Abstract
Among vertebrates, most of the largest genomes are found within the salamanders, a clade of amphibians that includes 613 species. Salamander genome sizes range from ∼14 to ∼120 Gb. Because genome size is correlated with nucleus and cell sizes, as well as other traits, morphological evolution in salamanders has been profoundly affected by genomic gigantism. However, the molecular mechanisms driving genomic expansion in this clade remain largely unknown. Here, we present the first comparative analysis of transposable element (TE) content in salamanders. Using high-throughput sequencing, we generated genomic shotgun data for six species from the Plethodontidae, the largest family of salamanders. We then developed a pipeline to mine TE sequences from shotgun data in taxa with limited genomic resources, such as salamanders. Our summaries of overall TE abundance and diversity for each species demonstrate that TEs make up a substantial portion of salamander genomes, and that all of the major known types of TEs are represented in salamanders. The most abundant TE superfamilies found in the genomes of our six focal species are similar, despite substantial variation in genome size. However, our results demonstrate a major difference between salamanders and other vertebrates: salamander genomes contain much larger amounts of long terminal repeat (LTR) retrotransposons, primarily Ty3/gypsy elements. Thus, the extreme increase in genome size that occurred in salamanders was likely accompanied by a shift in TE landscape. These results suggest that increased proliferation of LTR retrotransposons was a major molecular mechanism contributing to genomic expansion in salamanders.
Keywords: LTR retrotransposon, transposable element landscape, genomic expansion, TE age distributions, genome size evolution, plethodontid salamanders
Introduction
Genomes dictate phenotype via their gene and regulatory sequences, which control the production of proteins underlying organismal development and function. However, genomes also impact phenotype via their overall size, irrespective of their DNA sequence. Genome size can have profound effects on organismal biology, potentially affecting traits as diverse as nucleus size, cell size, duration of the cell cycle, cell differentiation rate, metabolic rate, embryonic developmental rate, limb regeneration rate, life history strategy, invasiveness, and extinction rate (Olmo and Morescalchi 1975; Sessions and Larson 1987; Jockusch 1997; Gregory 2003; Gregory 2005b), but see (Lynch 2007). Within animals, genome size varies 6,650-fold (0.02–130 Gb), with 530-fold variation within the vertebrates alone (0.34–130 Gb) (Gregory 2011). Understanding both the molecular mechanisms and the evolutionary forces shaping this variation remains a central goal in biology (Vinogradov 2004; Oliver et al. 2007).
Transposable elements (TEs) are mobile DNA sequences that can insert into new genomic locations, often replicating themselves during the process (Craig et al. 2002). Two classes of TEs exist that differ in the molecular mechanism by which they transpose from one genomic location to another: Class I TEs (retrotransposons) transpose via a “copy and paste” mechanism, utilizing an RNA intermediate, and generating a new TE copy that inserts into a novel genomic location. Most Class II TEs (DNA transposons) transpose via a “cut and paste” mechanism, utilizing a DNA intermediate and moving to a new genomic location without an obligate increase in copy number (Craig et al. 2002; Wicker et al. 2007). These two TE classes are further subdivided into subclasses, superfamilies, families, etc. based on structural features, details of the transposition mechanism, and sequence similarity (Wicker et al. 2007). Both TE classes coexist in a wide range of eukaryotes, suggesting their ancient evolutionary origins. However, extreme variation in the number, activity, and diversity of TEs occurs in the genomes of different species, both within and among the major eukaryotic clades (Goodier and Kazazian 2008).
TEs, and other types of repetitive DNA, make up the bulk of many eukaryotic genomes and are a major determinant of genome size and architecture (Pritham 2009; Venner et al. 2009). The effects of individual TE insertions on the “host” organism can also vary dramatically. Although some TE sequences have been domesticated by their hosts and now form critical components of genes and/or gene regulatory networks (Volff 2006; Feschotte 2008), TE insertions can be deleterious because they disrupt gene expression or protein function following insertion into coding or regulatory regions of the genome (Montgomery et al. 1987). More generally, TE insertions can negatively impact the host through 1) energetic costs of replication, transcription, and translation (Cavalier-Smith 2005); 2) disruptions of cellular processes by TE proteins (Nuzhdin 1999); 3) susceptibility to harmful gain-of-function mutations (De Gobbi et al. 2006); and 4) deletions and rearrangements caused by ectopic recombination between copies of the same TE family (Petrov et al. 2003). As a consequence, eukaryotic cells have evolved sophisticated machineries to silence TE proliferation and protect vital parts of the genome from TE insertion (Slotkin and Martienssen 2007; Lisch and Bennetzen 2011). However, the extreme variation in TE diversity and abundance among eukaryotic genomes suggests that the balance between TE proliferation and host silencing differs dramatically across the tree of life. The evolutionary processes affecting this balance remain poorly understood, despite the central role of TEs in shaping genome evolution (Venner et al. 2009).
TEs can also impact their host by affecting genome size. Proliferation and deletion of TEs cause genomic expansion and contraction, respectively (Petrov 2002; Bennetzen et al. 2005; Gregory 2005a; Vitte and Panaud 2005; Devos 2010), which can affect genome size's organism-level correlates (e.g., cell size and developmental rate) (Roth et al. 1997; Gregory 2005b). Such effects can be positive or negative, thereby enabling selection to act indirectly on TE content. The efficiency of such selection is determined by population genetic parameters such as effective population size (Lynch 2007; Lynch et al. 2011). Thus, genome size and content likely reflect a dynamic interaction between molecular processes (TE dynamics and host silencing) and selection acting on organismal traits (Bennetzen and Kellogg 1997; Agren and Wright 2011). Clades with extreme genome sizes provide critical test cases in which to explore this interaction; they represent instances where an unusual balance has been struck among these evolutionary forces.
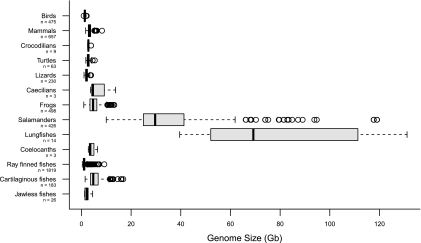
Among vertebrates, most of the largest genomes are found within salamanders, a clade of amphibians that includes 613 recognized species (AmphibiaWeb 2011) (fig. 1). Salamander genome sizes range from ∼14 to ∼120 Gb; these values are larger than all bird, mammal, reptile, and frog genomes, as well as most “fish” genomes (Gregory 2011), although extensive synteny conservation does exist between salamanders and other tetrapods (Voss et al. 2011). Karyotype and DNA reassociation kinetic studies have shown that salamanders' large genomes reflect high levels of repetitive DNA rather than polyploidy; however, such repeat elements remain almost completely uncharacterized, and TE silencing in salamanders remains unexplored (Green 1991; Sessions and Kezer 1991; Batistoni et al. 1995; Marracci et al. 1996). In contrast, organismal correlates of large genome size have been well characterized in salamanders, particularly in the Plethodontidae, the largest family (417 species, genome size range ∼14 to ∼74 Gb), where morphological evolution has been profoundly shaped by genomic gigantism (Hanken and Wake 1993). For example, constraints on the number of large cells that can fit into the braincase, as well as slow cell division and differentiation rates, have caused substantial simplification of the nervous and visual systems (e.g., low numbers of retinal and optic tectum neurons) (Sessions and Larson 1987; Roth et al. 1994; Roth et al. 1997). Such simplification reduces visual acuity (Hanken and Wake 1993; Roth et al. 1994); however, plethodontids have evolved compensatory visual adaptations (e.g., increased allocation of their brains to the optic tectum) (Wiggers and Roth 1991). Other compensatory adaptations are found in the circulatory system, where some miniaturized plethodontids have evolved enucleated red blood cells, likely to overcome physical constraints associated with circulating huge cells (Mueller et al. 2008). These examples suggest that plethodontids have evolved features that offset deleterious effects imposed by their expanding genomes (Wiggers and Roth 1991; Roth et al. 1997), indicating that an unusual balance between TE proliferation, host silencing, and selection on organism-level traits underlies the huge genome sizes in salamanders.
FIG. 1.—
Summary of nuclear genome sizes for 13 vertebrate clades. Data are compiled from the Animal Genome Size Database (Gregory 2011). Sample sizes (number of species summarized) are in parentheses following clade names.
