Abstract
Culture-independent sampling in conjunction with a functional cloning approach identified diverse antibiotic resistance genes for different classes of antibiotics in gut microbiomes from both conventionally raised and free-range chickens. Many of the genes are phylogenetically distant from known resistance genes. Two unique genes that conferred ampicillin and spectinomycin resistance were also functional in Campylobacter, a distant relative of the Escherichia coli host used to generate the genomic libraries.
TEXT
Zoonotic bacterial pathogens cause diseases in animals and/or humans, consequently threatening animal production, food safety, and public health (22, 27, 29). Increasing evidence indicates that antimicrobial usage in animals promotes the emergence of resistant zoonotic pathogens, greatly compromising the effectiveness of antibiotic treatments (7, 24). It has been proposed that normal intestinal microbiotas play a critical role in antibiotic resistance (AR) development and transmission via horizontal gene transfer (HGT) (13, 26). However, the majority of intestinal microbiotas cannot be cultivated using traditional culturing methods, greatly impeding our understanding of the AR reservoir in gut microbiotas. Recently, Sommer et al. (21) functionally characterized the AR reservoir in the human gut microbiota; many AR genes identified using a culture-independent approach are evolutionarily distant from known resistance genes, first demonstrating the immense diversity of AR genes in the human microbiome. Based on this recent human study, the gut microbiota in food animals may also contain diverse and novel AR genes that could be accessible to clinically relevant pathogens. However, the reservoirs of mobile AR genes in the gut microbiomes from food animals are still largely unknown. In addition, it is still not clear if the novel AR genes identified in the gut microbiome can function normally in a new host or a distant relative when HGT successfully occurs, a key issue for the emergence of AR in bacterial pathogens.
In this study, we examined AR genes in gut microbiomes of chickens from different production systems and determined the functional compatibility of the identified novel AR genes. The major bacterial strains and plasmids used in this study are listed in Table 1. We first isolated total genomic DNA directly from cecal contents of two free-range chickens that have never been treated with antimicrobials (kindly provided by a free-range farm in Dandridge, TN) and two conventionally raised chickens that have received antibiotic-containing feed (from Koch Foods, Inc., Morristown, TN). The sheared DNA fragments were cloned into expression vector pZE21-MCS (12) and transformed into Escherichia coli One Shot TOP10 electrocompetent cells (Epicentre) for library construction as described by Sommer et al. (21). The total sizes of the 4 expression libraries ranged from 0.4 × 108 bp to 2.4 × 108 bp. Using the same screening protocol as Sommer et al. (21), we selected AR clones from each library by plating culture on Luria broth agar plates containing the antibiotic of interest at the appropriate concentration (50 μg/ml of ampicillin, 10 μg/ml of tetracycline, 25 μg/ml of chloramphenicol, 75 μg/ml of spectinomycin, 12.5 μg/ml of norfloxacin, or 10 μg/ml of ciprofloxacin). Fifty inserts conferring resistance were sequenced by using primers pZE-F and pZE-R (Table 2); the sequences were annotated and analyzed as described by Sommer et al. (21).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| JL519 | DH5α containing pZE21-MCS | This study |
| JL48 | Conjugation helper strain; DH5α containing pRK2013 | 32 |
| JL860 | TOP10 containing pZE21-MCS-FRAmp1.1 | This study |
| JL861 | TOP10 containing pZE21-MCS-FRSpe1.1 | This study |
| JL850 | DH5α containing pZW | This study |
| JL851 | DH5α containing pZW1 | This study |
| JL852 | DH5α containing pZW2 | This study |
| JL853 | DH5α containing pZW3 | This study |
| C. jejuni strains | ||
| JL241 | NCTC 11168; human isolate | 16 |
| JL242 | C. jejuni 81-176; human isolate | 3 |
| JL854 | JL242 containing pZW | This study |
| JL855 | JL242 containing pZW1 | This study |
| JL856 | JL242 containing pZW2 | This study |
| JL857 | JL242 containing pZW3 | This study |
| Plasmids | ||
| pZE21-MCS | Cloning and expression vector; Kanr | 12 |
| pRY111 | E. coli-C. jejuni shuttle vector; Chlr | 30 |
| pRK2013 | Helper plasmid for triparental conjugation | 14 |
| pZE21-MCS-FRAmp1.1 | pZE21-MCS with cloned fragment containing ampicillin resistance gene FRAmp1.1 | This study |
| pZE21-MCS-FRSpe1.1 | pZE21-MCS with cloned fragment containing spectinomycin resistance gene FRSpe1.1 | This study |
| pZW | pRY111 containing flaA promoter | This study |
| pZW1 | pZW containing FRAmp1.1 | This study |
| pZW2 | pZW containing FRSpe1.1 | This study |
| pZW3 | pZW containing kanamycin resistance gene KAN-2 | This study |
Kanr, kanamycin resistant; Chlr, chloramphenicol resistant.
