels F, Backhaus S, Moore ERB, Timmis KN, Hofer B. 1999. Occurrence and expression of glutathione S-transferase-encoding bphK genes in Burkholderia sp. strain LB400 and other biphenyl-utilizing bacteria. Microbiology 145:2821–2834 [DOI] [PubMed] [Google Scholar]
- 5. Berggren K, et al. 1999. A luminescent ruthenium complex for ultrasensitive detection of proteins immobilized on membrane supports. Anal. Biochem. 276:129–143 [DOI] [PubMed] [Google Scholar]
- 6. Brühlmann F, Chen W. 1999. Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol. Bioeng. 63:544–551 [DOI] [PubMed] [Google Scholar]
- 7. Butler CS, Mason JR. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47–84 [DOI] [PubMed] [Google Scholar]
- 8. Cámara B, et al. 2004. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ. Microbiol. 6:842–850 [DOI] [PubMed] [Google Scholar]
- 9. Cámara B, et al. 2007. Generation by a widely applicable approach of a hybrid dioxygenase showing improved oxidation of polychlorobiphenyls. Appl. Environ. Microbiol. 73:2682–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Capodicasa S, et al. 2009. Terminal-restriction fragment length polymorphism analysis of biphenyl dioxygenase genes from a polychlorinated biphenyl-polluted soil. Res. Microbiol. 160:742–750 [DOI] [PubMed] [Google Scholar]
- 11. Dai S, et al. 2002. Identification and analysis of a bottleneck in PCB biodegradation. Nat. Struct. Biol. 9:934–939 [DOI] [PubMed] [Google Scholar]
- 11a. Eltis LD, Hofmann B, Hecht H-J, Lünsdorf H, Timmis KN. 1993. Purification and crystallization of 2,3-dihydroxybiphenyl 1,2-dioxygenase. J. Biol. Chem. 268:2727–2732 [PubMed] [Google Scholar]
- 12. Fava F. 1996. Aroclor 1221 aerobic dechlorination by a bacterial co-culture: role of chlorobenzoic acid degrading bacteria in the process. Chemosphere 32:1477–1483 [DOI] [PubMed] [Google Scholar]
- 13. Furukawa K, Hirose J, Suyama A, Zaiki T, Hayashida SJ. 1993. Gene components responsible for discrete substrate specificity in the metabolism of biphenyl (bph operon) and toluene (tod operon). J. Bacteriol. 175:5224–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furukawa K, Suenaga H, Goto M. 2004. Biphenyl dioxygenases: functional versatilities and directed evolution. J. Bacteriol. 186:5189–5196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goujon M, et al. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38(Suppl. 2):W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Havel J, Reineke W. 1992. Degradation of Aroclor 1221 and survival of strains in soil microcosms. Appl. Microbiol. Biotechnol. 38:129–134 [Google Scholar]
- 17. Iwai S, et al. 2010. Gene-targeted-metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment. ISME J. 4:279–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kahl S, Hofer B. 2003. A genetic system for the rapid isolation of aromatic-ring-hydroxylating dioxygenase activities. Microbiology 149:1475–1481 [DOI] [PubMed] [Google Scholar]
- 19. Kumamaru T, Suenaga H, Mitsuoka M, Watanabe T, Furukawa K. 1998. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenase. Nat. Biotechnol. 16:663–666 [DOI] [PubMed] [Google Scholar]
- 20. Kumar P, et al. 2011. Structural insight into the expanded PCB-degrading abilities of a biphenyl dioxygenase obtained by directed evolution. J. Mol. Biol. 405:531–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larkin MA, et al. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 22. Letunic I, Bork P. 2007. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128 [DOI] [PubMed] [Google Scholar]
- 23. Letunic I, Bork P. 2011. Interactive tree of life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39(Suppl. 2):W475–W478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lorenz P, Liebeton K, Niehaus F, Eck J. 2002. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr. Opin. Biotechnol. 13:572–577 [DOI] [PubMed] [Google Scholar]
