Abstract
Probiotics play an important role in the maintenance of the gastrointestinal barrier. In addition to direct effects on mucosal integrity, the interaction with the intestinal mucosa may have an active immunoregulatory effect. In the present work, we exposed HT29 intestinal epithelial cells to two Bifidobacterium species to determine their effect on gene expression profile, enterocyte monolayer integrity, and T-cell response. Bifidobacterium breve IPLA 20004 triggered a more pronounced increase in the transepithelial resistance of the enterocyte monolayer than Bifidobacterium bifidum LMG13195. The transcriptome profile of HT29 cells cultured in the presence of B. bifidum LMG13195 showed an increased expression of immune mediators and, interestingly, chemotactic molecules (CXCL10, CCL20, CXCL11 and CCL22) able to recruit lymphocytes. Since regulatory T cells (Treg cells) may express receptors for specific chemokines, we cultured peripheral blood mononuclear cells with supernatants of HT29 cells previously treated with Bifidobacterium strains and analyzed FOXP3 and CD25 Treg markers and CCR6, CXCR3, CCR4, and CCR3 expression on CD4+ lymphocytes. The proportion of CD25high FOXP3+ cells was significantly increased after culture with B. bifidum LMG13195-conditioned HT29 supernatant. Moreover, this treatment led to the largest amount of CCR6+ CXCR3− CCR4+ CCR3+ CD4+ cells expressing high levels of CD25, corresponding to the Treg population. These results suggest that soluble factors secreted after B. bifidum LMG13195 contact with intestinal epithelial cells favored the generation of CD4+ CD25high lymphocytes expressing chemokine receptor Treg markers, thus making possible their recruitment to the intestinal mucosa.
INTRODUCTION
Probiotics are live microorganisms which when administered in adequate amounts confer a health benefit on the host (33). The genus Bifidobacterium, a predominant member of the human gut microbiota, includes some strains which are widely used as probiotic bacteria (14). In this context, it is known that probiotics play an important role in the maintenance of the gastrointestinal barrier function (12, 18, 29), as well as exhibiting health properties through the modulation of both mucosal and systemic immunity under healthy or pathogenic conditions (6, 51).
Because of its strategically located anatomical situation, intestinal mucosa is of key importance for the communication between gut microbiota and immune cells of the gut-associated lymphoid tissue (GALT). Particularly, the layer of epithelial cells, mainly composed of enterocytes, plays an active role in the modulation of innate and adaptive immune responses (49). Intestinal epithelial cells can identify a wide variety of microorganisms, or their components, through membrane-bound pattern recognition receptors and could release chemokines, low-molecular-weight chemotactic cytokines relevant for the directional trafficking and the recruitment of specific immune cells. Specifically, chemokines have been described as being important in the recruitment and retention in different tissues of specific T-cell subsets expressing specific chemokine receptors (31). In addition, the chemokine receptor expression by T cells differs among the naïve, memory, effector, and regulatory T-cell subsets, depending on their activation status (37).
The immune cells localized in the GALT, such as dendritic cells (DCs) and T lymphocytes, constitute the first point of contact between gut commensals, or orally ingested probiotics, and our immune system (34, 50). In this sense, it has been described how distinct strains of Bifidobacterium species can induce different maturation and cytokine production patterns in DCs in a strain-specific manner (20, 27) that may direct the polarization of naïve CD4+ T cells toward different effector or regulatory T-cell subsets (4, 26, 52). In particular, nowadays there is increasing evidence regarding the ability of probiotic bacteria to induce CD25high FOXP3+ regulatory T cells (Treg cells) from naïve precursors (7, 10, 11, 26, 44). Since Treg cells can suppress uncontrolled effector responses to self and intraluminal antigens (5), the recruitment or induction of Treg cells by probiotics could have a beneficial effect on allergy and autoimmune diseases. Moreover, diverse studies have suggested that Treg cells could undergo selective migration, controlled by distinct signals from chemokines and their receptors, to sites where regulation is required (48).
Interestingly, we recently reported that exposing DCs to Bifidobacterium bifidum LMG13195 in vitro induces the polarization of naïve CD4+ lymphocytes into functional CD25high FOXP3+ Treg cells (26). However, nothing is known about the possible effect of this strain on intestinal mucosa. Thus, in the present work we wanted to study the response of human intestinal epithelial cells to this strain and its potential immune regulatory effect. For this purpose, we exposed HT29 cells to B. bifidum LMG13195 (and to Bifidobacterium breve IPLA 20004 as a control) to determine, first, the influence on HT29 gene expression, and second, the effect on chemokine receptors and Treg markers of human peripheral CD4+ lymphocytes. In addition, the effect on enterocyte monolayer integrity was evaluated.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. bifidum LMG13195 (LMG/BCCM [Belgian Co-ordinated Collections of Microorganisms], Brussels, Belgium) (36) and B. breve IPLA 20004 (2) were cultured in MRS medium (Difco, BD Biosciences, San Diego, CA) supplemented with 0.25% l-cysteine (Sigma Chemical Co., St. Louis, MO) (MRSc) at 37°C under anaerobic conditions (10% H2, 10% CO2, and 80% N2) in an MG500 chamber (Don Whitley Scientific, West Yorkshire, United Kingdom).
For the analysis of the effects on HT29 cell line monolayer integrity, UV-killed bacteria were obtained as previously described (26). The use of UV-killed bacteria was necessary in order to avoid acidification of the culture medium due to the long incubation times and the consequent monolayer damage. UV-treated bacterial suspensions were distributed in single-use aliquots, frozen in liquid N2, and stored at −80°C until use.
To evaluate the effects of the interaction between Bifidobacterium strains and HT29 cells on HT29 gene expression and Treg phenotype, live bacterial cells were used.
HT29 cell line culture conditions.
