Abstract
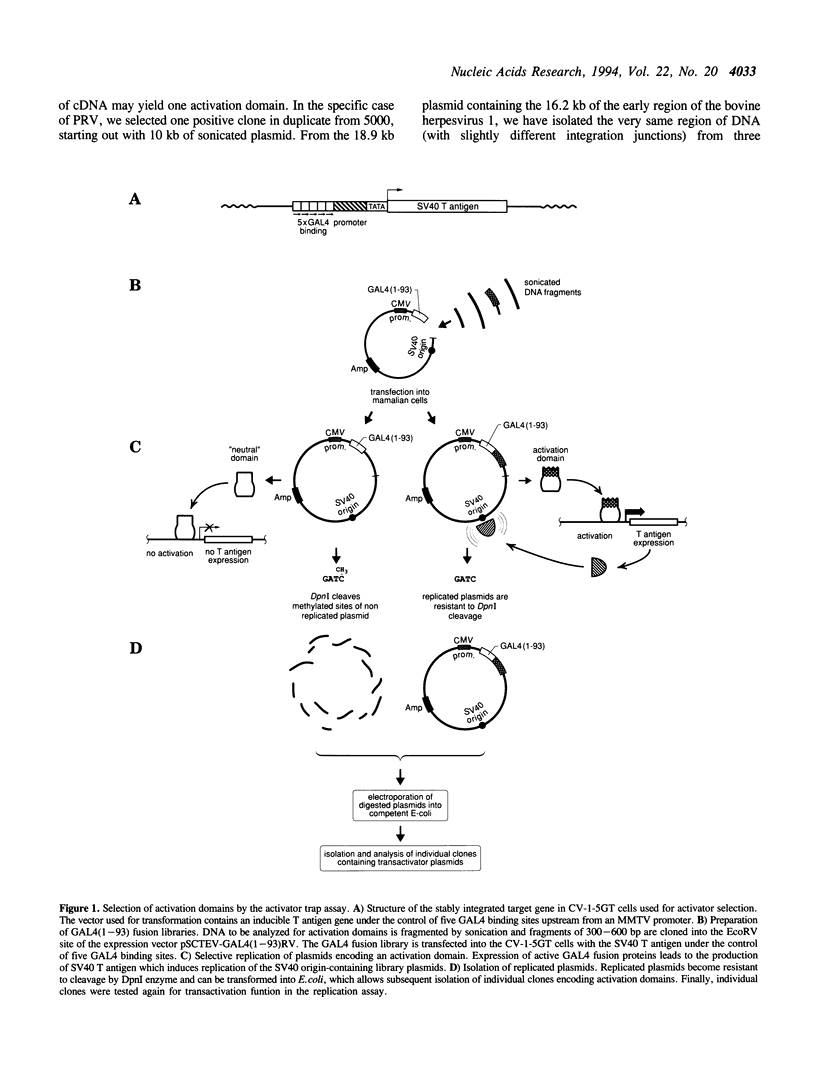
Transcription factors often contain activation domains that interact with the basic transcription machinery. We have developed a functional screening strategy in mammalian cells to selectively isolate activation domains from a library of random DNA inserts. For this, sonicated DNA fragments are cloned next to the DNA binding domain of GAL4 factor in a plasmid that also contains the SV40 origin of replication. Pools of fusion protein clones are transfected into CV-1-5GT monkey cells containing an SV40 T antigen gene under the control of a promoter with GAL4 binding sites. Plasmids that express functional transactivating fusion proteins activate the T antigen gene, thus promoting selective amplification of the plasmid in the mammalian host cell line. Using this method, we were able to select strong enhancer-type activation domains from the immediate early regions of two herpesviruses, namely pseudorabies virus and bovine herpesvirus 1. In both cases, the activation domains selected were homologues of the ICP4 regulatory protein of herpes simplex virus. The activation domain from pseudorabies virus is four times stronger than the activation domain of herpes simplex virus protein VP16 (Vmw65), making it the strongest activation domain characterized so far. This activator trap method should be useful for precisely localizing activation domain(s) in known factors, or to identify mammalian transcriptional adaptors that do not bind DNA and which may escape conventional detection methods.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abmayr S. M., Workman J. L., Roeder R. G. The pseudorabies immediate early protein stimulates in vitro transcription by facilitating TFIID: promoter interactions. Genes Dev. 1988 May;2(5):542–553. doi: 10.1101/gad.2.5.542. [DOI] [PubMed] [Google Scholar]
- Baker A., Schatz G. Sequences from a prokaryotic genome or the mouse dihydrofolate reductase gene can restore the import of a truncated precursor protein into yeast mitochondria. Proc Natl Acad Sci U S A. 1987 May;84(10):3117–3121. doi: 10.1073/pnas.84.10.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A., Hampl H. Functions of the major nonstructural DNA binding protein of a herpesvirus (pseudorabies). Virology. 1983 Jan 30;124(2):411–424. doi: 10.1016/0042-6822(83)90357-4. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Carey M., Leatherwood J., Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990 Feb 9;247(4943):710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- Cress W. D., Triezenberg S. J. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991 Jan 4;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Disney G. H., McKee T. A., Preston C. M., Everett R. D. The product of varicella-zoster virus gene 62 autoregulates its own promoter. J Gen Virol. 1990 Dec;71(Pt 12):2999–3003. doi: 10.1099/0022-1317-71-12-2999. [DOI] [PubMed] [Google Scholar]
