Abstract
In this paper we present a method for reconstructing D-MRI data on regular grids from sparse data without assuming specific diffusion models. This is particularly important when studying the fetal brain in utero, since registration methods applied for movement and distortion correction produce scattered data in spatial and angular (gradient) domains. We propose the use of a groupwise registration method, and a dual spatio-angular interpolation by using radial basis functions (RBF). Experiments performed on adult data showed a high accuracy of the method when estimating diffusion images in unavailable directions. The application to fetal data showed an improvement in the quality of the sequences according to criteria based on fractional anisotropy (FA) maps, and differences in the tractography results.
1 Introduction
Diffusion Magnetic Resonance Imaging (D-MRI) is an imaging modality increasingly used for studying the normal and pathological development of the fetal brain. Since the limited sensitivity of fetal ultrasound to detect and depict the maturational processes of the developing white matter, D-MRI can be considered as one of the most promising methods for studying in vivo the human white matter development.
Eddy current-induced image distortions and patient motion during prolonged acquisitions cause image misalignment in D-MRI sequences, invalidating the assumption of a consistent relationship between image space and anatomy in the image processing. Such distortions are generally assumed to be affine, and usually corrected by coregistration with the T2-weighted image ( ) by using affine transformation models and mutual information (MI) [1]. When imaging a fetus, there exists the additional problem of movement, and the acquired slices are no longer a regular sample of the volume [2][3]. This may be compensated by relaxing the 3D transformation model to a set of transformations applied to each slice independently. When these transformations are applied to the original sequence, scattered data are generated in the spatial and angular (gradient) domains, requiring an interpolation in both fields. Recently, Jiang et al. [3] have presented an interpolation method assuming that the local diffusion properties can be represented by a rank-2 tensor model. However, this model cannot describe voxels containing multiple fibers with different orientations, a condition referred to as intravoxel orientational hetereogeneity (IVOH) [4].
In this paper, we present a reconstruction method independent of the diffusion model than can be used with more complex diffusion models like Gaussian mixture models or higher-order diffusion tensors. Distortion corrections are performed by applying a groupwise registration of the D-MRI sequences with an affine slice-by-slice transformation model. A dual RBF-based interpolation in the spatial and angular domains was then used to reconstruct a regularly sampled D-MRI sequence.
2 Method
2.1 Origin of sparse data
Typically, a D-MRI sequence consists of a set of N + 1 regularly sampled images
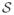 = {S0, S1, ··· SN } where S0 is the image obtained without diffusion weighting, and Si=1:N are diffusion-weighted (DW) images obtained with diffusion-sensitizing gradients Gi of direction Ui and strength b. Ideally, S0 and Si are related by the Stejskal-Tanner equation, but distortions caused by eddy currents and fetal motion invalidate this relationship. Image registration techniques can be applied to restore the lost spatial correspondence between S0 and Si, but a reconstruction from sparse data is then required.
= {S0, S1, ··· SN } where S0 is the image obtained without diffusion weighting, and Si=1:N are diffusion-weighted (DW) images obtained with diffusion-sensitizing gradients Gi of direction Ui and strength b. Ideally, S0 and Si are related by the Stejskal-Tanner equation, but distortions caused by eddy currents and fetal motion invalidate this relationship. Image registration techniques can be applied to restore the lost spatial correspondence between S0 and Si, but a reconstruction from sparse data is then required.
Let us express the original sequence as
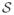 = {(X, Θ, S(X, Θ)}X∈ΩsΘ∈Ωa where Ωs is a regular spatial grid, and Ωa is a regular angular grid on the unit sphere. After correction,
= {(X, Θ, S(X, Θ)}X∈ΩsΘ∈Ωa where Ωs is a regular spatial grid, and Ωa is a regular angular grid on the unit sphere. After correction,
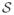 becomes
becomes
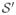 = {X′, Θ′, S(X, Θ))}X∈ΩsΘ∈Ωa, where
are the transformed spatial points, and
are the coordinates of Ui corrected with the rotational component
of
. In these equations, the superscript “z” has been added to indicate the slice-to-slice nature of the transformations. Differently of (X, Θ), the transformed coordinates (X′, Θ′) do not belong to regular grids, and
= {X′, Θ′, S(X, Θ))}X∈ΩsΘ∈Ωa, where
are the transformed spatial points, and
are the coordinates of Ui corrected with the rotational component
of
. In these equations, the superscript “z” has been added to indicate the slice-to-slice nature of the transformations. Differently of (X, Θ), the transformed coordinates (X′, Θ′) do not belong to regular grids, and
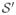 results sparse.
results sparse.