Table 2.
PCR primers used in this study
| Name | Sequencea |
|---|---|
| pZE-F | 5′-GAA TTC ATT AAA GAG GAG AAA GGT-3′ |
| pZE-R | 5′-TTT CGT TTT ATT TGA TGC CTC TAG-3′ |
| Sig28-F | 5′-GCT CTA GAG CGT AAA ATT GAA GAT GAA AGA GAG-3′ (XbaI) |
| Sig28-R | 5′-CGG GAT CCC GTT TTA AAT CCT TTT AAA TAA TTT C-3′ (BamHI) |
| CAm-BamHIF | 5′-CGG GAT CCC ATC GCA AGT GAA ATG ACA TCA GTA C-3′ (BamHI) |
| CAm-EcoRIR | 5′-CGG AAT TCC TCC TTA ACT CCT AAA ATT TAA CTT C-3′ (EcoRI) |
| CSp-BamHIF | 5′-CGG GAT CCC AAT GAA ATG ATA ATA TG-3′ (BamHI) |
| CSp-EcoRIR | 5′-CGG AAT TCC ATT TTA AGC AAA ACT TTA CAG CC-3′ (EcoRI) |
| CKa-F | 5′-CGG GAT CCG TAA TAC AAG GGG TGT TAT G-3′ (BamHI) |
| CKa-R | 5′-CGG AAT TCA TTA GAA AAA CTC ATC GAG C-3′ (EcoRI) |
Restriction sites are underlined in the primer sequences, and the corresponding names are in parentheses.
As shown in Table 3, 3 AR genes that conferred resistance to ampicillin or spectinomycin were identified in the gut microbiomes of two free-range chickens, while a total of 11 AR genes were identified from conventionally raised chickens (Table 3). The novel AR genes that shared low sequence similarity (58% to 76% at the amino acid level) with the known resistance genes constitute a substantial proportion of the AR genes identified in this study (6 of 14) (Table 3). Notably, among the 6 novel genes, 3 were unique and even shared low sequence similarities (<68% at the amino acid level) with the novel AR genes recently identified from the human gut microbiome using the same functional cloning approach (21); these 3 genes are FRAmp1.1, FRSpe1.1, and CRChl2.2 (Table 3). Discovery of the 3 AR genes from free-range chickens in this study strongly supports a recent theory that environmental bacteria harbor the readily available and diverse pool of AR genes regardless of antibiotic exposure (2). This finding is also consistent with a recent work directly showing that antibiotic resistance is a natural phenomenon that predates the modern selective pressure of clinical antibiotic use (4). It is not surprising that more AR genes, in terms of number and diversity, were identified in the conventionally raised chickens than in the free-range chickens. The extensive antimicrobial usage in conventionally raised chickens may enrich AR genes in the gut microbiome, consequently increasing likelihood and ease of identification of AR genes using the functional cloning approach. However, given the limitation of the small sample size of this study (2 birds for each production system), this finding needs to be confirmed in future large-scale experiments using animals from diverse sources.
Table 3.
Antibiotic resistance genes identified in the cecal contents of free-range and conventionally raised chickensa
| Antibioticb | Genec | Length (bp) | Annotation | % amino acid similarity to NCBId | % amino acid similarity to Sommer et al.e |
|---|---|---|---|---|---|
| AMP | FRAmp1.1 | 902 | β-Lactamase | 59 | 61 |
| FRAmp2.1 | 861 | β-Lactamase | 100 | 59 | |
| CRAmp1.1 | 861 | TEM β-lactamase | 99 | 59 | |
| CRAmp1.3 | 525 | TEM β-lactamase | 91 | 56 | |
| CRAmp2.1 | 504 | CbIA β-lactamase | 58 | 100 | |
| CRAmp2.2 | 642 | HGF-1 β-lactamase | 63 | 99 | |
| CRAmp2.3 | 510 | CbIA β-lactamase | 67 | 98 | |
| CRAmp1.4 | 756 | TEM β-lactamase | 100 | 62 | |
| TET | CRTet2.1 | 936 | TetW tetracycline resistance protein | 99 | 100 |
| CRTet2.2 | 660 | TetW tetracycline resistance protein | 99 | 100 | |
| CHL | CRChl2.1 | 804 | Chloramphenicol acetyltransferase | 92 | 77 |
| CRChl2.2 | 639 | Chloramphenicol acetyltransferase | 76 | 68 | |
| SPE | FRSpe1.1 | 786 | Adenyl transferase | 70 | NH |
| CRSpe2.1 | 774 | Adenyl transferase | 99 | NH | |
| NOR | – | – | – | – | – |
| CIP | – | – | – | – | – |
–, no genes identified or parameter not available.
