Abstract
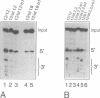
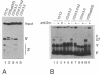
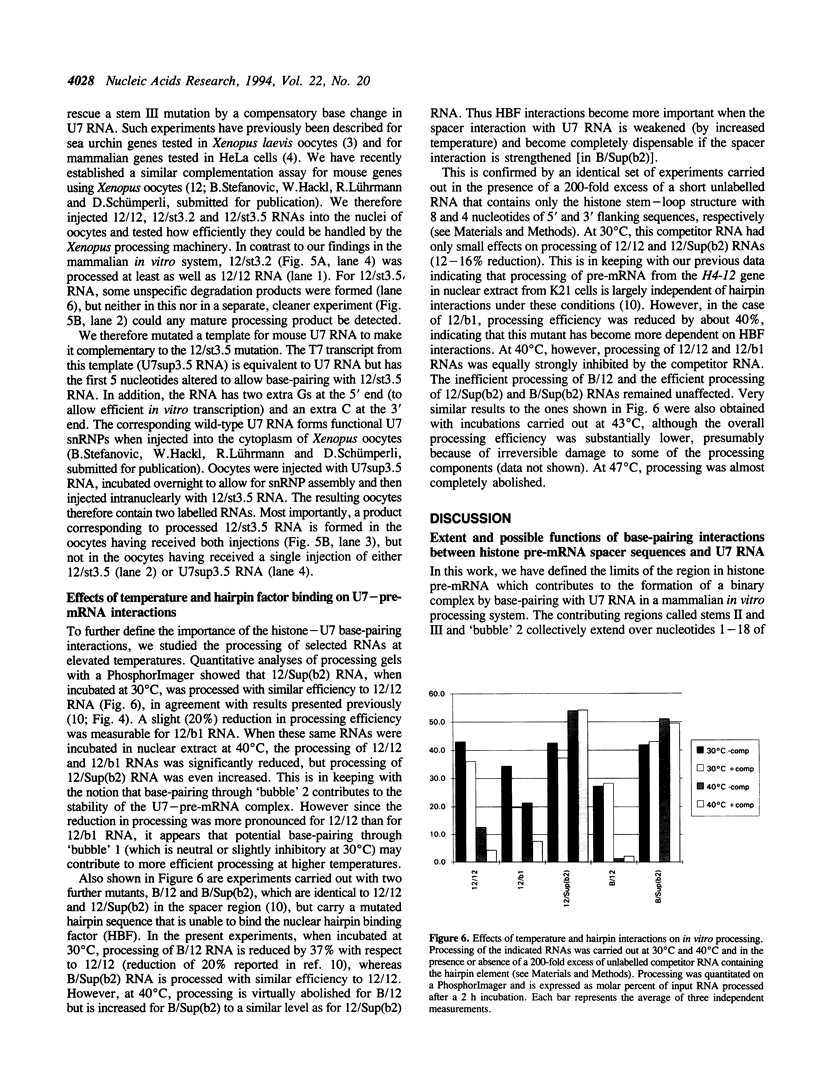
We have analysed the extent of base-pairing interactions between spacer sequences of histone pre-mRNA and U7 snRNA present in the trans-acting U7 snRNP and their importance for histone RNA 3' end processing in vitro. For the efficiently processed mouse H4-12 gene, a computer analysis revealed that additional base pairs could be formed with U7 RNA outside of the previously recognised spacer element (stem II). One complementarity (stem III) is located more 3' and involves nucleotides from the very 5' end of U7 RNA. The other, more 5' located complementarity (stem I) involves nucleotides of the Sm binding site of U7 RNA, a part known to interact with snRNP structural proteins. These potential stem structures are separated from each other by short internal loops of unpaired nucleotides. Mutational analyses of the pre-mRNA indicate that stems II and III are equally important for interaction with the U7 snRNP and for processing, whereas mutations in stem I have moderate effects on processing efficiency, but do not impair complex formation with the U7 snRNP. Thus nucleotides near the processing site may be important for processing, but do not contribute to the assembly of an active complex by forming a stem I structure. The importance of stem III was confirmed by the ability of a complementary mutation in U7 RNA to suppress a stem III mutation in a complementation assay using Xenopus laevis oocytes. The main role of the factor(s) binding to the upstream hairpin loop is to stabilise the U7-pre-mRNA complex. This was shown by either stabilising (by mutation) or destabilising (by increased temperature) the U7-pre-mRNA base-pairing under conditions where hairpin factor binding was either allowed or prevented (by mutation or competition). The hairpin dependence of processing was found to be inversely related to the strength of the U7-pre-mRNA interaction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birchmeier C., Folk W., Birnstiel M. L. The terminal RNA stem-loop structure and 80 bp of spacer DNA are required for the formation of 3' termini of sea urchin H2A mRNA. Cell. 1983 Dec;35(2 Pt 1):433–440. doi: 10.1016/0092-8674(83)90176-9. [DOI] [PubMed] [Google Scholar]
- Birchmeier C., Grosschedl R., Birnstiel M. L. Generation of authentic 3' termini of an H2A mRNA in vivo is dependent on a short inverted DNA repeat and on spacer sequences. Cell. 1982 Apr;28(4):739–745. doi: 10.1016/0092-8674(82)90053-8. [DOI] [PubMed] [Google Scholar]
- Bond U. M., Yario T. A., Steitz J. A. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991 Sep;5(9):1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- Cotten M., Gick O., Vasserot A., Schaffner G., Birnstiel M. L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3' RNA processing reaction. EMBO J. 1988 Mar;7(3):801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev O., Birnstiel M. L. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3' termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 1985 Feb;4(2):481–489. doi: 10.1002/j.1460-2075.1985.tb03654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer A., Keller W., Birnstiel M. L. Generation of histone mRNA 3' ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986 Jun;5(6):1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gick O., Krämer A., Vasserot A., Birnstiel M. L. Heat-labile regulatory factor is required for 3' processing of histone precursor mRNAs. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G. M., Schaufele F., Schaffner G., Birnstiel M. L. Functional analysis of the sea urchin U7 small nuclear RNA. Mol Cell Biol. 1988 Mar;8(3):1076–1084. doi: 10.1128/mcb.8.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Stefanovic B., Schümperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993 Mar;12(3):1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann I., Birnstiel M. L. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature. 1990 Aug 16;346(6285):665–668. doi: 10.1038/346665a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5495–5499. doi: 10.1073/pnas.76.11.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher B., Schümperli D. RNA 3' processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987 Jun;6(6):1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin L., Soldati D., Mital R., Streit A., Schümperli D. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992 Feb;11(2):691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Oh R., Steitz J. A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3'-end processing reaction. Mol Cell Biol. 1989 Jul;9(7):3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry K. L., Steitz J. A. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3' end formation in vitro. Mol Cell Biol. 1987 May;7(5):1663–1672. doi: 10.1128/mcb.7.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey N. B., Williams A. S., Sun J. H., Brown V. D., Bond U., Marzluff W. F. Point mutations in the stem-loop at the 3' end of mouse histone mRNA reduce expression by reducing the efficiency of 3' end formation. Mol Cell Biol. 1994 Mar;14(3):1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharl E. C., Steitz J. A. The site of 3' end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 1994 May 15;13(10):2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F., Gilmartin G. M., Bannwarth W., Birnstiel M. L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3' end of H3 messenger RNA. 1986 Oct 30-Nov 5Nature. 323(6091):777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- Schümperli D. Multilevel regulation of replication-dependent histone genes. Trends Genet. 1988 Jul;4(7):187–191. doi: 10.1016/0168-9525(88)90074-1. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Tabiti K., Schaffner G., Soldati D., Albrecht U., Birnstiel M. L. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2'-O-methyl oligoribonucleotides. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati D., Schümperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3' processing of histone pre-mRNA. Mol Cell Biol. 1988 Apr;8(4):1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C., Soldati D., Lüscher B., Schümperli D. Histone-specific RNA 3' processing in nuclear extracts from mammalian cells. Methods Enzymol. 1990;181:74–89. doi: 10.1016/0076-6879(90)81113-9. [DOI] [PubMed] [Google Scholar]
- Streit A., Koning T. W., Soldati D., Melin L., Schümperli D. Variable effects of the conserved RNA hairpin element upon 3' end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993 Apr 11;21(7):1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub K., Galli G., Busslinger M., Birnstiel M. L. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984 Dec 1;3(12):2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasserot A. P., Schaufele F. J., Birnstiel M. L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittop Koning T. H., Schümperli D. RNAs and ribonucleoproteins in recognition and catalysis. Eur J Biochem. 1994 Jan 15;219(1-2):25–42. doi: 10.1007/978-3-642-79502-2_3. [DOI] [PubMed] [Google Scholar]