2.2 Registration approach
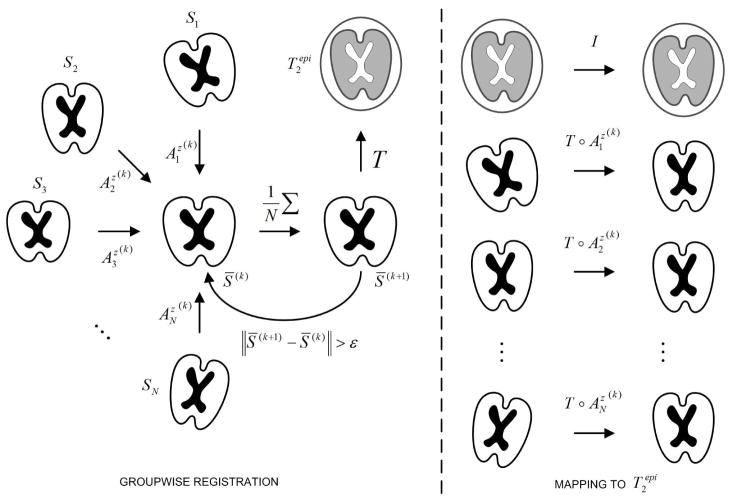
Distortion correction methods relying on the registration of Si to S0 may fail when applied to fetal D-MRI, probably a consequence of the large differences between both images [3]. In this paper, we propose a method that takes advantage of the similarity between DW images to first ensure their joint alignment. The mean of the registered images is characterized by a higher SNR than images Si, and provides a better depiction of the anatomical structure of the brain. These properties allow an accurate registration to S0, necessary to map all the sequence in its space of coordinates. This method is illustrated and explained in Figure 1. In essence, the method is quite similar to the one propose by Guimond et al. [5]. The main difference is the separation of the registration into (1) a groupwise registration of the DW images, and (2) the registration of the resulting mean image with the anatomical image.
Fig. 1.
Groupwise registration method. Initially all images Si are registered to an arbitrary DW image S* taken as reference. Then, all images are resampled and averaged to obtain a first average S̄(1), which becomes the new reference. At iteration k, the images Si are registered to the mean S̄(k) to obtain the transforms . After convergence, S̄(k) is registered to , and the composition of transformations is applied to map all images Si into the coordinate system of .
2.3 Sparse interpolation
To interpolate values on a regular grid from scattered data, we have used radial basis functions (RBF). RBFs have already been applied for interpolation on spherical geodesic grids in the context of numerical weather prediction, outperforming linear interpolation strategies [6]. The idea behind RBF interpolation is that every point has an influence on a neighborhood according to some functional φ(r), where r is the distance from the point. Then, the value of the function at the point P is given by a linear combination of the φ’s centered in the points Pi as:
| (1) |
where the weights wi are calculated by solving a linear system of equations for the function to agree with the observations at points Pi.
In this paper, we have used a Gaussian function as RBF since this function tends to zero for high r, and the influence of points Pi distant from P can be neglected. This allows considering only points in a neighborhood
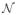 (P) of P for interpolation, which reduces the computational complexity of the method.
(P) of P for interpolation, which reduces the computational complexity of the method.
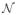 (P) was formed by points Pi falling inside the support region of the Gaussian function, defined in the context of this paper as the interval [−sφ, +sφ] so that φ (sφ) = 0.01 × φ(0).
(P) was formed by points Pi falling inside the support region of the Gaussian function, defined in the context of this paper as the interval [−sφ, +sφ] so that φ (sφ) = 0.01 × φ(0).