The epithelial intestinal cell line HT29 (ECACC no. 91072201), derived from human colon adenocarcinoma, was purchased from the European Collection of Cell Cultures (Salisbury, United Kingdom). The cell line was maintained in McCoy's medium supplemented with 3 mM l-glutamine, 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), and a mixture of antibiotics to give final concentrations of 50 μg/ml penicillin, 50 μg/ml streptomycin, 50 μg/ml gentamicin, and 1.25 μg/ml amphotericin B. All media and supplements were obtained from Sigma. The incubations took place at 37°C in 5% CO2 in an SL water-jacketed CO2 incubator (Sheldon Mfg., Inc., Cornelius, OR). Culture media were changed every 2 days, and the cell line was trypsinized with 0.25% trypsin-EDTA solution (Sigma) following standard procedures. For gene expression experiments, as well as for the collection of bifidobacterium-conditioned HT29 supernatants (SN), 105 cells/ml were seeded in 24-well plates, incubated to confluence (a monolayer, reaching about 107 cells/ml), and used after 13 ± 1 days. For monolayer integrity tests, the same procedure was used, but cells were grown in hanging cell culture inserts (0.4-μm pore size, polyethylene terephthalate; Millipore Corporation, Billerica, MA) placed in 12-well microplates (Millipore).
(i) HT29 cell gene expression analysis.
B. bifidum LMG13195 and B. breve IPLA 20004 were grown overnight in MRSc, harvested by centrifugation, washed twice with Dulbecco's phosphate-buffered saline (PBS) buffer, and resuspended in McCoy's medium without antibiotics. Five hundred microliters of bacterial suspension (5 × 107 bacteria) or McCoy's medium without bacteria (control) was added to each well containing HT29 monolayers, previously washed twice with Dulbecco's PBS to remove antibiotics. Microplates were then incubated for 6 h at 37°C in 5% CO2. After incubation, the culture media were collected and centrifuged at 4,000 rpm for 15 min to eliminate bacterial cells, and these bifidobacterium-conditioned supernatants were stored at −80°C until being used for stimulation of peripheral blood mononuclear cells (PBMCs). Then, the HT29 monolayers were released in 500 μl of RNA Protect cell reagent (Qiagen GmbH, Hilden, Germany) and kept frozen at −80°C until RNA extraction. Three independent experiments were carried out for each experimental condition.
RNA from HT29 cells was extracted by using the RNeasy Plus minikit (Qiagen) and QIAshredder homogenizer columns (Qiagen) following the manufacturer's instructions. RNA quality was monitored by gel electrophoresis, and it was quantified by using an Epoch apparatus (BioTek Instruments, Inc., Winoskii, VT). cDNA was synthesized using the Ambion WT expression kit (Applied Biosystems, Foster City, CA), and the hybridization was performed on Human Gene Expression arrays (GeneChip Human Gene 1.0 ST arrays; Affymetrix), following the protocols established by Affymetrix. Three independent experiments were carried out for each experimental condition. The microarray analyses were performed at the Parque Científico de Madrid (Campus de Cantoblanco, Madrid, Spain).
(ii) HT29 cell monolayer integrity.
Suspensions of UV-killed B. bifidum LMG13195 and B. breve IPLA 20004 cells were harvested by centrifugation and resuspended in McCoy's medium without antibiotics. Then, 500 μl of bacterial suspension (5 × 107 bacteria) was added to each HT29 monolayer grown on the apical insert compartment (bacterium/HT29 cell ratio, 10:1) and 2 ml of fresh medium was added to the basolateral compartment. Plates were incubated at 37°C in 5% CO2 in the Heracell 240 incubator (Thermo Electron LDD GmbH, Langenselbold, Germany) for 24 h. Transepithelial resistance (TER) was determined at different time points by using a Millicell ERS2 apparatus (Millipore). To calculate the resistance per area unit (Ω·cm2), the TER value obtained for the inserts with monolayer (with or without bifidobacteria) was subtracted with background TER from the insert alone (without monolayer) and multiplied by the area of the insert. Results were expressed with regard to that obtained for the control (medium without bifidobacteria), which was arbitrarily set to 100%. Experiments were carried out in duplicated HT29 microplates, and in each experiment, the strains were also tested in duplicate.
PBMC culture conditions.
Human peripheral blood mononuclear cells (PBMCs) were obtained from standard buffy coat preparations from routine blood donors (Asturian Blood Transfusion Center, Oviedo, Spain) by centrifugation over Ficoll-Hypaque gradients (Lymphoprep; Nycomed, Oslo, Norway) and extensive washing with PBS under sterile conditions. All blood donors (the numbers are specified in the figure legends) were healthy adult volunteers without any pathology or treatment. Approval for this study was obtained from the Regional Ethics Committee for Clinical Investigation.
(i) PBMC stimulation.
To determine the effect of bifidobacterium-conditioned HT29 supernatants on T-cell response, 2 × 104 PBMCs were cultured in 96-well plates in 200 μl of complete RPMI medium (RPMI 1640 containing 2 mM l-glutamine and 25 mM HEPES [Bio Whitaker, Verviers, Belgium] supplemented with 10% heat-inactivated FBS and the antibiotics streptomycin and ampicillin at 100 μg/ml each) at 37°C and 5% CO2. Additionally, complete RPMI medium was complemented with 10% of supernatants (SN) from cultures of HT29 cells incubated for 6 h with strain B. bifidum LMG13195 (LMG13195-HT29 SN) or B. breve IPLA 20004 (IPLA 20004-HT29 SN) or without any bacterial treatment (Control-HT29 SN), as previously explained above. After 5 days of culture, cells were collected and washed twice with PBS before cytometric analysis.
(ii) Flow cytometric analysis.