Abbreviations: AMP, ampicillin; TET, tetracycline; CHL, chloramphenicol; SPE, spectinomycin; NOR, norfloxacin; CIP, ciprofloxacin.
Each gene identified in this study was named with a two-letter code corresponding to the source (FR, free-range chicken; CR, conventionally raised chicken) followed by a three-letter code referring to the three initial letters of the antibiotic used for screening, the library number corresponding to a specific chicken, and a number used to distinguish each unique gene against the specific antibiotic.
The highest percent similarity when compared with the AR genes deposited in GenBank (excluding those identified by Sommer et al. [21]).
The highest percent similarity when compared with the AR genes discovered by Sommer et al. (21) using the same culture-independent sampling and functional cloning approach. NH, no homologs found based on alignment.
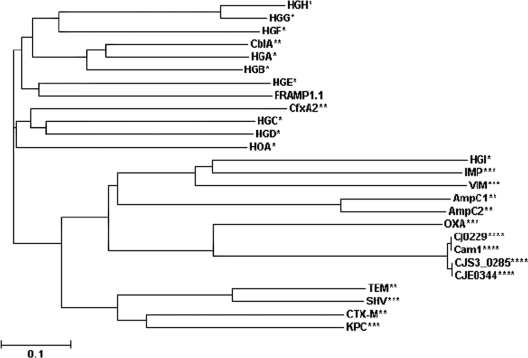
To better understand the uniqueness of the identified novel AR genes, we further analyzed the phylogenetic relationship of FRAmp1.1, a novel β-lactamase identified in the gut microbiome of a free-range chicken (Table 3), to 25 presently identified β-lactamases that represent 21 different families. As shown in Fig. 1, FRAmp1.1 and the β-lactamases recently identified in the human gut microbiome (21) were phylogenetically distant from most known β-lactamases that were characterized in cultivated organisms. In particular, FRAmp1.1 was very distantly related to the β-lactamases (Cj0229, Cam1, CJS_0285, and CJE0344) (Fig. 1) that have been identified in Campylobacter, an important food-borne bacterial pathogen with chicken as a major animal host (10, 11) and a distant relative of the E. coli host used here to generate genomic libraries. This finding prompted us to explore another significant issue of antibiotic resistance by taking advantage of the identified novel AR genes: can novel AR genes function normally in different bacterial species if the HGT barrier is successfully overcome? To address this issue, we first constructed an E. coli-Campylobacter shuttle vector (pZW) (Table 1) carrying a Campylobacter promoter according to the procedure described by Larsen et al. (8). The two selected novel AR gene products (FRAmp1.1, conferring ampicillin resistance, and FRSpe1.1, conferring spectinomycin resistance) shared low sequence similarity to the major determinants of ampicillin resistance (Cj0299) and spectinomycin resistance (AadA) in Campylobacter (6, 17). FRAmp1.1 and FRSpe1.1 were PCR amplified using proper primers pairs (Table 2), and the PCR fragments were cloned into pZW, creating pZW1 and pZW2, respectively. The promoterless kanamycin resistance gene KAN-2 from a Campylobacter jejuni strain (11) was also cloned into pZW to generate pZW3 (Table 1). All plasmids described above were transferred into C. jejuni 81-176 (JL242) (Table 1) using triparental conjugation as described previously (32). All the C. jejuni constructs were subjected to MIC tests for different antibiotics as determined by a standard microtiter broth dilution method (10). The JL857 strain (C. jejuni 81-176 containing pZW3) displayed a dramatically enhanced resistance to kanamycin (MIC > 128 μg/ml) compared to that of the control strain JL854 (MIC = 8 μg/ml), indicating that pZW was an effective expression vector for Campylobacter. The constructs harboring the novel AR genes (JL855 and JL856) (Table 1) also showed significantly higher levels of resistance to the corresponding antibiotics. Specifically, the JL856 strain containing the FRSpe1.1 gene displayed dramatically enhanced resistance to spectinomycin (MIC > 256 μg/ml) compared to that of the control strain JL854 (MIC = 8 μg/ml). Expression of FRAmp1.1 in JL855 also led to enhanced ampicillin resistance compared to that of the control JL854 strain (8-fold increase) (Table 4). The JL855 strain also displayed significantly increased resistance to a variety of β-lactams in C. jejuni, including those (e.g., cefotaxime) to which Cj0299 fails to confer high resistance (6) (Table 4). Together, these findings further support a previous hypothesis that if a barrier to gene transfer exists between the constituents of gut microbiomes and pathogens, it must be attributed to processes other than functional compatibility (21).