In our case, each point contains spatial and angular coordinates which must be considered separately because of the difference in scale between both types of coordinates. This situation is different from the problem dealt in [6] where only an interpolation in the sphere is required. To take into account these differences, we propose to modify Equation 1 by replacing the single RBF with the product of an spatial (φ) and an angular (ψ) RBF:
| (2) |
where X = (x, y, z) are the spatial coordinates, and Θ = (φ, θ) the spherical coordinates of the sampling vector Ui. Differently from Equation 1, Equation 2 allows a dual interpolation in two different unrelated spaces. In Equation 2, |X − Xi| represents the Euclidean distance, whereas |Θ − Θi| is the geodesic distance over the unit sphere.
3 Materials and experiments
3.1 Image data
Fetal MRI was performed on a 1.5 T Siemens Avanto MRI Scanner (SIEMENS, Erlangen, Germany) at the Hautepierre Hospital (Strasbourg, France) using an 6-channel phased array coil combined to the spine array positioned around the mother abdomen. An axial spin echo single-shot echo-planar sequence was acquired along 30 non-collinear diffusion gradient encoding directions with a b value of 700s/mm2. The following pulse sequence parameters were used: TR=6800 ms; TE=99 ms; FOV=250 × 250 mm2; matrix = 128 × 128; 41 contiguous axial slices of 3.5 mm thickness covering the whole fetal brain; no gap; number of excitations = 2. The total imaging time was 7mi 10s. Pregnant women were briefed before the exam and signed informed consent. To reduce motion artifacts, fetal sedation was obtained with 1 mg of flunitrazepam given orally to the mother 30 mi before the exam. The study was approved by the local ethics committee.
For validation purposes, three D-MRI sequences of the brain were acquired for an adult healthy subject in the following conditions: (i) static in supine position (
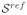 , the reference), and (ii) static with the head rotated with respect to the reference (
, the reference), and (ii) static with the head rotated with respect to the reference (
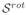 ).
).
3.2 Slice to volume registration accuracy
Initially we want to explore the ability to recover slice to volume alignment for typical but known motion, on typical anatomical structures. Fetal data are not suitable for assessing accuracy since motion artifacts are always present to some degree. Therefore only the adult dataset
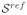 was used to this aim. Central slices of the volume were modified with a specific transformation and then registered independently to the whole volume. The displacements were chosen from a uniform distribution with a varying range of [−8, +8]mm for translations in each direction, and between [−10, +10]° for each rotation. These ranges of variation represent movements much larger than those observed in real fetal data.
was used to this aim. Central slices of the volume were modified with a specific transformation and then registered independently to the whole volume. The displacements were chosen from a uniform distribution with a varying range of [−8, +8]mm for translations in each direction, and between [−10, +10]° for each rotation. These ranges of variation represent movements much larger than those observed in real fetal data.
The accuracy was assessed by computing a registration error measured on a set of 4 points Pi within every slices as follows: , where TRE is the target registration error defined as TRE = ||Pi − T̂−1(T*(Pi))||2. T* denotes the known applied motion transformation, and T̂ is the estimated geometric transformation. Pi are the corners of the intersection between the bounding box containing the brain, and each slice. The error previously defined provides thus a maximum bound of the registration error for the region of interest.
3.3 Evaluation of RBF interpolation
Leave-one-out test
A leave-one-out test by using the adult data
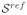 was performed for evaluating the capability of recovering non-acquired DW images from the available measurements. This test estimates the image
at a point (Xj, Θj) from
and
was performed for evaluating the capability of recovering non-acquired DW images from the available measurements. This test estimates the image
at a point (Xj, Θj) from
and
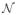 (Xj)\Xj, which in terms of RBF interpolation can be expressed as:
(Xj)\Xj, which in terms of RBF interpolation can be expressed as:
| (3) |
We have then computed the RMS error between
 (Xj, Θj) and
(Xj, Θj) and
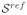 (Xj, Θj) for a set of random points distributed uniformly over the brain.
(Xj, Θj) for a set of random points distributed uniformly over the brain.
DW image estimation from a rotated sequence
In this experiment, DW images of
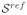 were estimated from
were estimated from
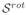 after performing a registration to put
after performing a registration to put
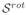 into the frame of reference of
into the frame of reference of
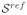 . Differently from the leave-one-out experiment, here all DW images of
. Differently from the leave-one-out experiment, here all DW images of
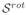 at all spatial positions were employed for estimation.
at all spatial positions were employed for estimation.
3.4 Diffusion descriptors
To evaluate the performance of the registration method, we have also considered two criteria for quality assessment of the resulting FA image.