Phenotypic studies of PBMCs were performed after staining with the appropriate monoclonal antibody (MAb) using a FACSCanto II flow cytometer (Becton Dickinson, BD Biosciences, San Diego, CA). Cells were stained with anti-CD25 (fluorescein isothiocyanate [FITC] conjugated), CCR3 (phycoerythrin [PE] conjugated), CCR4 (PE-Cy7 conjugated), CXCR3 (allophycocyanin [APC] conjugated), CD4 (APC-Cy7), CCR6 (peridinin chlorophyll protein [PerCP]-Cy5.5 conjugated), CD127 (PE-Cy7 conjugated) MAb or with the corresponding isotype-matched conjugated irrelevant MAb as a negative control. All MAbs were supplied by Pharmingen (BD Biosciences). Extracellular staining of CD4, CD25, CD127, and chemokine receptors was performed for 30 min at 4°C, and then cells were washed twice in staining buffer and resuspended in PBS. After that, cells were fixed, permeabilized, and intracellularly stained with anti-FOXP3 (PE conjugated) (clone PCH101) following the manufacturer's instructions (Foxp3/transcription factor staining buffer set; eBiosciences Inc., San Diego, CA). The analysis was based on cells of the living region defined using forward and side scatter. Cells were further gated according to the CD4 expression. A minimum of 10,000 CD4+ lymphocytes were acquired and analyzed using the FACSDiva software 6.1.2 (BD Biosciences). Positive cells for each marker were determined using fluorescence of cells treated with the corresponding isotype-matched conjugated irrelevant MAb as a negative control. The specific fluorescence intensity was quantified as the mean fluorescence intensity (MFI) calculated by subtracting the background of isotype-matched control staining from the total fluorescence. According to the MFI, CD4+ T cells expressing CD25 were subdivided into CD25low and CD25high populations. Because CD25 expression varies depending on the treatment applied to cells, a different MFI cutoff was used to determine CD25high populations. Freshly isolated human cells showed continuous CD25 expression, and then an MFI of 103 was used to determine CD25high population (corresponding to 1 to 2% of CD4+ lymphocytes). After PBMC culture, the CD25 activation marker increases and a population expressing high CD25 levels was detected presenting an MFI of >2.103 (extracellular staining) or >4.102 when cells were fixed and permeabilized (intracellular FOXP3 staining).
Statistical analysis.
The Kolmogorov-Smirnov test was used to assess the normal distribution of the data. The percentage of CD4+ CD25high FOXP3+ T cells, as well as the proportions of CD25high, CD25low, and CD25− lymphocytes out of CCR6+ CXCR3−CCR4+ CCR3+ population after culture with the different HT29 SN, was compared by using analysis of the variance (ANOVA); when a significant ANOVA test was obtained, Tukey's multiple comparisons tests were conducted to determine which groups' pairs had different means. The expression of chemokine receptors and Treg markers in CD25high with respect to CD25− T cells was analyzed by t test. The comparisons carried out between samples are described in the figure legend for each experiment. GraphPad Prism 5 software (GraphPad Software, San Diego, CA) and SPSS 18.0 software were used for all determinations, and a P value of <0.05 was considered significant.
RESULTS
B. bifidum LMG13195 affected the transcriptomic profile of HT29 cells.
In order to characterize the effect of B. bifidum LMG13195 and B. breve IPLA 20004 on human intestinal epithelial cells, we analyzed the transcriptome profile of HT29 cells cultured for 6 h in the presence of these strains, compared with control HT29 cells (not exposed to bacteria), by using a Human Gene Expression Array. In general, although changes in gene expression were modest, both strains showed the ability to modulate the transcriptome of HT29 cells. Overall, at a significance level P < 0.01, the treatment of HT29 cells with B. bifidum LMG13195 induced a differential expression of 121 genes with respect to the control culture, while the culture with B. breve IPLA 20004 modified the expression of 173 genes. Interestingly, the treatment with B. bifidum LMG13195 showed an increased expression of diverse genes associated with immune responses to a higher degree than B. breve IPLA 20004 (see Table S1 in the supplemental material). Specifically, among the genes showing higher induction (Table 1), we identified a number of genes coding for chemokines; thus, B. bifidum LMG13195 was able to increase the expression of CXCL10, CCL20, CXCL11, and CCL22 in HT29 cells. In addition, genes induced by interferon (IFN), involved in microbial defense in human mucosa, were also found to be increased, like those coding for IL-28 and IL-29, two IFN type III molecules.
Table 1.
Genes upregulated in HT29 cells treated with Bifidobacterium strains
| Gene product designation | GenBank accession no. |
Bifidobacterium-cultured/control HT29 cell expression ratioa |
Biological process | |
|---|---|---|---|---|
| B. bifidum LMG13195 | B. breve IPLA 20004 | |||
| MX2 | NM_002463 | 1.741 | 1.187 | Antiviral defense |
| RSAD2 | NM_080657 | 2.333 | 1.355 | Antiviral defense |
| NUPR1 | NM_001042483 | 1.807 | 1.581 | Cellular growth factor |
| CCL20 | NM_004591 | 2.033 | 1.232 | Chemokine |
| CCL22 | NM_002990 | 1.754 | 1.591 | Chemokine |
| CCL5 | NM_002985 | 1.889 | 1.060 | Chemokine |
| CX3CL1 | NM_002996 | 1.746 | 1.327 | Chemokine |
| CXCL10 | NM_001565 | 3.063 | 1.546 | Chemokine |
| CXCL11 | NM_005409 | 1.953 | 1.293 | Chemokine |
| LAMP3 | NM_014398 | 1.511 | 1.056 | Dendritic cell function |
| SLFN5 | NM_144975 | 1.536 | 1.173 | Differentiation |