Although studies integrating organismal biology and TE dynamics have recently been initiated in the avian clade, which has experienced genome size reduction (e.g., Organ et al. 2007), relatively little attention has been paid to vertebrate genome size evolution at the large end of the size spectrum (but for notable exceptions, see Smith et al. 2009; Voss et al. 2011). The repetitive landscapes of salamanders' huge genomes remain largely uncharacterized, and hypotheses integrating TE dynamics and organism-level selection remain untested. Here, we begin to fill this gap by using low-coverage high-throughput shotgun sequencing to generate genomic data for six species of salamanders and leveraging these data to perform the first comprehensive analysis of TE landscapes in the salamander clade. We developed a pipeline to mine TE sequences from low-coverage shotgun reads and estimate TE abundance and diversity, allowing us to make comparisons 1) between salamanders and other vertebrates with more “typical” (i.e., smaller) genome sizes, as well as 2) among the different salamander species. Our results show that salamander genomes contain all of the main TE superfamilies identified in well-annotated eukaryotic genomes. Across our six focal species, the most abundant TE superfamilies are very similar, and Ty3/gypsy elements (Class I retrotransposons) are by far the most abundant in all species examined. However, our results demonstrate a substantial difference between salamanders and other vertebrates: salamander genomes accumulate much larger amounts of long terminal repeat (LTR) retrotransposons. More generally, our results emphasize the importance of studying “outlier” taxa to generate a more comprehensive picture of vertebrate genome evolution.
Materials and Methods
Taxon Selection
We chose to generate low-coverage data from multiple taxa, rather than deep coverage data from a single taxon, in order to identify shared genomic features characteristic of the salamander clade. We focused our analyses on the family Plethodontidae, which contains more than two-thirds of extant salamander species. Plethodontids have been the focus of much genome size evolution research (Sessions and Larson 1987; Roth et al. 1994, 1997; Jockusch 1997), providing context for our genomic analyses. Six species of plethodontids were chosen that span the deepest phylogenetic split within the family: subfamily Plethodontinae (Aneides flavipunctatus and Desmognathus ochrophaeus) and Hemidactyliinae (Batrachoseps nigriventris, Bolitoglossa occidentalis, Bolitoglossa rostrata, and Eurycea tynerensis) (Vieites et al. 2011). These taxa encompass a range of the smaller genome sizes found in the clade (∼15 to ∼ 47 Gb; the largest plethodontid genome is ∼70 Gb) (Gregory 2011). The phylogenetic relationships among the six species are ((((B. occidentalis, B. rostrata), B. nigriventris), E. tynerensis), (A. flavipunctatus, D. ochrophaeus)). Divergence dates in salamanders remain the topic of much debate. The basal split within plethodontids has been dated at ∼ 40 to ∼130 Myr, depending on data set and analytical technique; divergence time estimates for our six focal taxa are similarly varied, but all are ≥25 Myr (Mueller 2006; Marjanovic and Laurin 2007; Kozak et al. 2009; Zhang and Wake 2009; Zheng et al. 2011). Genome size estimates and voucher specimen information is summarized in table 1.
Table 1.
Specimen Information and Shotgun Sequencing Results
| Species | Voucher Information | Genome Size (Gb) | Number of Reads | Total Number of base pairs | Percentage of Coverage |
| Aneides flavipunctatus | RLM172 | 44 | 1,044,399 | 308,615,225 | 0.70 |
| Batrachoseps nigriventris | ELJ 1556 | 25 | 1,131,828 | 487,538,903 | 1.91 |
| Eurycea tynerensis | RMB3457 | 25a | 1,089,945 | 389,972,620 | 1.59 |
| Desmognathus ochrophaeus | UAHC 16065 | 15 | 845,984 | 227,156,262 | 1.49 |
| Bolitoglossa rostrata | SMR 360 | 47 | 183,143 | 40,553,103 | 0.09 |
| Bolitoglossa occidentalis | GP1395 | 43 | 124,242 | 28,841,057 | 0.07 |
Represents an average of nine other Eurycea species.
Shotgun Library Creation and Sequencing
Total DNA was extracted from liquid-nitrogen snap-frozen liver or tail tissue by standard phenol–chloroform–isoamyl alcohol extraction methods or the Gentra Puregene tissue kit (Qiagen). 454 FLX–LR and 454 Titanium–XLR genomic shotgun libraries were prepared using the 454 shotgun library preparation kits and protocols (Roche) for FLX and Titanium sequencing, respectively. Libraries for Bolitoglossa occidentalis and B. rostrata were sequenced on the Roche 454-FLX sequencing platform using FLX–LR sequencing kits, whereas all other species were sequenced on the Roche 454-FLX platform with FLX–XLR Titanium reagents. Based on previous studies of complex plant genomes (e.g., barley and pea), we scaled our data collection efforts to produce ∼1% genomic coverage (i.e., 0.01× of the genome at 1× depth), as this sequencing depth has been shown to yield reasonable summaries of TE abundance for elements present at ≥1,000 copies/genome (Macas et al. 2007; Wicker et al. 2009). Library preparation and sequencing were performed by the Consortium for Comparative Genomics at the University of Colorado School of Medicine (B. rostrata, B. occidentalis, and Desmognathus ochrophaeus) and the University of Idaho Institute for Bioinformatics and Evolutionary Studies Genomics Resources Core facility (Aneides flavipunctatus, Batrachoseps nigriventris, and Eurycea tynerensis).
Initial Data Processing
Mitochondrial reads were screened out from all data sets using Blast with reference mitochondrial genome sequences from the same or closely related taxa (Mueller et al. 2004, 2008). Next, shotgun reads from each data set were checked for sequencing artifacts generated by the presence of multiple beads and a single template in emPCR drops, which can potentially produce multiple identical sequences that can skew estimates of repeat element abundance (Gomez-Alvarez et al. 2009; Niu et al. 2010). For data sets with <350 Mb of shotgun reads, the online 454 Replicate Filter (http://microbiomes.msu.edu/replicates/ [date last accessed 17 Nov 2011]) was used to filter out exact replicates (cutoff = 1.0, length requirement = 1.00, and initial base pair match = 3). For data sets with >350 Mb of shotgun reads, the locally installed cdhit-454 (http://weizhong-lab.ucsd.edu/cdhit_454/cgi-bin/index.cgi?cmd=Introduction [date last accessed 26 Sep 2011]) was used to filter out exact replicates (-c 1.00 -aS 0.9 -aL 0.6, other parameters set to default values). In total, 0.70–4.89% of shotgun reads were identified as potential sequencing artifacts in each data set, and all such reads were removed from further analysis. Finally, repeat elements with significant sequence similarity to elements identified from well-annotated genomes were identified using RepeatMasker, with RepBase (version 16.04) (http://www.girinst.org/ [date last accessed 26 Sep 2011]) as a reference library.
We developed a pipeline to mine TE sequences from low-coverage shotgun sequence data representing unexplored genomes. The pipeline includes five main steps, outlined below, and is summarized in supplementary file 1, Supplementary Material online. Most of the pipeline was automated by custom Perl scripts, which are available upon request.
TE Mining Step 1: Identify and Classify Repeat Sequences from Shotgun Reads
We used RepeatModeler (http://www.repeatmasker.org/RepeatModeler.html [date last accessed 26 Sep 2011]) to identify de novo repetitive sequences for each species. To identify repeats, RepeatModeler combines de novo repeat detection programs RepeatScout (Price et al. 2005) and RECON (Bao and Eddy 2002), which use self-comparison and k-mer approaches, respectively. To classify de novo repeats, RepeatModeler generates consensus sequences from alignments of similar reads and attempts to classify them using RepBase. Such consensus sequences from RepeatModeler were further classified using REPCLASS, software that automates the classification of TEs based on homology, structure, and target-site duplication (Feschotte et al. 2009). Following REPCLASS analyses, all de novo repeats initially identified by RepeatModeler were classified as “TE-derived repeats” or “unknown repeats.”
TE Mining Step 2: Assemble Shotgun Reads into Contigs
We assembled shotgun reads from each of our focal genomes into contigs using Phrap (http://phrap.org/ [date last accessed 26 Sep 2011]) (minmatch = 20, other parameters set to default values) and PCAP (Huang et al. 2003) (all parameters set to default values). Although our data provide only ≤1.9% coverage, TEs present in high copy number, with low sequence divergence, should be represented by composite contigs that span much of their length, including both coding and noncoding sequences (Macas et al. 2007; Swaminathan et al. 2007).