Mean FA
The first criterion we have considered is the average value of FA over the cerebrospinal fluid (CSF), . This value is expected to be close to zero because of the isotropic diffusion properties of the CSF. Registration errors may induce an increase of this measure, since voxels belonging to the CSF in some DW images may be matched with voxels belonging to the gray or white matter in others.
Entropy
The second considered criterion was the entropy of the FA image (HFA) over the brain. Nielsen et al. [7] have compared polynomial and affine distortion correction, and observed a reduction in erroneous regions of FA maps along with a more spiky FA distribution in favor of the polynomial registration. Netsch and van Muiswinkel [8] have reported an increase in the sharpness of FA maps after distortion correction. In both cases, the observations should be manifested as lower HFA values.
4 Results
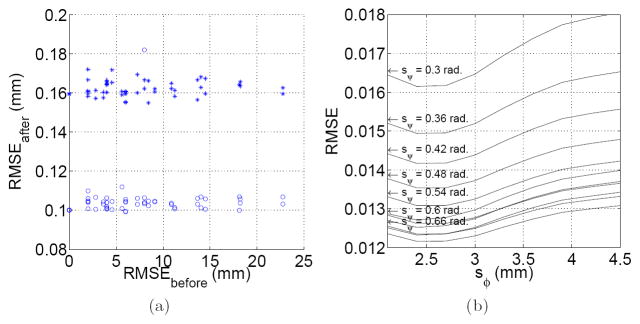
The slice-to-volume test was applied to and . RMS values higher than 0.2 mm were considered as registration failures, and discarded for analysis. Under this criterion, successful registrations were obtained in 95% of the applied transformations. The obtained RMS errors were 0.162 ± 0.004 mm for , and 0.105 ± 0.011 mm for . Figure 2(a) shows this error distribution.
Fig. 2.
(a) Slice to volume registration accuracy. This Figure shows the RMS error before and after registration for Si (○) and
(+). Each point corresponds to a specific applied transformation. (b) Estimation of
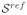 from
from
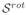 . The figure shows the normalized RMS error for different support regions of the RBF functions φ and ψ.
. The figure shows the normalized RMS error for different support regions of the RBF functions φ and ψ.
Th RMS error between the original sequence
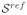 and its leave-one-out estimation
and its leave-one-out estimation
 was lower than 0.001 for sφ ∈ [2.0, 4.5] mm, and sψ ∈ [0.6, 0.9] rad. The minimum values were obtained for sφ = 3.5 mm (the inter-plane resolution), suggesting that the use of only in – plane neighbors is not sufficient for interpolation purposes, and adjacent slices must be included. Higher values of sφ make the interpolated value dependent of remote regions which could not present the same diffusion properties, and the error starts increasing. No dependence of sψ was found in this experiment.
was lower than 0.001 for sφ ∈ [2.0, 4.5] mm, and sψ ∈ [0.6, 0.9] rad. The minimum values were obtained for sφ = 3.5 mm (the inter-plane resolution), suggesting that the use of only in – plane neighbors is not sufficient for interpolation purposes, and adjacent slices must be included. Higher values of sφ make the interpolated value dependent of remote regions which could not present the same diffusion properties, and the error starts increasing. No dependence of sψ was found in this experiment.
Figure 2(b) shows the results when estimating
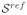 form
form
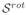 . In this case the error does depends on sψ, being lower for higher sψ values. After a given value (0.72 in our case) the error starts increasing again, because of the influence of distant gradient directions.
. In this case the error does depends on sψ, being lower for higher sψ values. After a given value (0.72 in our case) the error starts increasing again, because of the influence of distant gradient directions.
Table 1 compares the values of and for the original sequences, the values provided by the scanner’s manufacturer, and after applying the reconstruction method presented in this paper.
Table 1.