| AKR1C2 | NM_001354 | 1.710 | 1.244 | Enzymatic activity |
| CMPK2 | NM_207315 | 1.695 | 1.214 | Enzymatic activity |
| MMP13 | NM_002427 | 1.773 | 1.298 | Enzymatic activity |
| ND6 | ENST00000361681 | 1.629 | 0.661 | Enzymatic activity |
| UBE2L6 | NM_004223 | 1.548 | 1.148 | Enzymatic activity |
| USP18 | NM_017414 | 1.736 | 1.333 | Enzymatic activity |
| RARRES3 | NM_004585 | 1.649 | 1.333 | Enzymatic activity |
| XAF1 | NM_017523 | 1.655 | 1.201 | Inhibitor of apoptosis protein |
| IL-28A | NM_172138 | 1.876 | 1.089 | Inmune function |
| IL-29 | NM_172140 | 2.872 | 1.157 | Inmune function |
| RAET1L | NM_130900 | 1.629 | 1.071 | Inmune function |
| SECTM1 | NM_003004 | 1.840 | 1.440 | Inmune function |
| NLRC5 | NM_032206 | 1.595 | 1.129 | Interferon signaling |
| TRIM22 | NM_006074 | 1.630 | 1.162 | Interferon-induced antiviral protein |
| TRIM31 | NM_007028 | 1.801 | 1.337 | Interferon-induced antiviral protein |
| OAS2 | NM_002535 | 1.580 | 1.119 | Interferon-mediated signaling |
| OAS3 | NM_006187 | 1.584 | 1.163 | Interferon-mediated signaling |
| OASL | NM_003733 | 2.212 | 1.338 | Interferon-mediated signaling |
| IFIT1 | NM_001548 | 1.547 | 1.116 | Interferon-mediated signaling |
| IFIT2 | NM_001547 | 2.864 | 1.569 | Interferon-mediated signaling |
| IFIT3 | NM_001031683 | 1.560 | 1.097 | Interferon-mediated signaling |
| APOL6 | NM_030641 | 1.615 | 1.227 | Lipid transporter protein |
| MT2A | NM_005953 | 1.506 | 1.261 | Metal binding protein |
| SERPINE1 | NM_000602 | 2.037 | 1.274 | mRNA stability |
| SLC15A3 | NM_016582 | 1.641 | 1.265 | Oligopeptide transport |
| RBMY1A1 | NM_005058 | 1.252 | 1.577 | RNA-binding protein |
| KRT18P49 | XR_018216 | 1.523 | 1.329 | Structural protein |
| TMEM140 | NM_018295 | 1.531 | 1.209 | Structural protein |
| SP110 | NM_080424 | 1.784 | 1.339 | Transcription factor |
| BATF2 | NM_138456 | 1.739 | 1.258 | Transcription factor |
| TAP1 | NM_000593 | 1.509 | 1.130 | Transporter protein |
| ZBP1 | NM_030776 | 1.519 | 1.134 | Transporter protein |
| PML | NM_033240 | 1.661 | 1.356 | Tumor suppressor |
| SAMD9L | NM_152703 | 1.680 | 1.169 | Unknown function |
Boldface values represent ratios of >1.5 between gene expression in HT29 cells cultured with each Bifidobacterium strain and gene expression in HT29 cells without bacterial treatment.
B. bifidum LMG13195-conditioned HT29 supernatants influenced T-cell recruitment responses.
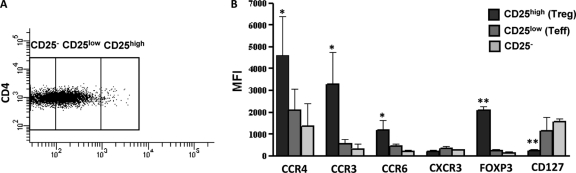
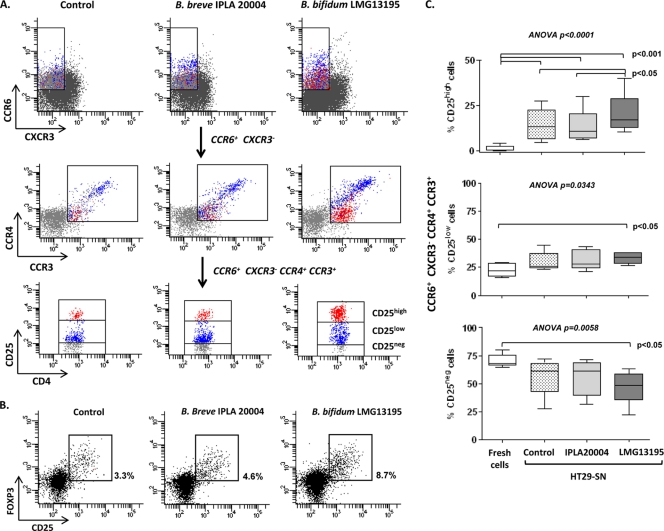
We recently reported that DCs exposed to B. bifidum LMG13195 in vitro induce the polarization of naïve CD4+ lymphocytes into functional CD25high FOXP3+ Treg cells (26). Since HT29 cells increased the expression of immune mediators and chemotactic molecules after exposure to live bifidobacteria, we wanted to determine their possible involvement in the generation or recruitment of Treg cells, given that these lymphocytes may express receptors for specific chemokines. Thus, PBMCs from healthy individuals were cultured with the supernatants of HT29 cells previously incubated with B. bifidum LMG13195 (LMG13195-HT29 SN), B. breve IPLA 20004 (IPLA 20004-HT29 SN), or medium alone (Control-HT29 SN) for 5 days, and the expression of CD25, FOXP3, and CD127, as well as that of the chemokine receptors CCR3, CCR4, CCR6, and CXCR3, was analyzed by flow cytometry in CD4+ lymphocytes before and after culture. Figure 1 shows that before culture, freshly isolated CD4+ CD25high cells, the natural Treg (nTreg) cell population, in addition to showing high FOXP3 and low CD127 levels, expressed CCR6, CCR4, and CCR3, but not CXCR3, so we used the CCR6+ CCR4+ CCR3+ CXCR3− phenotype as a chemokine receptor Treg marker (Fig. 2A). After 5 days of culture with the different HT29 SN, the proportion of CD4+ CD25high FOXP3+ T cells increased significantly (P < 0.05) in all treatments (Control-HT29 SN, 5.44% ± 1.92%; LMG13195-HT29 SN, 6.20% ± 1.59%; IPLA 20004-HT29 SN, 3.68% ± 1.05%) with respect to freshly isolated cells (1.10% ± 3.35%), as well as being significantly higher after LMG13195-HT29 SN compared with IPLA 20004-HT29 SN treatment (P < 0.05) (Fig. 2A and B). Similarly, the CCR6+ CCR4+ CCR3+ CXCR3− population, regardless of CD25 expression, increased after culture compared with freshly isolated cells (2.76% ± 2.39%), but no significant differences were observed between the two bifidobacterium-HT29 SN treatments (LMG13195-HT29 SN, 4.54% ± 0.81%; IPLA 20004-HT29 SN, 4.58% ± 1.31%). Given that these chemokine receptors could also be expressed by CD25low effector T cells (24), which were also generated after stimulation with the different HT29 SN (Fig. 2A), we determined the amount of CCR6+ CCR4+ CCR3+ CXCR3− cells expressing high levels of CD25 (Fig. 2A, red cells), the putative Treg population. Figures 2A and C show that stimulation with LMG13195-HT29 SN was significantly the most efficient at inducing the production of CD25high cells expressing the chemokine receptor Treg markers compared with IPLA 20004-HT29 SN or Control-HT29 SN. No significant differences in the percentages of these cells were observed in the CD25low subset among different cultures, but stimulation with LMG13195-HT29 SN resulted in the lower levels of resting CD25− cells. Interestingly, CD25high CCR6+ CCR4+ CCR3+ CXCR3− cells corresponded with a population expressing intermediate levels of CCR6, CCR4, and CCR3 (Fig. 2A, red cells), whereas CCR4high CCR6high cells (a Th17-associated phenotype) (24) were mostly included in the CD25low subset (Fig. 2A, blue cells), thus probably being effector T cells.