Fig 1.
Phylogenetic relationship of β-lactamases from different sources. Unrooted phylogenetic trees were generated from ClustalW2 alignments using the neighbor-joining algorithm. FRAmp1.1, discovered in this study, was analyzed together with 25 identified β-lactamases. *, the novel β-lactamases belonging to different families that were discovered from the human gut microbiome by Sommer et al. (21); **, the β-lactamases that were discovered from the human microbiome by Sommer et al. (21) that share high similarity to the previously characterized β-lactamase families; ***, the β-lactamases representing previously characterized common β-lactamase families that were not analyzed by Sommer et al. (21) (SHV [25], OXA-10 [accession number AY841859], IMP [15], VIM [9], and KPC [31]); ****, the β-lactamases identified in different C. jejuni strains, including Cj0299 (accession number YP_002343737), CJE0344 (accession number AAW34933), CJS3_0285 (accession number ADT72049), and Cam1 (accession number AAT01092).
Table 4.
MICs of different β-lactam antibiotics for the C. jejuni JL865 strain that carries resistance gene FRAmp1.1 and for the control strain JL854
| β-Lactam agent | MIC (μg/ml) |
|
|---|---|---|
| JL855 | JL854 | |
| Ampicillin | 8 | 1 |
| Amoxicillin | 128 | 16 |
| Penicillin G | 64 | 4 |
| Ticarcillin | 32 | 8 |
| Carbenicillin | 32 | 8 |
| Cloxacillin | >256 | 256 |
| Cefotaxime | >256 | 128 |
Characterization of novel AR genes from animal gut microbiomes is important in several ways. First, the genes that are divergent in sequence can help to assemble a more complete image of the evolutionary history of AR genes (2). Second, further functional and structural study of novel AR genes will improve our understanding of the relationship between sequence diversity and the resistance spectrum/level (28). In particular, identification of novel AR genes using the functional cloning approach would greatly complement modern high-throughput pyrosequencing of the metagenome by improving annotation power. Third, since the novel AR genes may function effectively in zoonotic pathogens, thorough examination of novel AR gene pools in food animals and even other agriculture ecosystems (e.g., manure) will greatly facilitate development of more-powerful molecular diagnostic tools to monitor AR in zoonotic pathogens and examine the development and transmission of AR (20). Finally, considering the fact that genes for antibiotic biosynthesis were sometimes clustered with AR genes, identification of novel AR genes may lead to the discovery of potentially novel antibiotics (19).
Although the novel gut AR genes have been successfully identified in both conventionally raised and free-range chickens in this study, the following limitations and issues should be addressed for future large-scale investigation of the AR reservoir. First, the sizes of expression libraries are a limiting factor of the likelihood of identifying more novel AR genes. The sizes of the libraries used in both a recent human study (∼109 bp per library) (21) and this study (∼108 bp per library) cover only a proportion of the gut microbiome (18). Thus, increasing library size, for instance by cloning larger insertions into the vector, will provide a greater chance to identify more novel AR genes. Second, we used chickens as a model here to analyze the gut AR reservoir in food animals. The functional cloning method can be extended into the investigation of the AR gene reservoir in other habitats, including the guts of other food animals as well as specific niches of agricultural ecosystems (e.g., manure, soil, water, lagoon). Recent studies for the identification of specific AR genes in soils also showed the feasibility of the similar methodology in revealing a natural AR gene reservoir, which exhibited a higher level of diversity than previously expected (1, 5, 19). Finally, the complex mechanisms of HGT should be elucidated in parallel to the effort for identification of novel AR gene reservoirs. The novel AR genes need to overcome multiple HGT barriers (23) before being able to confer resistance to the pathogens. AR gene reservoirs and HGT are equally important issues to be addressed in the future to improve our understanding of the development, transmission, and evolution of AR genes.