TE Mining Step 3: Identify TE-Containing Contigs
Following assembly, we used Blast to query the repeats identified in Step 1 against the contigs generated in Step 2 to identify contigs that include transposition-associated protein-coding sequences. Specifically, we started by using each TE-derived repeat from Step 1 (with the exception of SINEs, which encode no transposition-associated proteins) as a query to BlastN against the assembled contigs with an e-value threshold cutoff of e−10. The top 20 hits for each such repeat were parsed to a file, and the sequence of each hit was used to BlastX against the amino acid sequences of TE-encoded proteins (http://www.repeatmasker.org/RepeatProteinMask.html#database [date last accessed 26 Sep 2011]) to verify that the contig contained the expected target transposition-associated protein-encoding sequences. Then, the three longest contigs that met these criteria were chosen to represent the query repeat, and these contigs were assigned to the same TE superfamily as the query repeat.
We also analyzed repeats identified by RepeatModeler, but classified as “unknown” in Step 1, in order to determine whether we could classify them successfully using our assembled contigs. We began by using all of the TE sequence contigs identified above to mask, using RepeatMasker, the set of unknown repeats identified in Step 1; reads that remained unmasked were extracted. Then, each unmasked repeat was queried using BlastN against the contigs generated in Step 2 with an e-value threshold cutoff of e−10. The top 3 hits were collected to represent the unknown repetitive sequence. Finally, these collected sequences were queried using BlastX (e-value threshold cutoff of e−4) against the amino acid sequences of TE-encoded proteins to identify contigs that contained sequences encoding transposition-associated proteins, and each identified contig was assigned to the same TE superfamily as its first hit.
TE Mining Step 4: Verify and Refine TE-Containing Contigs
All the contigs we identified that contained transposition-associated protein-coding sequences were combined and sorted by length. We then examined each sequence to determine if it represented a complete full-length TE based on the following criteria: 1) Does the sequence contain intact coding regions for all relevant transposition-related proteins? This was determined using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html [date last accessed 26 Sep 2011]), coupled with BlastX against the amino acid sequences of TE-encoded proteins. For elements (e.g., non-LTR retrotransposons and Helitrons) that lack diagnostic structural features associated with their boundaries (e.g., LTRs or terminal inverted repeats [TIRs]), this was our sole criterion. 2) Does the sequence contain the hallmarks of TE sequence boundaries (e.g., LTRs or TIRs), indicating that then contig represents a full-length TE? This was determined using NCBI-Blast2 (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch&PROG_DEF=blastn&BLAST_PROG_DEF=megaBlast&BLAST_SPEC=blast2seq [date last accessed 26 Sep 2011]). Additionally, contigs were checked to ensure that they lacked endogenous (non-TE) gene fragments and nested TE insertions using TBlastX against the amino acid sequences of frog annotated proteins (ftp://ftp.ncbi.nih.gov/genomes/Xenopus_Silurana_tropicalis/protein/ [date last accessed 26 Sep 2011]) and Repbase, respectively. Contigs were also checked to ensure that they were not dimers or other assembly artifacts formed by joining intact element sequences with additional partial, or complete, elements through misassembled LTR or TIR sequences. Finally, as a reference, we searched for full-length TEs from the 16 bacterial artificial chromosomes (BAC) clones of the salamander Ambystoma mexicanum available in GenBank (Smith et al. 2009). Ambystoma mexicanum is a representative of the salamander family Ambystomatidae, which last shared a common ancestor with plethodontid salamanders ∼85–200 Ma (Marjanovic and Laurin 2007; Zhang and Wake 2009; Zheng et al. 2011). Candidate full-length TEs were identified using the amino acid sequences of TE-encoded proteins (http://www.repeatmasker.org/RepeatProteinMask.html#database [date last accessed 26 Sep 2011]) as queries to TBlastN against the BAC clone sequences. All regions that produced significant hits (e-values < e−10) were excised with 5 kb of flanking regions. TIRs or LTRs were identified by NCBI-Blast2.
TE Mining Step 5: Summarize the Overall TE Landscape of Each Species
All of the refined contigs that encode transposition-related proteins (Step 4), all of the repeats derived from TEs that were not represented by any contigs (Steps 1 and 4), all of the unknown repeats (Step 1), and all of the repeats classified as SINEs (Step 1) were combined to produce a species-specific repeat library for each of our focal taxa. Because none of our focal species is particularly closely related to any other (≤25 Myr since common ancestry), masking species with the repeat libraries of other species did not improve our results. Using these libraries, we masked each genome with RepeatMasker to yield a comprehensive summary of the TE landscape of each species. The annotation file produced by RepeatMasker was used to determine the TE diversity and abundance within each species. All elements comprising ≥0.01% of our shotgun reads were ranked by abundance in each genome. Next, for each species, we calculated the total proportion of shotgun data annotated to the three main TE orders: 1) LTR retrotransposons, 2) non-LTR retrotransposons (including SINEs), and 3) DNA transposons.
Comparison of the Salamander TE Landscape with Other Vertebrate Genomes
To test whether salamanders' large genomes reflect a fundamentally different TE landscape than is found in the genomes of other vertebrates with more typical genome sizes, we obtained summaries of TE content from five complete vertebrate genomes (Homo sapiens, Gallus gallus, Danio rerio, Anolis carolinensis, and Xenopus tropicalis) and compared the proportions of each genome composed of 1) LTR retrotransposons, 2) non-LTR retrotransposons, and 3) DNA transposons. TE summaries for Homo sapiens, Gallus gallus, Anolis carolinensis, and Xenopus tropicalis were obtained from their genome publications (International Human Genome Sequencing Consortium 2001; Hillier et al. 2004; Hellsten et al. 2010; Alföldi et al. 2011). The summary for Danio rerio was obtained using the out file of RepeatMasker from the University of California Santa Cruz genome browser (http://hgdownload.cse.ucsc.edu/goldenPath/danRer7/bigZips/ [date last accessed 26 Sep 2011]) and the genome assembly from the Danio rerio Sequencing Project (http://www.sanger.ac.uk/Projects/D_rerio/; Wellcome Trust Sanger Institute).
Comparison of TE Landscapes among Salamanders
Although our primary goal was to compare salamander genomes with those of other vertebrates, we also compared TE content among our six focal taxa. To this end, we performed principal component analysis (PCA) on the relative abundances of different elements present in the genomes of Desmognathus ochrophaeus, Eurycea tynerensis, Aneides flavipunctatus, and Batrachoseps nigriventris, as these data sets represent fairly equivalent coverage (0.7–1.9%); the final two species (Bolitoglossa rostrata and B. occidentalis) were excluded from this analysis because their coverage is much lower (0.07–0.09%), limiting our power to estimate TE abundance.
TE Age Distributions and Element Proliferation History in Salamanders
We analyzed the proliferation history of the most abundant superfamily from each TE class: the Gypsy superfamily (LTR retrotransposon), the L2/CR1 superfamily (non-LTR retrotransposon), and the Harbinger superfamily (DNA transposon). All shotgun reads masked by each superfamily were collected from the four species for which we had 0.7–1.9% genome coverage (Desmognathus ochrophaeus, Eurycea tynerensis, Aneides flavipunctatus, and Batrachoseps nigriventris). RepeatScout was used to construct consensus sequences representing fragments of ancestral elements from all shotgun reads masked by each family; multiple divergent consensus sequences mapping to the same TE region represent different subfamilies (Macas et al. 2007). Such consensus sequences were used as a repeat library to mask the relevant reads with RepeatMasker, generating percent divergence estimates for each read from its ancestral sequence. Corrected percent sequence divergences were then estimated using the Jukes–Cantor model of nucleotide substitution. Results were summarized as frequency histograms and represent summaries of superfamily-wide proliferation history.
TE Proliferation Dynamics, TE Content, and Genome Size Comparisons across Salamander Species
Our six focal taxa differ in genome size (table 1), encompassing a range of the large sizes found across the salamander clade (fig. 1). To test whether such differences reflect any aspect(s) of TE proliferation dynamics, we tested whether larger genomes showed evidence of either 1) more recent or 2) more frequent bursts of proliferation than smaller genomes by comparing the shapes of the element age distributions across taxa. We also tested whether genome size differences primarily reflect variation in the abundance of specific TEs by testing whether PC scores for each PC axis were related to genome size.
Results
Shotgun Library Summary Statistics and Initial Data Processing
The sequence data obtained for our six focal salamander species are summarized in table 1. Sequences have been deposited in the GenBank short read archive (accession numbers SRA046114.1, SRA046116.1, SRA046118.1, SRA046119.1, SRA046120.1, and SRA046121.1) and the DRYAD repository (doi:10.5061/dryad.308g1h54). The number of reads obtained per species ranges from 124,242 to 1,131,828, and the total amount of sequence generated per species ranges from 28 to 487 Mb. Sequencing coverage per species ranges from 0.07% to 1.91% of the genome; 0.01–0.06 % of this was screened out as mitochondrial sequence and 0.70–4.89% of this was filtered out as identical reads, likely sequencing artifacts generated during emPCR.