- Donaldson L., Capone J. P. Purification and characterization of the carboxyl-terminal transactivation domain of Vmw65 from herpes simplex virus type 1. J Biol Chem. 1992 Jan 25;267(3):1411–1414. [PubMed] [Google Scholar]
- Gerster T., Matthias P., Thali M., Jiricny J., Schaffner W. Cell type-specificity elements of the immunoglobulin heavy chain gene enhancer. EMBO J. 1987 May;6(5):1323–1330. doi: 10.1002/j.1460-2075.1987.tb02371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. A., Hoey T., Thut C. J., Admon A., Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993 Nov 5;75(3):519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Green M. R., Treisman R., Maniatis T. Transcriptional activation of cloned human beta-globin genes by viral immediate-early gene products. Cell. 1983 Nov;35(1):137–148. doi: 10.1016/0092-8674(83)90216-7. [DOI] [PubMed] [Google Scholar]
- Hahn S. Structure(?) and function of acidic transcription activators. Cell. 1993 Feb 26;72(4):481–483. doi: 10.1016/0092-8674(93)90064-w. [DOI] [PubMed] [Google Scholar]
- He X., Rosenfeld M. G. Mechanisms of complex transcriptional regulation: implications for brain development. Neuron. 1991 Aug;7(2):183–196. doi: 10.1016/0896-6273(91)90257-z. [DOI] [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Kaiser C. A., Preuss D., Grisafi P., Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987 Jan 16;235(4786):312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Young R. A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994 Mar 31;368(6470):466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Künzler M., Braus G. H., Georgiev O., Seipel K., Schaffner W. Functional differences between mammalian transcription activation domains at the yeast GAL1 promoter. EMBO J. 1994 Feb 1;13(3):641–645. doi: 10.1002/j.1460-2075.1994.tb06302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. A new class of yeast transcriptional activators. Cell. 1987 Oct 9;51(1):113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- Martin K. J., Lillie J. W., Green M. R. Transcriptional activation by the pseudorabies virus immediate early protein. Genes Dev. 1990 Dec;4(12B):2376–2382. doi: 10.1101/gad.4.12b.2376. [DOI] [PubMed] [Google Scholar]