Nucleotide sequence accession numbers.
The full coding sequences of the 14 AR genes identified in this study were deposited in the GenBank database under accession numbers JN625754, JN625755, JN625756, JN625757, JN625758, JN625759, JN625760, JN625761, JN625762, JN625763, JN625764, JN625765, JN625766, and JN625767.
ACKNOWLEDGMENTS
We thank Andree A. Hunkapiller for technical support.
This study was supported by Microbiology across Campuses Educational and Research Venture (M-CERV) and AgResearch at The University of Tennessee.
Footnotes
Published ahead of print 27 January 2012
REFERENCES
- 1. Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. 2009. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME J. 3:243–251 [DOI] [PubMed] [Google Scholar]
- 2. Aminov RI, Mackie RI. 2007. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 271:147–161 [DOI] [PubMed] [Google Scholar]
- 3. Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472–479 [DOI] [PubMed] [Google Scholar]
- 4. D'Costa VM, et al. 2011. Antibiotic resistance is ancient. Nature 477:457–461 [DOI] [PubMed] [Google Scholar]
- 5. Donato JJ, et al. 2010. Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl. Environ. Microbiol. 76:4396–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Griggs DJ, et al. 2009. β-Lactamase-mediated β-lactam resistance in Campylobacter species: prevalence of Cj0299 (blaOXA-61) and evidence for a novel β-lactamase in C. jejuni. Antimicrob. Agents Chemother. 53:3357–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gyles CL. 2008. Antimicrobial resistance in selected bacteria from poultry. Anim. Health Res. Rev. 9:149–158 [DOI] [PubMed] [Google Scholar]
- 8. Larsen JC, Szymanski C, Guerry P. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186:6508–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lauretti L, et al. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin J, Wang Y, Hoang KV. 2009. Systematic identification of genetic loci required for polymyxin resistance in Campylobacter jejuni using an efficient in vivo transposon mutagenesis system. Foodborne Pathog. Dis. 6:173–185 [DOI] [PubMed] [Google Scholar]
- 12. Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marshall BM, Ochieng DJ, Levy SB. 2009. Commensals: underappreciated reservoirs of resistance. Microbe 4:8 [Google Scholar]
- 14. Miller WG, et al. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osano E, et al. 1994. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38:71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parkhill J, et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 17. Pinto-Alphandary H, Mabilat C, Courvalin P. 1990. Emergence of aminoglycoside resistance genes aadA and aadE in the genus Campylobacter. Antimicrob. Agents Chemother. 34:1294–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qin J, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Riesenfeld CS, Goodman RM, Handelsman J. 2004. Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ. Microbiol. 6:981–989 [DOI] [PubMed] [Google Scholar]
- 20. Shamputa IC, Rigouts L, Portaels F. 2004. Molecular genetic methods for diagnosis and antibiotic resistance detection of mycobacteria from clinical specimens. APMIS 112:728–752 [DOI] [PubMed] [Google Scholar]
- 21. Sommer MO, Dantas G, Church GM. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor LH, Latham SM, Woolhouse ME. 2001. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3:711–721 [DOI] [PubMed] [Google Scholar]
- 24. van den Bogaard AE, Stobberingh EE. 1999. Antibiotic usage in animals: impact on bacterial resistance and public health. Drugs 58:589–607 [DOI] [PubMed] [Google Scholar]
- 25. Veras DL, et al. 2011. Prevalence of the bla (SHV) gene in Klebsiella pneumoniae isolates obtained from hospital and community infections and from the microbiota of healthy individuals in Recife, Brazil. Curr. Microbiol. 62:1610–1616 [DOI] [PubMed] [Google Scholar]
- 26. Witte W. 2000. Ecological impact of antibiotic use in animals on different complex microflora: environment. Int. J. Antimicrob. Agents 14:321–325 [DOI] [PubMed] [Google Scholar]
- 27. Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wolter DJ, et al. 2009. Phenotypic and enzymatic comparative analysis of the novel KPC variant KPC-5 and its evolutionary variants, KPC-2 and KPC-4. Antimicrob. Agents Chemother. 53:557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Woolhouse ME, Gowtage-Sequeria S. 2005. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11:1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao R, Alm RA, Trust TJ, Guerry P. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127–130 [DOI] [PubMed] [Google Scholar]
- 31. Yigit H, et al. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45:1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zeng X, Xu F, Lin J. 2009. Molecular, antigenic, and functional characteristics of ferric enterobactin receptor CfrA in Campylobacter jejuni. Infect. Immun. 77:5437–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]