Efficiency of Our TE-Mining Method for Low-Coverage Shotgun Read Data
More than 260 Myr have elapsed since salamanders last shared a common ancestor with Xenopus, the most closely related organism with annotated TEs in RepBase (Marjanovic and Laurin 2007; Roelants et al. 2007). Thus, we anticipated low success identifying TEs based on sequence similarity to TEs known from other organisms. Consistent with this, our RepeatMasker analyses, using RepBase (16.04) as the repeat library, were largely unsuccessful; only ∼0.2–1.9% of our data were recognized as TEs (table 2). Furthermore, because 454 shotgun data consist of only short (<400 bp) reads, TE identification based on structural features and target site sequence information is not feasible. Thus, we relied on de novo repeat detection methods (RepeatModeler) to identify/classify candidate TE sequences in our data set and further classified them using REPCLASS. De novo salamander repeats classified as TEs were then used as repeat libraries to mask the shotgun reads of each species with RepeatMasker. Although these results were a significant improvement over our initial RepeatMasker runs (3.5–23.9% of each genome was classified as TEs, table 2), the majority of our shotgun reads remained unclassified. Examination of our repeat classification results showed that almost all classified repeats were derived from the conserved protein-coding portions of TEs. However, full-length TEs may also include large amounts of less conserved coding and noncoding sequences. Thus, our results suggested that the classification performed by RepeatModeler/REPCLASS was unable to identify shotgun reads mapping to less conserved TE regions, likely leading to severe underestimation of TE content in these largely unexplored genomes.
Table 2.
Percentage of 454 Shotgun Data Classified Using Different Methods
| Species | RepeatMasker/RepBase (%) | RepeatModeler/REPCLASS (%) | Our TE-Mining Method (%) | Our TE-Mining Method (% unclassified repeats)a |
| Aneides flavipunctatus | 1.91 | 23.90 | 47.52 | 15.01 |
| Batrachoseps nigriventris | 1.29 | 16.92 | 39.39 | 7.57 |
| Eurycea tynerensis | 1.15 | 9.81 | 25.18 | 8.09 |
| Desmognathus ochrophaeus | 0.16 | 9.41 | 39.69 | 11.98 |
| Bolitoglossa rostrata | 1.15 | 3.50 | 30.18 | 17.79 |
| Bolitoglossa occidentalis | 1.64 | 4.35 | 33.19 | 8.38 |
For comparison, we also show the percentage of data identified by our method as nonsimple repeats, but not classified as known TE sequence.
To address this issue, we assembled all 454 shotgun reads for each species into contigs and identified those harboring sequences encoding transposition-related proteins. Such contigs, in turn, allowed us to classify sequences derived from less conserved coding and noncoding regions of TEs through their location on the same contig as classifiable TE-coding sequences. When we used such contigs as repeat libraries to mask our shotgun reads, we were able to classify 25.18–47.52% of each data set as known TE sequences, representing a 20- to 200-fold increase over RepeatMasker analyses using RepBase as a library and a 2- to 9-fold increase over RepeatModeler/REPCLASS-based classification methods (table 2). Thus, our TE-mining pipeline is an improvement in analytical tools available to characterize the repeat element landscape of large unexplored genomes using low-coverage shotgun sequences.
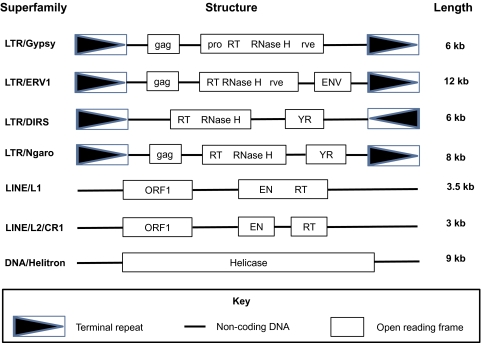
In addition, the assembly step of our TE-mining pipeline allowed us to successfully generate seven putatively full-length elements, composite sequences representative of salamander TE superfamilies. After verification and refinement, we confirmed contigs representing full-length sequences of several superfamilies of Class I TEs: Ty3/gypsy, ERV1, DIRS, and Ngaro elements (LTR retrotransposons), as well as L1 and L2/CR1 elements (non-LTR retrotransposons). In addition, we confirmed contigs representing a full-length rolling circle Helitron (Class II TE). The structures of the seven full-length TEs we assembled are summarized in figure 2, and each is largely consistent with the structure reported for the same superfamily from other eukaryotic genomes. Sequences of these complete elements, as well as the full-length elements identified from Ambystoma mexicanum BAC clones, are available as supplementary file 2, Supplementary Material online. To our knowledge, this is the first description of the structure of full-length TEs in salamander genomes. Our successful assembly of full-length contigs from ∼1% genome coverage (using a stringent assembly algorithm) indicates that all seven elements are present in very high copy number, and that little sequence divergence (<5–8% based on assembly parameters) exists among individual copies. This suggests that all seven TE superfamilies have been recently active and/or continue to be active in our focal salamander species. We tested whether ongoing transcription of these same superfamilies was also occurring in Ambystoma mexicanum using TBlastX against the A. mexicanum transcriptome (http://www.ambystoma.org/genome-resources/21-blast [date last accessed 26 Sep 2011]) and confirmed transcripts of all seven superfamilies.
FIG. 2.—
The structures of seven full-length TE sequences mined from salamander shotgun reads. Abbreviations: gag, capsid-like protein; pro, protease; RT, reverse transcriptase; rve, integrase; ENV, envelope protein; YR, tyrosine recombinase; EN, endonuclease.
Summary of TE Landscapes across Salamander Species
The proportion of 454 shotgun data classified as TEs in each species is summarized in table 2. Because we are working with low-coverage shotgun reads of largely unexplored genomes, all of these numbers are underestimates of total TE content; they do not necessarily reflect proportions of the genome made up of low–copy-number TEs, TEs with no recent proliferation activity, or TE boundary sequences (see Discussion). Regardless, our results clearly demonstrate that TEs have played a substantial role in generating salamanders enormous genomes. For example, 47.52% of the shotgun reads of Aneides flavipunctatus represent recognizable TEs. Note that an additional 15.01% of this genome is unclassifiable, but falls within the category of nonsimple repetitive sequence, suggesting that they are interspersed repeats likely derived from transposition activity. These results are consistent with earlier DNA–DNA hybridization analyses, which showed high levels of repetitive sequence in salamander genomes, as well as with limited recombinant DNA-based studies identifying select TEs active in salamanders (Baldari and Amaldi 1976; Batistoni et al. 1995; Marracci et al. 1996).
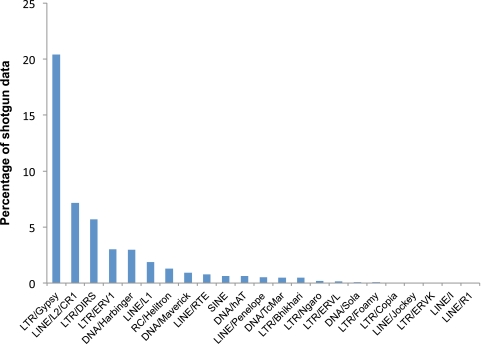
Our results show that salamander genomes harbor almost all of the major TE types reported in previously characterized eukaryotic genomes. We identified 29 different TE superfamilies in total across the 6 species, 22 of which were present in two or more species (supplementary file 3, Supplementary Material online). The percentage of shotgun data mapping to each superfamily is depicted in figure 3 (Aneides flavipunctatus) and summarized numerically in supplementary file 3, Supplementary Material online (all species) and depicted in supplementary file 4, Supplementary Material online. Across all six species, the most abundant elements are Ty3/gypsy retrotransposons, comprising 7–20% of the data set for each species. Ty3/gypsy elements were previously shown to exist at high copy numbers in the plethodontid genus Hydromantes based on cloning/hybridization analyses, although such methods failed to recover them from the genus Desmognathus (Marracci et al. 1996). Three other elements are also consistently among the most abundant across species: LINE/L2 non-LTR retrotransposons (1.7–8.8% of the genome), DIRS retrotransposons (2.0–5.7% of the genome), and LTR/ERV1 endogenous retroviruses (0.5–11.3% of the genome).
FIG. 3.—
The TE landscape of the Aneides flavipunctatus genome. Element superfamilies are ranked from most to least abundant along the x axis.