Diffusion descriptors based on FA maps. HFA = Entropy of FA, . For each column, the best value is shown in bold.
| Method | Fetus | #1 | Fetus | #2 | Fetus | #3 | Fetus | #4 |
|---|---|---|---|---|---|---|---|---|
| Original | 2.04 | 0.14 | 2.17 | 0.26 | 2.15 | 0.23 | 2.31 | 0.24 |
| Manufacturer | 2.11 | 0.12 | 2.09 | 0.21 | 2.20 | 0.20 | 2.33 | 0.19 |
| Our approach | 1.93 | 0.08 | 1.78 | 0.13 | 1.72 | 0.16 | 2.20 | 0.10 |
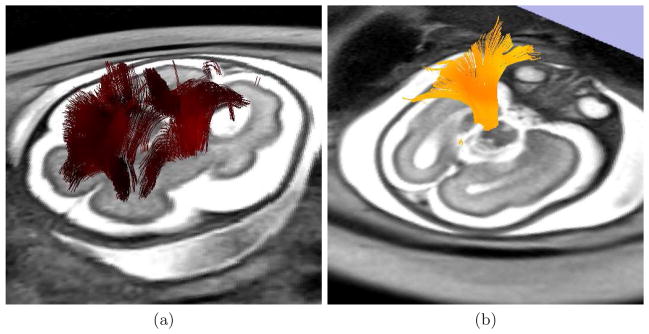
To perform the tractography, an expert radiologist traced regions containing the following bundles on images: (i) corpus callosum, (ii) inferior fronto-occipital tract, and (iii) pyramidal tract. These regions were used for seeding the tractography after propagation to the image by using affine registration, and to check the presence of specific bundles. Tensor was estimated by using a standard least squares method, and tractography was performed by applying a streamline method. In both cases we have used the algorithms implemented in Slicer4. Figure 3 shows some examples of the fibers recovered after application of the reconstruction method.
Fig. 3.
Tractography performed on a reconstructed sequence for a fetus of 28 weeks of gestational age. (a) Fibers recovered by seeding the streamline method with voxels presenting a linear measure higher than 0.3. (b) A bundle found for reconstructed sequences that is completely missing for in the original sequence.
5 Discussion and conclusions
In this paper, we have presented a method for reconstructing fetal D-MRI sequences from sparse data. A groupwise registration method based on slice-by-slice affine transformations was applied to compensate motion and eddy-current distortions, and a dual spatio-angular interpolation based on RBFs was used to estimate signal values on regular sampling grids. As the proposed method does not assume any diffusion model, the generated data can be used to study diffusion patterns even in IVOH. Experiments with adult data showed a high accuracy for the slice-to-volume registration, and for the estimation of DW images along unavailable gradient directions. In fetuses, the method improved the quality of the sequences as evidenced by the lower values of and HFA with respect to the original ones. The tractography provided different results for the original and reconstructed sequences, but they must be quantified and compared with objective criteria in order to assess their clinical significance. This is the current line of research.
Footnotes
References
- 1.Rohde GK, Barnett AS, Basser PJ, Marenco S, Pierpaoli C. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn Reson Med. 2004 Jan;51(1):103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- 2.Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza C, Vigneron DB, Barkovich JA, Studholme C. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Acad Radiol. 2006 Sep;13(9):1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Jiang S, Xue H, Counsell S, Anjari M, Allsop J, Rutherford M, Rueckert D, Hajnal JV. Diffusion tensor imaging (DTI) of the brain in moving subjects: application to in-utero fetal and ex-utero studies. Magn Reson Med. 2009 Sep;62(3):645–655. doi: 10.1002/mrm.22032. [DOI] [PubMed] [Google Scholar]
- 4.Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magn Reson Med. 2002 Oct;48(4):577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- 5.Guimond A, Meunier J, Thirion J-P. Average brain models: a convergence study. Comput Vis Image Understand. 2000;77:192210. [Google Scholar]
- 6.Carfora MF. Interpolation on spherical geodesic grids: a comparative study. J Comput Appl Math. 2007 Dec;210(1–2):99–105. [Google Scholar]
- 7.Nielsen JF, Ghugre NR, Panigrahy A. Affine and polynomial mutual information coregistration for artifact elimination in diffusion tensor imaging of newborns. Magn Reson Imaging. 2004 Nov;22(9):1319–1323. doi: 10.1016/j.mri.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 8.Netsch T, van Muiswinkel A. Quantitative evaluation of image-based distortion correction in diffusion tensor imaging. IEEE Trans Med Imaging. 2004 Jul;23(7):789–798. doi: 10.1109/TMI.2004.827479. [DOI] [PubMed] [Google Scholar]