Fig 1.
Differential chemokine receptor expression on CD25high, CD25low, and CD25− CD4+ T cells. Freshly isolated human PBMCs were stained extracellularly for CD4, CD25, CCR3, CCR4, CXCR3, CCR6, and CD127 and intracellularly for FOXP3 markers and analyzed by flow cytometry. (A) CD4+ T lymphocytes were divided according to CD25 expression. (B) Gated CD25high (nTreg), CD25low (effector Th [Teff]), and CD25− (resting) CD4+ T cells were analyzed for the expression of chemokine receptors and Treg markers. Bars represent the mean and standard deviation of MFI obtained in five independent experiments performed with different blood donors. Statistical differences between CD25high and CD25− cells were evaluated by t test. *, P < 0.05; **, P < 0.01.
Fig 2.
Supernatants from Bifidobacterium-treated HT29 cells influence chemokine receptor expression on T lymphocytes. PBMCs were incubated for 5 days with a 10% pool of supernatants from cultures of HT29 cells (HT29 SN) with B. bifidum LMG13195 (LMG13195-HT29 SN), B. breve IPLA 20004 (IPLA 20004-HT29 SN), or without any bacterial treatment (Control-HT29 SN). Cultured cells were recovered, stained for chemokine receptors and Treg markers, and analyzed by flow cytometry. (A) A sequential gating strategy was used to select CD4+ T cells with the phenotype CCR6+ CCR4+ CCR3+ CXCR3− and afterwards to determine the expression level of the CD25 marker. Red cells represent the CD25high subset among the CCR6+ CCR4+ CCR3+ CXCR3− population, whereas blue cells are the CD25low subset presenting this chemokine receptor phenotype. Dot plots are representative of 5 independent experiments performed with different blood donors. (B) CD4+ lymphocytes stained after culture were gated, and the percentage of CD25high FOXP3+ cells induced by LMG13195-HT29 SN, IPLA 20004-HT29 SN, or Control-HT29 SN was determined. Dot plots correspond to a representative example of 8 independent blood donors. (C) Box plots represent the percentage of CD25high, CD25low, and CD25− lymphocytes out of the CCR6+ CCR4+ CCR3+ CXCR3− population before (fresh cells) and after PBMC culture with the different HT29 SN. Statistical differences among groups were assessed by ANOVA for repeated measures; Tukey's posttests for multiple comparisons were conducted to determine which groups' pairs had different means.
B. bifidum LMG13195 and B. breve IPLA 20004 enhanced intestinal barrier function.
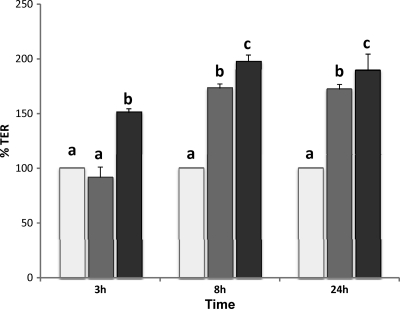
Finally, to characterize the effect of B. bifidum LMG13195 and B. breve IPLA 20004 on intestinal barrier function, we determined the TER of HT29 intestinal epithelia cell monolayers exposed to the UV-inactivated bifidobacteria at different times (Fig. 3). After 3 h of coculture with the bacteria, an increase in the monolayer TER was already observed for the strain B. breve IPLA 20004, while strain B. bifidum LMG13195 did not show differences with regard to the control (medium without bacteria). However, at later incubation times (8 and 24 h), a significant increase in TER was observed for both microorganisms, this increase being significantly higher for B. breve IPLA 20004 than for B. bifidum LMG13195.
Fig 3.
Influence of B. bifidum LMG13195 and B. breve IPLA 20004 on HT29 monolayer integrity. Transepithelial resistance of HT29 cell monolayers was determined after 3, 8, and 24 h of cocultivation with B. bifidum LMG13195 (dark gray bars) and B. breve IPLA 20004 (black bars) or in culture media without bacteria (control; light gray bars). TER obtained for the control (HT-29 cells cultured in medium without bifidobacteria) was normalized to 100% and used as a reference. Statistical analysis was assessed by means of ANOVA. Samples that do not share equal letters are statistically different (P < 0.05).