Comparison of the Salamander TE Landscape with Other Vertebrate Genomes
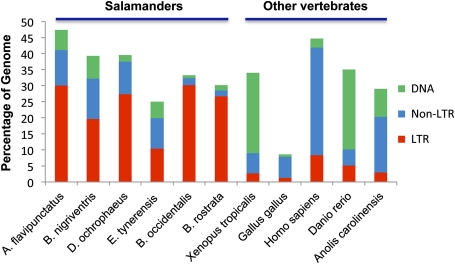
Although the same TEs are present in salamanders as in most other vertebrates, our results indicate that the proportion of LTR retrotransposons is much higher in all six species of salamanders than it is in any of the other vertebrate genomes we examined (fig. 4). This pattern holds, despite substantial differences in both genome size and percentage of genomic coverage across our six focal salamander species. We emphasize that this difference is an underestimate of the true difference in LTR levels, as our analyses of low-coverage shotgun data underestimate the total TE content of salamanders (see Discussion). Thus, LTR retrotransposons underlie genomic gigantism in extant plethodontid salamanders. This result, in turn, suggests expansion of LTR retrotransposons as a likely molecular mechanism underlying genomic expansion at the base of the salamander clade. Further analyses that include basal salamander lineages, as well as analytical tools designed to identify highly divergent TE copies (Gu et al. 2008; Singh et al. 2010), will allow an even more rigorous test of this hypothesis.
FIG. 4.—
The TE landscape of salamanders compared with that of other vertebrates. Salamanders have higher relative levels of LTR retrotransposons. For Danio rerio, we did not include the 11% of the genome identified as repetitive, but classified only as “DNA.”
Notably, genome content in salamanders differs most dramatically from Xenopus, the only other amphibian for which comparable data exist. The Xenopus TE landscape is largely composed of DNA transposons (fig. 4). Such extensive divergence in TE content, coupled with the extreme genomic expansion seen in salamanders, points to amphibians as an interesting clade to target for more detailed analysis of genome evolution.
Comparison of TE Landscapes among Salamanders
Our PCA analyses summarize the main differences in TE landscape among four of our six focal taxa. All three PC axes are composed of TEs from all three classes (LTR retrotransposons, non-LTR retrotransposons, and DNA transposons), indicating that differences in genome content among taxa are not limited to differences in a specific type of TE (fig. 5). More generally, these results allow us to test whether genome content is similar among taxa with more recent shared ancestry, similar genome sizes, or neither. Species show no clustering based on phylogenetic relationships, indicating that species are sufficiently diverged from one another (≥25 Myr) that their TE landscapes retain no pattern of shared ancestry. Finally, no PC scores for any axis were related to genome size, indicating that groups of different TEs that vary in a correlated fashion do not explain genome size variation among these four species.
FIG. 5.—
PCA results summarizing differences in TE landscape across four species. Phylogenetic relationships are (Batrachoseps nigriventris, Eurycea tynerensis), (Aneides flavipunctatus, Desmognathus ochrophaeus).
Overall, we note that the total TE content estimated for our six focal species (table 2) does not match predictions based on genome size; for example, Desmognathus ochrophaeus has the smallest genome but does not show the smallest proportion of TEs in our analyses. Although this pattern may reflect true differences in TE content, suggesting that genome size variation within salamanders reflects differences in non-TE DNA content, we conservatively attribute this discrepancy to limitations in our ability to detect TEs from low-coverage 454 shotgun data. For example, if the D. ochrophaeus genome contains a greater number of low-frequency TEs, or TEs with no recent proliferation activity, we would fail to detect them in our analysis, leading to a greater underestimation of total TE content in this species.
TE Proliferation History in Salamanders
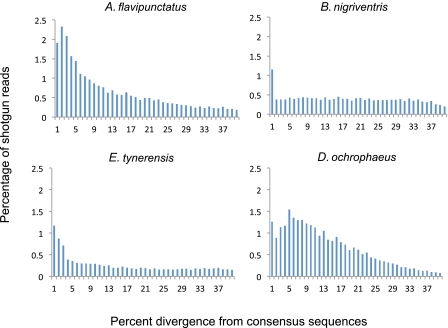
Sequence divergence distributions representing proliferation history of the most abundant superfamilies in each TE class are shown in figure 6 (Ty3/gypsy elements), supplementary file 5, Supplementary Material online (LINE/L2 elements), and supplementary file 6, Supplementary Material online (DNA/Harbinger). Under the assumption of a constant substitution rate, sequence divergence distributions are equivalent to age distributions. All distributions suggest ongoing TE proliferation, indicated by element copies with sequence divergence ≤1% from the consensus (Novick et al. 2010). Transcripts of all such superfamilies were detected in the Ambystoma mexicanum transcriptome database (http://www.ambystoma.org/genome-resources/21-blast [date last accessed 26 Sep 2011]), suggesting that they are also transcriptionally active in this ambystomatid salamander species. In addition, all distributions include copies with high (≤40%) sequence divergence, suggesting that elements reach fixation in salamander populations and are subsequently preserved in the genome for long periods of time; this pattern is consistent with a low negative impact of TE insertions on the host (Novick et al. 2009; Novick et al. 2010). However, we emphasize that this pattern may also reflect our inability to assemble consensus sequences representing all families/subfamilies within each superfamily from low-coverage shotgun data. Thus, additional data collection will be required to rigorously test this hypothesis.
FIG. 6.—
Age distribution of Ty3/gypsy elements in four species of salamanders.
TE Proliferation Dynamics, TE Content, and Genome Size Comparisons among Salamander Species
In total, we generated sequence divergence distributions for the most abundant superfamily in each TE class from the four species for which we have 0.7–1.9% coverage. The genome sizes of these four species range from 15 to 44 Gb. We examined these distributions to determine whether this variation in genome size reflects 1) the frequency of bursts of TE proliferation and/or 2) how recently such bursts occurred. Our results show no such correlations; larger salamander genomes show no consistent pattern of having more frequent, or more recent, proliferation bursts. This result, coupled with the results of our PCA showing that no PC scores for any axis are related to genome size, suggests that evolutionary changes in genome size among these four taxa have not been dictated solely by the tempo and mode of proliferation of any of the most abundant elements. However, we emphasize that our sampling was designed to identify differences between salamanders and other vertebrates; increased phylogenetic breadth, and sequencing depth, is required to test whether TE dynamics correlate with evolutionary changes in genome size within the salamander clade.
Discussion
Our results represent the first in-depth comparative analyses of the repetitive landscape of salamander genomes, the largest among the tetrapods and, with the exception of lungfish, among vertebrates as a whole (Gregory 2011). We demonstrate that 1) salamander genomes have fairly high TE content, including representatives of all of the major types of TEs found in well-annotated eukaryotic genomes, 2) many TEs show evidence of recent and/or ongoing proliferation, and 3) Ty3/gypsy elements are the most abundant TE superfamily. Furthermore, we show that salamanders are unique among vertebrates in their overall genome composition; although LTR retrotransposon abundance varies among salamanders, LTR retrotransposon levels are higher in all sampled salamanders than in other vertebrates (fig. 4). This pattern holds, despite 3-fold differences in genome size among our focal salamander species, as well as limitations in our ability to identify TE-derived sequences from low-coverage shotgun data (see below). Thus, LTR retrotransposons underlie genomic gigantism in extant plethodontid salamanders and increased LTR proliferation is a candidate molecular mechanism underlying genomic expansion at the base of the salamander clade.