- Muller M. T. Binding of the herpes simplex virus immediate-early gene product ICP4 to its own transcription start site. J Virol. 1987 Mar;61(3):858–865. doi: 10.1128/jvi.61.3.858-865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera L. P., Mosca J. D., Ruyechan W. T., Hayward G. S., Straus S. E., Hay J. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J Virol. 1993 Aug;67(8):4474–4483. doi: 10.1128/jvi.67.8.4474-4483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier J. L., Shen F., Triezenberg S. J. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. G., Ha I., Maldonado E., Reinberg D., Green M. R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993 Jun 24;363(6431):741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- Rusconi S., Severne Y., Georgiev O., Galli I., Wieland S. A novel expression assay to study transcriptional activators. Gene. 1990 May 14;89(2):211–221. doi: 10.1016/0378-1119(90)90008-f. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
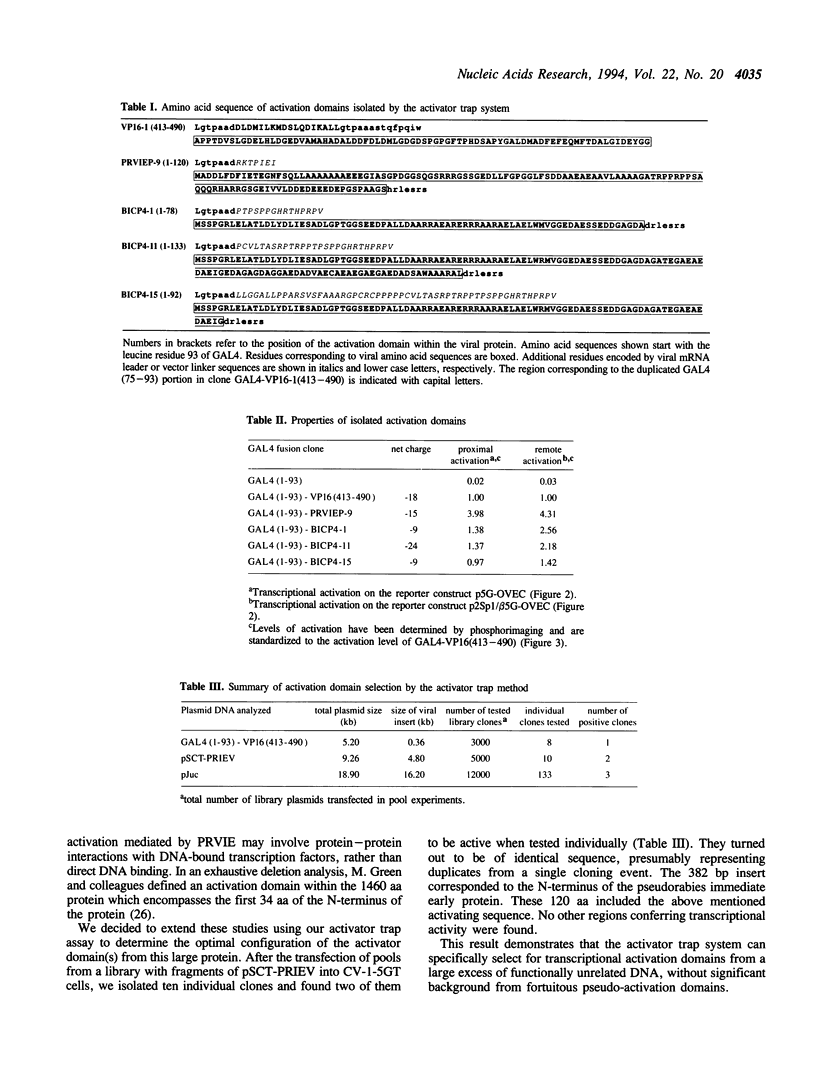
- Schwyzer M., Vlcek C., Menekse O., Fraefel C., Paces V. Promoter, spliced leader, and coding sequence for BICP4, the largest of the immediate-early proteins of bovine herpesvirus 1. Virology. 1993 Nov;197(1):349–357. doi: 10.1006/viro.1993.1596. [DOI] [PubMed] [Google Scholar]
- Seipel K., Georgiev O., Gerber H. P., Schaffner W. C-terminal domain (CTD) of RNA-polymerase II and N-terminal segment of the human TATA binding protein (TBP) can mediate remote and proximal transcriptional activation, respectively. Nucleic Acids Res. 1993 Dec 11;21(24):5609–5615. doi: 10.1093/nar/21.24.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
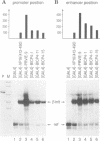
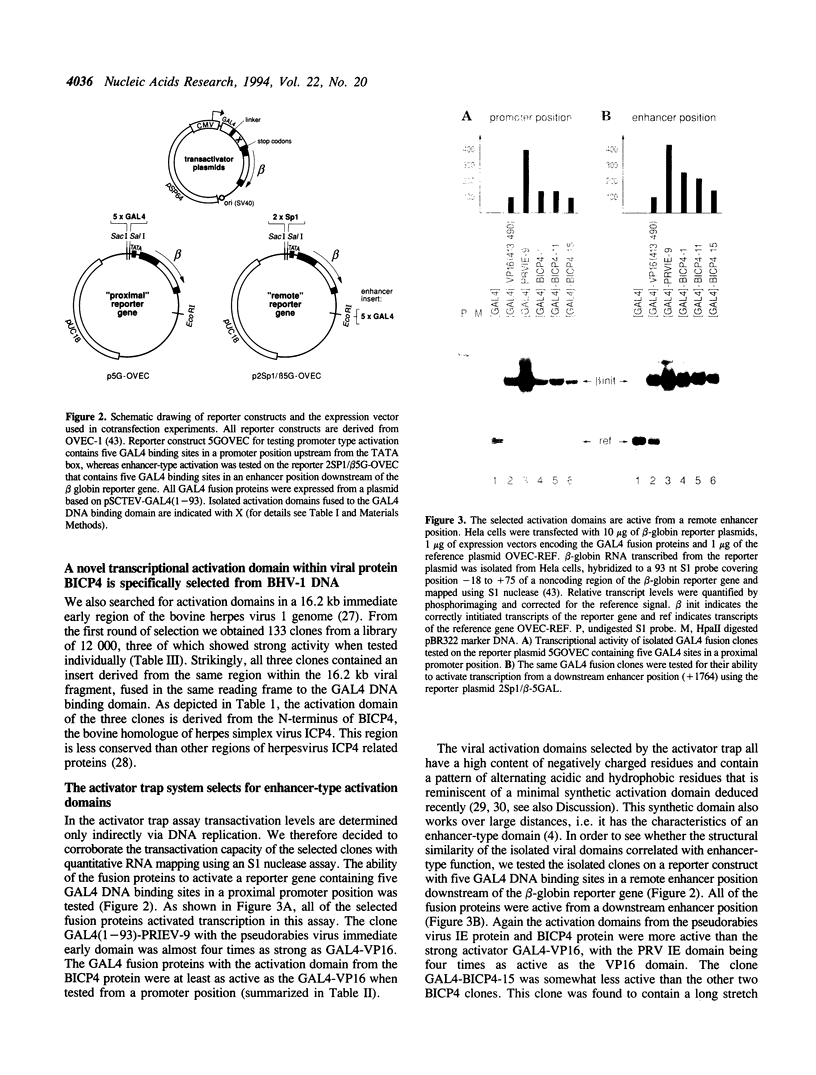
- Seipel K., Georgiev O., Schaffner W. Different activation domains stimulate transcription from remote ('enhancer') and proximal ('promoter') positions. EMBO J. 1992 Dec;11(13):4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfling E., Lübbe A., Dorsch-Häsler K., Schaffner W. Metal-dependent SV40 viruses containing inducible enhancers from the upstream region of metallothionein genes. EMBO J. 1985 Dec 30;4(13B):3851–3859. doi: 10.1002/j.1460-2075.1985.tb04157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Bates P., Rivera-Gonzalez R., Gu B., DeLuca N. A. ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB. J Virol. 1993 Aug;67(8):4676–4687. doi: 10.1128/jvi.67.8.4676-4687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Stern S., Tanaka M., Herr W. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989 Oct 19;341(6243):624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- Strum K. Acid connections. Curr Biol. 1991 Jun;1(3):188–191. doi: 10.1016/0960-9822(91)90230-t. [DOI] [PubMed] [Google Scholar]