DISCUSSION
Commonly used probiotics include Bifidobacterium species, commensal microorganisms usually present in the gut of adult individuals where they could interact with intestinal epithelial cells. This interplay may modulate the local immune environment through, for instance, the modification of gene expression and the production of chemokines and other immune molecules by intestinal cells under both physiological and pathological conditions (15, 21, 38, 40). However, the extent to which commensal bacteria regulate the expression of immune molecules by epithelial cells is poorly understood. For this reason, we analyzed the transcriptome profile of the colonic epithelial HT29 cell line after short-term treatment with B. bifidum LMG13195, a Treg inducer strain in in vitro studies, or B. breve IPLA 20004, a control bifidobacterium strain. Interestingly, although both strains induced gene expression changes in the HT29 cells compared with cells cultured without bacteria, B. bifidum LMG13195, rather than B. breve IPLA 20004, was able to increase the expression of a number of genes associated with immune responses. Specifically, B. bifidum LMG13195 upregulated the HT29 gene expression of diverse chemokines (CCL20, CCL22, CXCL10, and CXCL11) and two type III interferon molecules (IL-28 and IL-29), important immune mediators that play key roles in host mucosa homeostasis and defense (47). These data suggest that soluble factors and chemotactic cytokines derived from the combined effect of intestinal epithelial cells and B. bifidum LMG13195 contact could influence mucosal immunity and attract specific subsets to gut mucosa.
Studies on transcriptional responses of human epithelial cells to probiotic bifidobacteria are scarce and inconclusive (35, 43). More studies, however, have been performed with Lactobacillus strains, observing differences both in vitro (28, 30, 39) and in vivo (8, 45, 46) after bacterial treatment. In fact, the inducing effect on a number of genes coding for chemotactic cytokines observed in this work has also been described in in vivo studies with probiotic lactobacilli (8).
Due to its relevant role in intestinal immunity, we considered especially interesting the finding about the increased expression of CCL20, previously reported after Lactobacillus johnsonii N6.2 stimulation of the epithelial cell line Caco-2 (22). CCL20 (also known as macrophage inflammatory protein-3α [MIP-3α]) is a chemokine constitutively expressed at a low basal level by a variety of normal human mucosa-associated tissues, especially in the gut mucosal epithelial cells (19, 32). During normal development and immune homeostasis, CCL20 selectively attract CCR6-expressing lymphocytes and DCs (3, 13) to the mucosal surfaces, organizing lymphoid tissues, such as Peyer's patches, mesenteric lymph nodes, and GALT (9, 19). Additionally, CCL20/CCR6-mediated signals can be strongly induced by proinflammatory stimuli, including cytokines (e.g., tumor necrosis factor alpha [TNF-α]) and Toll-like receptor (TLR) agonists, originating from microbes (41), thus contributing to the recruitment of target cells to epithelial mucosal surfaces. Therefore, intestinal epithelial cells might have the capacity to link innate and acquired mucosal immunity through the upregulation of CCL20, which in turn recruited CCR6-expressing T cells, specifically, Th1, Th17, and Treg subsets.
In view of the capability of B. bifidum LMG13195 to increase HT29 gene transcription of chemotactic molecules that could activate and attract specific immune cell subsets to gut mucosa, and its ability to induce the polarization of naïve CD4+ lymphocytes into functional CD25high FOXP3+ Treg cells through its effect on DCs (26), we wanted to determine the possible contribution of epithelial cell exposure to this strain to the generation of Treg cells able to be recruited to the intestinal mucosa. The Treg population can be identified by its high expression of CD25 and the transcription factor FOXP3 and lack of the IL-7R alpha chain (CD127) (25, 42). Additionally, CD4+ CD25high Treg cells may express several chemokine receptors, some of them shared with CD25−/low effector Th cells. In fact, we observed that freshly isolated CD4+ CD25high Treg cells (nTreg), in addition to high FOXP3 and low CD127 levels, express CCR6, CCR4, and CCR3 but not CXCR3. Thus, we considered CCR6+ CCR4+ CCR3+ CXCR3− a chemokine receptor Treg marker for the CD25high population, in accordance with the reported expression of CCR4 (16), CCR6 (23), and CCR3 (1) in nTreg cells. Then, we observed that stimulation of PBMCs with all the HT29 SN increased the expression of these chemokine receptors on CD4+ T cells. An unexpected and interesting result was that the highest expression of CCR4, CCR6, and CCR3 corresponded to the CD25low population (enhanced after activation), while CD25high lymphocytes presented intermediate levels of these molecules. In this respect, it is known that Th17 effector cells express CCR6 and CCR4 (24), whereas expression of CCR3, associated with Th2 profile, has been reported to be higher in CD25low than in CD25high cells (1).
After culture with the different HT29 SN, no differences were detected in the amount of total CCR6+ CXCR3− CCR4+ CCR3+ CD4+ T cells. However, the percentage of CD25high cells expressing chemokine receptor Treg markers was significantly higher after LMG13195-HT29 SN treatment, whereas IPLA 20004 SN and Control-HT29 SN showed similar results. Moreover, the highest proportion of total and CD25high FOXP3+ cells was also obtained after stimulation with LMG13195-HT29 SN. Our results indicate that soluble factors secreted after B. bifidum LMG13195 contact with intestinal epithelial cells favored the generation of CD4+ CD25high cells expressing chemokine receptor Treg markers, thus making possible their recruitment to the intestinal mucosa. Although some bacterial products could be present in supernatants without HT29 cells, in a previous work, we reported that diverse cell-free bifidobacterial culture supernatants were unable to induce significant responses in PBMCs (27), suggesting that the results observed in this work came from the combined effects of the bacteria and the epithelial cells. These data are in agreement with the proposed Treg inducer ability of B. bifidum LMG13195 and support that an effect on the intestinal mucosa could be a mechanism by which this strain plays an active immunoregulatory role. In line with our findings, how the cross talk between human intestinal epithelial and immune cells helped in maintaining gut immune homeostasis was recently described. Iliev et al. reported that monocyte-derived DCs, conditioned with supernatants from Caco-2 or intestinal epithelial cells isolated from healthy donors, promoted the differentiation of tolerogenic DCs able to drive the development of gut-homing Treg cells, which were effective in suppressing T-cell proliferation in vitro and extremely potent in protecting against colitis in vivo (17). This evidence supports epithelial cells not only being a physical barrier but also playing an active role in the production of diverse factors which control DC function, Treg differentiation, and intestinal tolerance. In this sense, the effect of probiotics on Treg induction/recruitment to the mucosal surface would be therapeutically beneficial in controlling the excessive immune responses involved in chronic inflammation, mucosal allergies, and autoimmune diseases.