Among our six focal species, however, no clear relationship exists between genome size and TE content or proliferation dynamics. There are both biological and analytical possible explanations for this lack of correlation. First, genome size evolution within plethodontids may be shaped by factors other than TE proliferation dynamics. For example, selection for smaller genome size has been proposed in lineages experiencing metamorphosis, where slow rates of cell division and differentiation associated with large genomes would extend a vulnerable stage of ontogeny (Wake and Marks 1993). Such indirect selection against TE expansion could impact relative TE abundance (the variable we measured) in many different ways. Second, our analytical method may have obscured a true correlation between TE content and genome size. Our analysis of low-coverage shotgun data underestimates true TE content in predictable ways: 1) We miss low–copy-number repeats; RepeatModeler requires a minimum number of four sequence copies per data set to identify a sequence as repetitive (http://www.repeatmasker.org/RepeatModeler.html [date last accessed 26 Sep 2011]); 2) We miss noncoding sequence of superfamilies with higher levels of sequence divergence. Our analysis requires ≥92% sequence identity during contig assembly (Huang et al. 2003). Thus, we will not obtain full-length or near–full-length contigs of older divergent elements and such element abundances will be underestimated. Therefore, if genomes of our focal taxa differ in the proportion of low frequency or highly divergent TEs, we will differentially underestimate TE content across species. Finally, comparison of the LTR sequences from our putative full-length LTR retrotransposons with those we mined from Ambystoma BAC clones shows that the LTRs of Ty3/gypsy are much shorter in our contigs (supplementary file 2, Supplementary Material online); thus, even under the “best” conditions, when elements exist in high copy number with low sequence divergence, we will still underestimate their relative abundance. This underestimate is likely to be uniform across all six species, but nonetheless contributes to the imprecision in our estimates of TE content. More generally, our analyses do not take into account TE deletion. Removal of TE sequences via both small deletions mediated by replication slippage and larger deletions mediated by ectopic recombination between TE copies is a critical component of TE dynamics that clearly impacts genome size evolution (Petrov 2002; Bennetzen et al. 2005). Using low-coverage shotgun data, the tempo and mode of DNA/TE loss is much more difficult to estimate than that of DNA gain through TE proliferation; however, future research aimed at understanding DNA loss is required. Finally, we note that other studies have shown a disconnect between TE dynamics and evolutionary changes in genome size (e.g., Wicker et al. 2009), supporting the view that integration of molecular, organismal, and population-level analyses is critical for generating a comprehensive picture of genome size evolution (Gregory 2003; Cavalier-Smith 2005).
Our results complement recent work describing the genic component of the genome of Ambystoma mexicanum (Smith et al. 2009), a representative of the salamander family Ambystomatidae and a major model system for laboratory studies in a number of biomedical and basic research disciplines (Smith et al. 2005). Ambystomatid salamanders diverged from plethodontid salamanders, the focal clade of this study, ∼85–200 Ma (Marjanovic and Laurin 2007; Zhang and Wake 2009; Zheng et al. 2011). BAC sequencing in Ambystoma demonstrated that salamander introns are substantially longer than human, chicken, and frog introns. Thus, increased intron length also contributes to genomic expansion in salamanders (Smith et al. 2009), although longer introns may reflect TE accumulation. Combining analyses that target the genic component of the Ambystoma genome (Salamander Genome Project: http://www.ambystoma.org/research/salamander-genome-project [date last accessed 26 Sep 2011]), as well as the nongenic component from diverse salamander species (current study), will ultimately yield a comprehensive picture of the molecular processes underlying genomic gigantism in salamanders, as it has in other taxa (Bennetzen et al. 2005).
Recent work has stressed the importance of considering the role of population genetic parameters in shaping genome size evolution; specifically, in organisms with smaller effective population sizes, natural selection is less effective at purging slightly deleterious “extra” DNA, which may lead to genome size increases (Lynch 2007, 2011; but see, Whitney et al. 2011). Under this hypothesis, salamanders are predicted to have much smaller effective population sizes than other vertebrates. However, there is no evidence that this is the case (Frankham 1995). Furthermore, using body size as a rough proxy for effective population size refutes this hypothesis (Organ and Shedlock 2009); salamanders are small relative to many other vertebrate taxa. Thus, although stronger genetic drift in smaller populations may underlie broad patterns of genome size evolution across the tree of life, it does not appear to explain genomic gigantism in salamanders.
Across eukaryotes, only a limited number of larger genomes have been analyzed in detail because of obvious technological and analytical challenges (Ambrozova et al. 2011). The majority of such studies have been performed in angiosperms, reflecting both their great agricultural importance and their enormous diversity of genome sizes (Bennett and Leitch 2010); however, even such angiosperm studies have emphasized genomes toward the smaller end of the size range. LTR retrotransposons appear to form the majority of most angiosperm genomes (Vitte and Bennetzen 2006; Huo et al. 2008), and their increased abundance is correlated with genome expansion in diverse plant taxa (Vitte and Panaud 2005), including Gossypium (cotton) (Hawkins et al. 2006), Oryza (rice) (Zuccolo et al. 2007; Gill et al. 2010), Eleocharis (family Cyperaceae) (Zedek et al. 2010), Vicia (family Fabaceae) (Neumann et al. 2006), maize (Sanmiguel et al. 1998), and Helianthus (sunflower) (Staton et al. 2009). Finally, Ty3/gypsy LTR retrotransposons are the most abundant elements found in the extremely large genomes of Fritillaria species (Liliaceae), although the vast majority of those ∼44 Gb–sized genomes remains uncharacterized (Ambrozova et al. 2011). Fungi, in contrast, have small nuclear genomes with comparably limited size variation across taxa; only a few outliers reach even 400–700 Mbp (Kullman et al. 2005). Such “outliers” that have been partially characterized (e.g., Gigaspora margarita) contain both LTR and non-LTR retrotransposons (Gollotte et al. 2006). Limited examples of genomic expansion exist from the other main eukaryotic clades; for example, the genome of Phytophthora infestans, the chromalveolate pathogen responsible for the Irish potato famine in the 1800s, shows genomic expansion (genome size 240 Mb) caused by proliferation of Ty3/gypsy retrotransposons (Haas et al. 2009). Within animals, limited cases of genomic gigantism are found not only in the deuterostomes (e.g., salamanders, lungfishes; see fig. 1) but also within several protostome clades; certain lineages of grasshoppers (e.g., genus Podisma), flatworms (e.g., genus Otomesostoma), and amphipods (e.g., genus Ampelisca) have genome sizes estimated at 64, 21, and 64 Gb, respectively (Gregory 2011), but the molecular mechanisms underlying such genomic expansion remain largely unknown (Parchem et al. 2010) (but see Bensasson et al. 2001 for evidence of slower DNA loss in Podisma). Our results in salamanders, coupled with results from several angiosperm taxa, indicate that extreme increases in genome size may be more likely to reflect expansion of LTR retrotransposons than other TEs, which could suggest a different balance between TE proliferation and silencing among the main TE classes. Alternatively, it could suggest that LTR retrotransposons may more effectively mitigate their deleterious effects on the host genome through the targeting of “safe havens” for insertion (Gao et al. 2008). Analysis of diverse eukaryotic taxa with large genomes is required to rigorously test this hypothesis. More generally, extending genomic analyses to phylogenetically diverse lineages with large genomes will be critical for generating a more complete picture of eukaryotic genome evolution (Ambrozova et al. 2011; Voss et al. 2011). Our work, as well as other recent studies using low-coverage data to characterize repeat element landscapes, suggests that such analyses are now feasible, despite the fact that assembling large repetitive genomes remains intractable (Macas et al. 2007; Castoe et al. 2011).
Although the TE landscape of salamanders is the focus of our work (as it provides a potential mechanism for genomic expansion), many researchers target the single- or low-copy sequences within a genome for analyses ranging from protein function to phylogenetic history. Such studies are hampered by unknown repetitive landscapes; without a database of known TEs, homology-based repeat-masking analyses are ineffective. Our work will benefit researchers targeting the single- or low-copy sequences within salamanders by providing such a database of TEs. More generally, the pipeline we developed can be used by any researcher to generate a similar database in an unexplored genome, provided the TEs exist in sufficiently high copy number with sufficient sequence identify. Thus, our work also contributes to other fields (e.g., phylogenetic systematics and population genetics) transitioning to large-scale genomic data sets (Thomson et al. 2010).
For decades, evolutionary biologists have inferred that salamanders' huge genomes relative to other vertebrates are related to the clade's extremely low metabolic rates, just as the compact genomes of birds and flying mammals are linked to high metabolic rates (Olmo and Morescalchi 1975; Szarski 1983; Burton et al. 1989; Cavalier-Smith 1991; Gatten et al. 1992; Waltari and Edwards 2002). Mechanistically, this inverse relationship between genome size and metabolic rate has been explained in several subtly different ways that build on the positive correlation between genome size and cell size and, more specifically, the low cell surface-to-volume ratios associated with large cells (Olmo and Morescalchi 1975; Szarski 1983; Lay and Baldwin 1999; Kozlowski et al. 2003; Kozlowski et al. 2010). Within salamanders, however, no strong correlation exists between metabolic rate and genome size, suggesting that other factors drive among-lineage genome size variation within the clade (Gregory 2003). A mechanistic link between large genomes and low metabolic rate remains the topic of debate, as does the adaptive significance of genomic expansion in salamanders (Cavalier-Smith 1991; Roth et al. 1994). However, we emphasize that a full understanding of the forces shaping genome expansion in this clade requires integrating detailed analyses of molecular mechanisms into tests of these long-standing physiological hypotheses. Our results represent a first step toward such a comprehensive picture of salamander genomics that considers evolutionary forces acting at the genome, cell, organism, and population levels. Future studies aimed at the balance between host-mediated TE silencing and TE proliferation in salamanders, particularly for LTR retrotransposons, will add to this picture, as will analyses integrating genomic and organismal data in an explicit phylogenetic context.