- Thali M., Rusconi S., Schaffner W. Immediate early protein of pseudorabies virus is a general transactivator but stimulates only suboptimally utilized promoters. A clue to specificity? J Mol Biol. 1990 Sep 20;215(2):301–311. doi: 10.1016/S0022-2836(05)80348-1. [DOI] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- Tognoni A., Cattaneo R., Serfling E., Schaffner W. A novel expression selection approach allows precise mapping of the hepatitis B virus enhancer. Nucleic Acids Res. 1985 Oct 25;13(20):7457–7472. doi: 10.1093/nar/13.20.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triezenberg S. J., Kingsbury R. C., McKnight S. L. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 1988 Jun;2(6):718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Van Hoy M., Leuther K. K., Kodadek T., Johnston S. A. The acidic activation domains of the GCN4 and GAL4 proteins are not alpha helical but form beta sheets. Cell. 1993 Feb 26;72(4):587–594. doi: 10.1016/0092-8674(93)90077-4. [DOI] [PubMed] [Google Scholar]
- Vlcek C., Kozmík Z., Paces V., Schirm S., Schwyzer M. Pseudorabies virus immediate-early gene overlaps with an oppositely oriented open reading frame: characterization of their promoter and enhancer regions. Virology. 1990 Nov;179(1):365–377. doi: 10.1016/0042-6822(90)90304-a. [DOI] [PubMed] [Google Scholar]
- Weber F., Schaffner W. Simian virus 40 enhancer increases RNA polymerase density within the linked gene. Nature. 1985 May 2;315(6014):75–77. doi: 10.1038/315075a0. [DOI] [PubMed] [Google Scholar]
- Weber F., de Villiers J., Schaffner W. An SV40 "enhancer trap" incorporates exogenous enhancers or generates enhancers from its own sequences. Cell. 1984 Apr;36(4):983–992. doi: 10.1016/0092-8674(84)90048-5. [DOI] [PubMed] [Google Scholar]
- Westin G., Gerster T., Müller M. M., Schaffner G., Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987 Sep 11;15(17):6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. C., LaMarco K., Peterson M. G., Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993 Jul 16;74(1):115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- Wirth U. V., Vogt B., Schwyzer M. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J Virol. 1991 Jan;65(1):195–205. doi: 10.1128/jvi.65.1.195-205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Rungger D., Georgiev O., Seipel K., Schaffner W. Different potential of cellular and viral activators of transcription revealed in oocytes and early embryos of Xenopus laevis. Biol Chem Hoppe Seyler. 1994 Feb;375(2):105–112. doi: 10.1515/bchm3.1994.375.2.105. [DOI] [PubMed] [Google Scholar]