Finally, in addition to the beneficial activities on the immune system, in the present work, we wanted to extend the understanding of the interaction between bifidobacteria and the gut mucosa. One previously described probiotic-related beneficial mechanism is to increase the physical resistance of the mucosa. In the present study, we observed that both B. bifidum LMG13195 and B. breve IPLA 20004 strains were able to increase the integrity of the HT29 monolayer in vitro, thus contributing to strengthening the gut barrier, this effect being more pronounced for IPLA 20004. Additionally, the adherence to human epithelial cells and cell lines is one of the most exhaustive tests used to indicate the increased ability of a probiotic strain to have transitory persistence in the colon. Interestingly, this trait could also be a way to connect the interaction between bifidobacteria and intestinal immune homeostasis. Previous experiments carried out by our group showed an extremely high adhesion rate of B. bifidum LMG13195 to HT29 cells (adhesion rate to HT29 cells of about 75%; González-Rodriguez et al., unpublished data) compared to B. breve IPLA 20004 (about 5.5% adhesion to HT29 cells) (2). Thus, the greater capacity of B. bifidum LMG13195 to stay in contact with the epithelium could partially explain its ability to induce different characteristics on the HT29 supernatant, which, in turn, could influence the generation of CD4+ CD25high cells expressing chemokine receptor Treg markers, thus making possible their recruitment to the intestinal mucosa.
In summary, our results suggested a strengthening of the gut barrier through the interaction of the B. bifidum LMG13195 strain with colonocytes. This may lead to a specific environment at local epithelium promoting the induction of Treg cells expressing chemokine receptors that favor mucosal homing, an attractive goal in the prevention and treatment of diseases characterized by an overreaction of the immune system, such as autoimmune diseases, asthma, and allergy.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by European Union FEDER funds, the Spanish Plan Nacional de I+D (projects AGL2007-61805, AGL2009-09445, and AGL2010-14952), and the Consejo Superior de Investigaciones Científicas (CSIC PIE 200870I049).
Footnotes
Published ahead of print 17 February 2012
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1. Ahern D, Lloyd CM, Robinson DS. 2009. Chemokine responsiveness of CD4+ CD25+ regulatory and CD4+ CD25− T cells from atopic and nonatopic donors. Allergy 64:1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arboleya S, et al. 2011. Characterization and in vitro properties of potentially probiotic Bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 149:28–36 [DOI] [PubMed] [Google Scholar]
- 3. Baba M, et al. 1997. Identification of CCR6, the specific receptor for a novel lymphocyte-directed CC chemokine LARC. J. Biol. Chem. 272:14893–14898 [DOI] [PubMed] [Google Scholar]
- 4. Banchereau J, et al. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767–811 [DOI] [PubMed] [Google Scholar]
- 5. Belkaid Y, Rouse BT. 2005. Natural regulatory T cells in infectious disease. Nat. Immunol. 6:353–360 [DOI] [PubMed] [Google Scholar]
- 6. Boirivant M, Strober W. 2007. The mechanism of action of probiotics. Curr. Opin. Gastroenterol. 23:679–692 [DOI] [PubMed] [Google Scholar]
- 7. de Roock S, et al. 2010. Lactic acid bacteria differ in their ability to induce functional regulatory T cells in humans. Clin. Exp. Allergy 40:103–110 [DOI] [PubMed] [Google Scholar]
- 8. Di Caro S, et al. 2005. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig. Liver Dis. 37:320–329 [DOI] [PubMed] [Google Scholar]
- 9. Dieu MC, et al. 1998. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 188:373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. 2005. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J. Immunol. 174:3237–3246 [DOI] [PubMed] [Google Scholar]
- 11. Foligne B, et al. 2007. A key role of dendritic cells in probiotic functionality. PLoS One 2:e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gionchetti P, Rizzello F, Campieri M. 2001. Probiotics and antibiotics in inflammatory bowel disease. Curr. Opin. Gastroenterol. 17:331–335 [DOI] [PubMed] [Google Scholar]
- 13. Greaves DR, et al. 1997. CCR6, a CC chemokine receptor that interacts with macrophage inflammatory protein 3alpha and is highly expressed in human dendritic cells. J. Exp. Med. 186:837–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guarner F, Malagelada JR. 2003. Bacterial flora of the digestive tract. Gastroenterol. Hepatol. 26(Suppl 1):1–512585997 [Google Scholar]
- 15. Haller D, Blum S, Bode C, Hammes WP, Schiffrin EJ. 2000. Activation of human peripheral blood mononuclear cells by nonpathogenic bacteria in vitro: evidence of NK cells as primary targets. Infect. Immun. 68:752–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iellem A, et al. 2001. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4(+)CD25(+) regulatory T cells. J. Exp. Med. 194:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iliev ID, et al. 2009. Human intestinal epithelial cells promote the differentiation of tolerogenic dendritic cells. Gut 58:1481–1489 [DOI] [PubMed] [Google Scholar]
- 18. Isolauri E, Juntunen M, Rautanen T, Sillanaukee P, Koivula T. 1991. A human Lactobacillus strain (Lactobacillus casei sp strain GG) promotes recovery from acute diarrhea in children. Pediatrics 88:90–97 [PubMed] [Google Scholar]
- 19. Iwasaki A, Kelsall BL. 2000. Localization of distinct Peyer's patch dendritic cell subsets and their recruitment by chemokines macrophage inflammatory protein (MIP)-3alpha, MIP-3beta, and secondary lymphoid organ chemokine. J. Exp. Med. 191:1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joffre O, Nolte MA, Spörri R, Reis e Sousa C. 2009. Inflammatory signals in dendritic cell activation and the induction of adaptive immunity. Immunol. Rev. 227:234–247 [DOI] [PubMed] [Google Scholar]