Supplementary Material
Supplementary files 1–6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Science Foundation grant NSF-DEB 1021489 to R.L.M. and by Colorado State University. We gratefully acknowledge tissues from S.M. Rovito, G. Parra, R.M. Bonett, E.L. Jockusch, L.J. Rissler, the University of Alabama Herpetology Collection, and the Museum of Vertebrate Zoology (University of California Berkeley). M. Settles provided critical assistance with 454 data collection. Two anonymous reviewers gave useful comments that improved the manuscript. Work on vertebrate TEs in the Feschotte lab is supported by the National Institutes of Health grant GM077582 to C.F.
References
- Agren JA, Wright SI. Co-evolution between transposable elements and their hosts: a major factor in genome size evolution? Chromosome Res. 2011;19:777–786. doi: 10.1007/s10577-011-9229-0. [DOI] [PubMed] [Google Scholar]
- Alföldi J, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrozova K, et al. Diverse retrotransposon families and an AT-rich satellite DNA revealed in giant genomes of Fritillaria lilies. Ann Bot. 2011;107:255. doi: 10.1093/aob/mcq235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AmphibiaWeb . AmphibiaWeb: information on amphibian biology and conservation. 2011. [Internet]. Berkeley (CA): AmphibiaWeb. [cited 2011 Sep 26]. Available from: http://amphibiaweb.org. [Google Scholar]
- Baldari CT, Amaldi F. DNA reassociation kinetics in relation to genome size in four amphibian species. Chromosoma. 1976;59:13–22. doi: 10.1007/BF00327706. [DOI] [PubMed] [Google Scholar]
- Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistoni R, Pesole G, Marracci S, Nardi I. A tandemly repeated DNA family originated from SINE-related elements in the European plethodontid salamanders (Amphibia, Urodela) J Mol Evol. 1995;40:608–615. doi: 10.1007/BF00160508. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Angiosperm DNA C-values database [Internet]. (release 7.0) [cited 2011 Sep 26] 2010. Available from: http://www.kew.org/cvalues/ [Google Scholar]
- Bennetzen JL, Kellogg EA. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann Bot. 2005;95:127. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson D, et al. Genomic gigantism: dNA loss is slow in mountain grasshoppers. Mol Biol Evol. 2001;18:246. doi: 10.1093/oxfordjournals.molbev.a003798. [DOI] [PubMed] [Google Scholar]
- Burton DW, Bickham JW, Genoways HH. Flow-cytometric analyses of nuclear DNA content in four families of neotropical bats. Evolution. 1989;43:756–765. doi: 10.1111/j.1558-5646.1989.tb05174.x. [DOI] [PubMed] [Google Scholar]
- Castoe TA, et al. Discovery of highly divergent repeat landscapes in snake genomes using high throughput sequencing. Genome Biol Evol. 2011 doi: 10.1093/gbe/evr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Coevolution of vertebrate genome, cell, and nuclear sizes. In: Ghiara G, Angelini F, Olmo E, Varano L, editors. Symposium on the evolution of terrestrial vertebrates. Selected symposia and monographs U.Z.I. Modena (Italy): Mucchi; 1991. pp. 51–86. [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Ann Bot. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Herndon (VA): American Society for Microbiology Press; 2002. [Google Scholar]
- De Gobbi M, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;26:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- Devos KM. Grass genome organization and evolution. Curr Opin Plant Biol. 2010;13:139–145. doi: 10.1016/j.pbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Gent. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, et al. Exploring repetitive DNA landscapes using REPCLASS, a tool that automates the classification of transposable elements in eukaryotic genomes. Genome Biol Evol. 2009;1:205–220. doi: 10.1093/gbe/evp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Effective population size/adult population size ratios in wildlife: a review. Genet Res. 1995;66:95–107. doi: 10.1017/S0016672308009695. [DOI] [PubMed] [Google Scholar]
- Gao X, et al. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatten REJ, Miller K, Full RJ. Energetics at rest and during locomotion. In: Feder ME, editor. Environmental physiology of the amphibians. Chicago (IL): University of Chicago Press; 1992. pp. 314–377. [Google Scholar]
- Gill N, et al. Dynamic Oryza genomes: repetitive DNA sequences as genome modeling agents. Rice. 2010;3:251–269. [Google Scholar]
- Gollotte A, et al. Repetitive DNA sequences include retrotransposons in genomes of the Glomeromycota. Genetica. 2006;128:455–469. doi: 10.1007/s10709-006-0019-0. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH., Jr Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Green DM. Supernumerary chromosomes in amphibians. In: Green DM, Sessions SK, editors. Amphibian cytogenetics and evolution. San Diego (CA): Academic Press, Inc; 1991. p. 333–355. [Google Scholar]
- Gregory TR. Variation across amphibian species in the size of the nuclear genome supports a pluralistic, hierarchical approach to the C-value enigma. Biol J Linn Soc. 2003;79:329–339. [Google Scholar]
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Gent. 2005a;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- Gregory TR. The evolution of the genome. San Diego (CA): Elsevier; 2005b. [Google Scholar]
- Gregory TR. Animal genome size database [Internet]. [cited 2011 Sep 26] 2011. Available from: http://www.genomesize.com. [Google Scholar]
- Gu W, et al. Identification of repeat structure in large genomes using repeat probability clouds. Anal Biochem. 2008;380:77–83. doi: 10.1016/j.ab.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Hanken J, Wake DB. Miniaturization of body size: organismal consequences and evolutionary significance. Annu Rev Ecol Syst. 1993;24:501–519. [Google Scholar]
- Hawkins JS, et al. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006;16:1252. doi: 10.1101/gr.5282906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, et al. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LDW, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Huang X, et al. PCAP: a whole-genome assembly program. Genome Res. 2003;13:2164. doi: 10.1101/gr.1390403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo N, et al. The nuclear genome of Brachypodium distachyon: analysis of BAC end sequences. Funct Integr Genomics. 2008;8:135–147. doi: 10.1007/s10142-007-0062-7. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jockusch EL. An evolutionary correlate of genome size change in plethodontid salamanders. Proc R Soc Lond B Biol Sci. 1997;264:597. [Google Scholar]
- Kozak KH, Mendyk RW, Wiens JJ. Can parallel diversification occur in sympatry? Repeated patterns of body-size evolution in coexisting clades of North American salamanders. Evolution. 2009;63:1769–1784. doi: 10.1111/j.1558-5646.2009.00680.x. [DOI] [PubMed] [Google Scholar]
- Kozlowski J, Konarzewski M, Gawelczyk AT. Cell size as a link between noncoding DNA and metabolic rate scaling. Proc Natl Acad Sci U S A. 2003;100:14080–14085. doi: 10.1073/pnas.2334605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski J, et al. Cell size is positively correlated between different tissues in passerine birds and amphibians, but not necessarily in mammals. Biol Lett. 2010;6:792–796. doi: 10.1098/rsbl.2010.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullman B, Tamm H, Kullman K. Fungal genome size database [Internet]. [cited 2011 Sep 26] 2005. Available from: http://www.zbi.ee/fungal-genomesize. [Google Scholar]
- Lay PA, Baldwin J. What determines the size of teleost erythrocytes? Correlations with oxygen transport and nuclear volume. Fish Physiol Biochem. 1999;20:31–35. [Google Scholar]
- Lisch D, Bennetzen JL. Transposable element origins of epigenetic gene regulation. Curr Opin Plant Biol. 2011;14:156–161. doi: 10.1016/j.pbi.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of genome architecture. Sunderland (MA): Sinauer Associates, Inc; 2007. [Google Scholar]
- Lynch M. Statistical inference on the mechanisms of genome evolution. PLoS Genet. 2011;7:e1001389. doi: 10.1371/journal.pgen.1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, et al. The repatterning of eukaryotic genomes by random genetic drift. Annu Rev Genomics Hum Genet. 2011;12:347–366. doi: 10.1146/annurev-genom-082410-101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navratilova A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanovic D, Laurin M. Fossils, molecules, divergence times, and the origin of lissamphibians. Syst Biol. 2007;56:369. doi: 10.1080/10635150701397635. [DOI] [PubMed] [Google Scholar]
- Marracci S, et al. Gypsy/Ty3-like elements in the genome of the terrestrial salamander Hydromantes (Amphibia, Urodela) J Mol Evol. 1996;43:584–593. doi: 10.1007/BF02202106. [DOI] [PubMed] [Google Scholar]
- Montgomery EA, Charlesworth B, Langley CH. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 1987;49:31–41. doi: 10.1017/s0016672300026707. [DOI] [PubMed] [Google Scholar]