- 25. Nogales B, Moore ERB, Abraham W-R, Timmis KN. 1999. Identification of the metabolically active members of a bacterial community in a polychlorinated biphenyl-polluted moorland soil. Environ. Microbiol. 1:199–212 [DOI] [PubMed] [Google Scholar]
- 26. Nogales B, et al. 2001. Combined use of 16S ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parnell JJ, et al. 2006. Coping with polychlorinated biphenyl (PCB) toxicity: physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl. Environ. Microbiol. 72:6607–6614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patil GS. 1991. Correlation of aqueous solubility and octanol-water partition coefficient based on molecular structure. Chemosphere 22:723–738 [Google Scholar]
- 29. Sakai M, Miyauchi K, Kato N, Masai E, Fukuda M. 2003. 2-Hydroxypenta-2,4-dienoate metabolic pathway genes in a strong polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 69:427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sambrook J, Russel DW. 2001. Molecular cloning a laboratory manual, 3rd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 31. Schloss PD, Handelsman J. 2003. Biotechnological prospects from metagenomics. Curr. Opin. Biotechnol. 14:303–310 [DOI] [PubMed] [Google Scholar]
- 32. Seah SY, et al. 2000. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 275:15701–15708 [DOI] [PubMed] [Google Scholar]
- 33. Seeger M, Timmis KN, Hofer B. 1995. Conversion of chlorobiphenyls into phenylhexadienoates and benzoates by the enzymes of the upper pathway for polychlorobiphenyl degradation encoded by the bph locus of Pseudomonas sp. strain LB400. Appl. Environ. Microbiol. 61:761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seeger M, Zielinski M, Timmis KN, Hofer B. 1999. Regiospecificity of dioxygenation of di- to pentachlorobiphenyls and their degradation to chlorobenzoates by the bph-encoded catabolic pathway of Burkholderia sp. strain LB400. Appl. Environ. Microbiol. 65:3614–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suenaga H, Goto M, Furukawa K. 2001. Emergence of multifunctional oxygenase activities by random priming recombination. J. Biol. Chem. 276:22500–22506 [DOI] [PubMed] [Google Scholar]
- 36. Suenaga H, Ohnuki T, Miyazaki K. 2007. Functional screening of a metagenomic library for genes involved in microbial degradation of aromatic compounds. Environ. Microbiol. 9:2289–2297 [DOI] [PubMed] [Google Scholar]
- 37. Suenaga H, Watanabe T, Sato M, Ngadiman Furukawa K. 2002. Alteration of regiospecificity in biphenyl dioxygenase by active-site engineering. J. Bacteriol. 184:3682–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Suenaga H, et al. 2009. Novel organization of aromatic degradation pathway genes in a microbial community as revealed by metagenomic analysis. ISME J. 3:1335–1348 [DOI] [PubMed] [Google Scholar]
- 39. Wei X-Y, Ge Z-G, Wang Z-Y, Xu J. 2007. Estimation of aqueous solubility (-lgSw) of all polychlorinated biphenyl (PCB) congeners by density functional theory and position of Cl substitution (NPCS) method. Chinese J. Struct. Chem. 26:519–528 [Google Scholar]
- 40. Witzig R, et al. 2007. Molecular detection and diversity of novel diterpenoid dioxygenase DitA1 genes from proteobacterial strains and soil samples. Environ. Microbiol. 9:1202–1218 [DOI] [PubMed] [Google Scholar]
- 41. Zielinski M, Backhaus S, Hofer B. 2002. The principal determinants for the structure of the substrate-binding pocket are located within a central core of a biphenyl dioxygenase alpha subunit. Microbiology 148:2439–2448 [DOI] [PubMed] [Google Scholar]
- 42. Zielinski M, Kahl S, Hecht H-J, Hofer B. 2003. Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction. J. Bacteriol. 185:6976–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zielinski M, et al. 2006. Generation of novel-substrate-accepting biphenyl dioxygenases through segmental random mutagenesis and identification of residues involved in enzyme specificity. Appl. Environ. Microbiol. 72:2191–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]