- 21. Jung HC, et al. 1995. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J. Clin. Invest. 95:55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kingma SDK, et al. 2011. Lactobacillus johnsonii N6.2 stimulates the innate immune response through Toll-like receptor 9 in Caco-2 cells and increases intestinal crypt Paneth cell number in biobreeding diabetes-prone rats. J. Nutr. 141:1023–1028 [DOI] [PubMed] [Google Scholar]
- 23. Kleinewietfeld M, et al. 2005. CCR6 expression defines regulatory effector/memory-like cells within the CD25(+)CD4+ T-cell subset. Blood 105:2877–2886 [DOI] [PubMed] [Google Scholar]
- 24. Lim HW, Lee J, Hillsamer P, Kim CH. 2008. Human Th17 cells share major trafficking receptors with both polarized effector T cells and FOXP3+ regulatory T cells. J. Immunol. 180:122–129 [DOI] [PubMed] [Google Scholar]
- 25. Liu W, et al. 2006. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J. Exp. Med. 203:1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. López P, González-Rodríguez I, Gueimonde M, Margolles A, Suárez A. 2011. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One 6:e24776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. López P, Gueimonde M, Margolles A, Suárez A. 2010. Distinct Bifidobacterium strains drive different immune responses in vitro. Int. J. Food Microbiol. 138:157–165 [DOI] [PubMed] [Google Scholar]
- 28. Mack DR, Ahrne S, Hyde L, Wei S, Hollingsworth MA. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52:827–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Madsen KL. 2001. The use of probiotics in gastrointestinal disease. Can. J. Gastroenterol. 15:817–822 [DOI] [PubMed] [Google Scholar]
- 30. Paolillo R, Romano Carratelli C, Sorrentino S, Mazzola N, Rizzo A. 2009. Immunomodulatory effects of Lactobacillus plantarum on human colon cancer cells. Int. Immunopharmacol. 9:1265–1271 [DOI] [PubMed] [Google Scholar]
- 31. Pease JE, Williams TJ. 2006. Chemokines and their receptors in allergic disease. J. Allergy Clin. Immunol. 118:305–318 [DOI] [PubMed] [Google Scholar]
- 32. Reibman J, Hsu Y, Chen LC, Bleck B, Gordon T. 2003. Airway epithelial cells release MIP-3alpha/CCL20 in response to cytokines and ambient particulate matter. Am. J. Respir. Cell Mol. Biol. 28:648–654 [DOI] [PubMed] [Google Scholar]
- 33. Reid G, et al. 2003. New scientific paradigms for probiotics and prebiotics. J. Clin. Gastroenterol. 37:105–118 [DOI] [PubMed] [Google Scholar]
- 34. Rescigno M. 2008. Intestinal epithelial cells control dendritic cell function. J. Pediatr. Gastroenterol. Nutr. 46(Suppl 1):E17–E19 [DOI] [PubMed] [Google Scholar]
- 35. Riedel CU, Foata F, Goldstein DR, Blum S, Eikmanns BJ. 2006. Interaction of bifidobacteria with Caco-2 cells—adhesion and impact on expression profiles. Int. J. Food Microbiol. 110:62–68 [DOI] [PubMed] [Google Scholar]
- 36. Sakata S, et al. 2006. Characterization of the genus Bifidobacterium by automated ribotyping and 16S rRNA gene sequences. Microbiol. Immunol. 50:1–10 [DOI] [PubMed] [Google Scholar]
- 37. Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. 1998. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 187:875–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Savkovic SD, Koutsouris A, Hecht G. 1997. Activation of NF-kappaB in intestinal epithelial cells by enteropathogenic Escherichia coli. Am. J. Physiol. 273:C1160–C1167 [DOI] [PubMed] [Google Scholar]
- 39. Schlee M, et al. 2008. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin. Exp. Immunol. 151:528–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schuerer-Maly CC, Eckmann L, Kagnoff MF, Falco MT, Maly FE. 1994. Colonic epithelial cell lines as a source of interleukin-8: stimulation by inflammatory cytokines and bacterial lipopolysaccharide. Immunology 81:85–91 [PMC free article] [PubMed] [Google Scholar]
- 41. Schutyser E, Struyf S, Van Damme J. 2003. The CC chemokine CCL20 and its receptor CCR6. Cytokine Growth Factor Rev. 14:409–426 [DOI] [PubMed] [Google Scholar]
- 42. Seddiki N, et al. 2006. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J. Exp. Med. 203:1693–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shima T, et al. 2008. Differential effects of two probiotic strains with different bacteriological properties on intestinal gene expression, with special reference to indigenous bacteria. FEMS Immunol. Med. Microbiol. 52:69–77 [DOI] [PubMed] [Google Scholar]
- 44. Smits HH, et al. 2005. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 115:1260–1267 [DOI] [PubMed] [Google Scholar]
- 45. Troost FJ, et al. 2008. Identification of the transcriptional response of human intestinal mucosa to Lactobacillus plantarum WCFS1 in vivo. BMC Genomics 9:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Baarlen P, et al. 2011. Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4562–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vilcek J. 2003. Novel interferons. Nat. Immunol. 4:8–9 [DOI] [PubMed] [Google Scholar]
- 48. Wei S, Kryczek I, Zou W. 2006. Regulatory T-cell compartmentalization and trafficking. Blood 108:426–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wen L, et al. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Westendorf AM, Fleissner D, Hansen W, Buer J. 2010. T cells, dendritic cells and epithelial cells in intestinal homeostasis. Int. J. Med. Microbiol. 300:11–18 [DOI] [PubMed] [Google Scholar]
- 51. Yan F, Polk DB. 2011. Probiotics and immune health. Curr. Opin. Gastroenterol. 27:496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu J, Paul WE. 2008. CD4 T cells: fates, functions, and faults. Blood 112:1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.