- Mueller RL. Evolutionary rates, divergence dates, and the performance of mitochondrial genes in Bayesian phylogenetic analysis. Syst Biol. 2006;55:289. doi: 10.1080/10635150500541672. [DOI] [PubMed] [Google Scholar]
- Mueller RL, et al. Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. Proc Natl Acad Sci U S A. 2004;101:13820–13825. doi: 10.1073/pnas.0405785101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RL, et al. Genome size, cell size, and the evolution of enucleated erythrocytes in attenuate salamanders. Zoology. 2008;111:218–230. doi: 10.1016/j.zool.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, Koblizkova A, Navratilova A, Macas J. Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics. 2006;173:1047–1056. doi: 10.1534/genetics.106.056259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Fu L, Sun S, Li W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics. 2010;11:187. doi: 10.1186/1471-2105-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick PA, et al. The evolutionary dynamics of autonomous non-LTR retrotransposons in the lizard Anolis carolinensis shows more similarity to fish than mammals. Mol Biol Evol. 2009;26:1811. doi: 10.1093/molbev/msp090. [DOI] [PubMed] [Google Scholar]
- Novick PA, et al. The evolution and diversity of DNA transposons in the genome of the lizard Anolis carolinensis. Genome Biol Evol. 2010;3:1–14. doi: 10.1093/gbe/evq080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin SV. Sure facts, speculations, and open questions about the evolution of transposable element copy number. Genetica. 1999;107:129–137. [PubMed] [Google Scholar]
- Oliver MJ, et al. The mode and tempo of genome size evolution in eukaryotes. Genome Res. 2007;17:594–601. doi: 10.1101/gr.6096207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo E, Morescalchi A. Evolution of the genome and cell sizes in salamanders. Experientia. 1975;31:804–806. doi: 10.1007/BF01938475. [DOI] [PubMed] [Google Scholar]
- Organ CL, Shedlock AM. Palaeogenomics of pterosaurs and the evolution of small genome size in flying vertebrates. Biol Lett. 2009;5:47. doi: 10.1098/rsbl.2008.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organ CL, et al. Origin of avian genome size and structure in non-avian dinosaurs. Nature. 2007;446:180–184. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- Parchem RJ, et al. BAC library for the amphipod crustacean, Parhyale hawaiensis. Genomics. 2010;95:261–267. doi: 10.1016/j.ygeno.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov DA. Mutational equilibrium model of genome size evolution. Theor Popul Biol. 2002;61:533–546. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- Petrov DA, et al. Size matters: non-LTR retrotransposable elements and ectopic recombination in Drosophila. Mol Biol Evol. 2003;20:880–892. doi: 10.1093/molbev/msg102. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics 21(S1):i351–i358. [DOI] [PubMed] [Google Scholar]
- Pritham EJ. Transposable elements and factors influencing their success in eukaryotes. J Hered. 2009;100:648. doi: 10.1093/jhered/esp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelants K, et al. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci U S A. 2007;104:887. doi: 10.1073/pnas.0608378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth G, Blanke J, Wake DB. Cell size predicts morphological complexity in the brains of frogs and salamanders. Proc Natl Acad Sci U S A. 1994;91:4796–4800. doi: 10.1073/pnas.91.11.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth G, Nishikawa KC, Wake DB. Genome size, secondary simplification, and the evolution of the brain in salamanders. Brain Behav Evol. 1997;50:50–59. doi: 10.1159/000113321. [DOI] [PubMed] [Google Scholar]
- Sanmiguel P, et al. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- Sessions SK, Kezer J. Evolutionary cytogenetics of bolitoglossine salamanders (family Plethodontidae) In: Green DM, Sessions SK, editors. Amphibian cytogenetics and evolution. San Diego (CA): Academic Press, Inc; 1991. pp. 89–130. [Google Scholar]
- Sessions SK, Larson A. Developmental correlates of genome size in plethodontid salamanders and their implications for genome evolution. Evolution. 1987;41:1239–1251. doi: 10.1111/j.1558-5646.1987.tb02463.x. [DOI] [PubMed] [Google Scholar]
- Singh A, Keswani U, Levine D, Feschotte C. An algorithm for the reconstruction of consensus sequences of ancient segmental duplications and transposon copies in eukaryotic genomes. Int J Bioinform Res Appl. 2010;6:147–162. doi: 10.1504/IJBRA.2010.032118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Gent. 2007;8:272. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Smith J, et al. Sal-Site: integrating new and existing ambystomatid salamander research and informational resources. BMC Genomics. 2005;6:181. doi: 10.1186/1471-2164-6-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, et al. Genic regions of a large salamander genome contain long introns and novel genes. BMC Genomics. 2009;10:19. doi: 10.1186/1471-2164-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton SE, Ungerer MC, Moore RC. The genomic organization of Ty3/gypsy-like retrotransposons in Helianthus (Asteraceae) homoploid hybrid species. Am J Bot. 2009;96:1646. doi: 10.3732/ajb.0800337. [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Varala K, Hudson M. Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics. 2007;8:132. doi: 10.1186/1471-2164-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szarski H. Cell size and the concept of wasteful and frugal evolutionary strategies. J Theor Biol. 1983;105:201–209. doi: 10.1016/s0022-5193(83)80002-2. [DOI] [PubMed] [Google Scholar]
- Thomson RC, Wang IANJ, Johnson JR. Genome enabled development of DNA markers for ecology, evolution and conservation. Mol Ecol. 2010;19:2184–2195. doi: 10.1111/j.1365-294X.2010.04650.x. [DOI] [PubMed] [Google Scholar]
- Venner S, Feschotte C, Bièmont C. Dynamics of transposable elements: towards a community ecology of the genome. Trends Genet. 2009;25:317–323. doi: 10.1016/j.tig.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieites DR, Rom·N SN, Wake MH, Wake DB. A multigenic perspective on phylogenetic relationships in the largest family of salamanders, the Plethodontidae. Mol Phylogent Evol. 2011;59:623–635. doi: 10.1016/j.ympev.2011.03.012. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. Evolution of genome size: multilevel selection, mutation bias or dynamical chaos? Curr Opin Genet Dev. 2004;14:620–626. doi: 10.1016/j.gde.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Vitte C, Bennetzen JL. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci U S A. 2006;103:17638. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitte C, Panaud O. LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet Genome Res. 2005;110:91–107. doi: 10.1159/000084941. [DOI] [PubMed] [Google Scholar]
- Volff J-N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Voss SR, et al. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. 2011;21:306–312. doi: 10.1101/gr.116491.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake DB, Marks SB. Development and evolution of plethodontid salamanders: a review of prior studies and a prospectus for future research. Herpetologica. 1993;49:194–203. [Google Scholar]
- Waltari E, Edwards SV. Evolutionary dynamics of intron size, genome size, and physiological correlates in archosaurs. Am Nat. 2002;160:539–552. doi: 10.1086/342079. [DOI] [PubMed] [Google Scholar]
- Whitney KD, Boussau B, Baack EJ, Garland T., Jr Drift and genome complexity revisited. PLoS Genet. 2011;7:e1002092. doi: 10.1371/journal.pgen.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Gent. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wicker T, et al. A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 2009;59:712–722. doi: 10.1111/j.1365-313X.2009.03911.x. [DOI] [PubMed] [Google Scholar]
- Wiggers W, Roth G. Anatomy, neurophysiology and functional aspects of the nucleus isthmi in salamanders of the family Plethodontidae. J Comp Physiol A. 1991;169:165–176. [Google Scholar]
- Zedek F, Ämerda J, Ämarda P, Bureö P. Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Plant Biol. 2010;10:265. doi: 10.1186/1471-2229-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wake DB. Higher-level salamander relationships and divergence dates inferred from complete mitochondrial genomes. Mol Phylogenet Evol. 2009;53:492–508. doi: 10.1016/j.ympev.2009.07.010. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Peng R, Kuro-O M, Zeng X. Exploring patterns and extent of bias in estimating divergence time from mitochondrial DNA sequence data in a particular lineage: a case study of salamanders (Order Caudata) Mol Biol Evol. 2011;28:2521–2535. doi: 10.1093/molbev/msr072. [DOI] [PubMed] [Google Scholar]
- Zuccolo A, et al. Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol. 2007;7:152. doi: 10.1186/1471-2148-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]