Abstract
Intramuscular fat content and water-holding capacity are important traits in livestock as they influence meat quality, nutritive value of the muscle, and animal health. As a model for livestock, two inbred lines of the Berlin Muscle Mouse population, which had been long-term selected for high muscle mass, were used to identify genomic regions affecting intramuscular fat content and water-holding capacity. The intramuscular fat content of the Musculus longissimus was on average 1.4 times higher in BMMI806 than in BMMI816 mice. This was accompanied by a 1.5 times lower water-holding capacity of the Musculus quadriceps in BMMI816 mice. Linkage analyses with 332 G3 animals of reciprocal crosses between these two lines revealed quantitative trait loci for intramuscular fat content on chromosome 7 and for water-holding capacity on chromosome 2. In part, the identified loci coincide with syntenic regions in pigs in which genetic effects for the same traits were found. Therefore, these muscle-weight-selected mouse lines and the produced intercross populations are valuable genetic resources to identify genes that could also contribute to meat quality in other species.
Introduction
Meat is a major protein source in human nutrition and the consumption of meat products is growing worldwide. Understanding the genetic and metabolic backgrounds of muscle tissue characteristics is important for the sustainable improvement of meat quality but also for human and livestock health. The aim of this study was to localize regions in the mouse genome that regulate muscle traits that are important for meat production in livestock. The results contribute to improving the knowledge of genomic regions that are involved in economically important traits in animal production.
Intramuscular fat content (IMF) affects glucose uptake and the metabolism and health of humans and animals. Elevated levels of intramyocellular lipids can be a sign of impaired utilization of fatty acids as energy fuel or even insulin resistance (Phillips et al. 1996; Yaspelkis et al. 2004). Besides the health aspect, IMF greatly impacts the taste and tenderness of meat products (Cannata et al. 2010; Fernandez et al. 1999). In farm animal species, the amount of IMF and its distribution in the muscle differ between breeds, which suggests genetic components for this trait.
So far, only a few quantitative trait loci (QTL) for IMF have been mapped in sheep, pigs, and cattle (de Koning et al. 1999; Lambe et al. 2010; Underwood et al. 2007), and some candidate genes, e.g., porcine delta-like 1 homolog (DLK1), fatty acid binding protein 3 (FABP3), and the leptin receptor (LEPR), have shown associations with that trait (Schwab et al. 2009; Tyra et al. 2010; Zhao et al. 2010). In mice, for example, Ccr2 was described as an IMF-influencing gene in a knockout model (Ochoa et al. 2006).
Genetic determinants that have an impact on postmortem muscle traits are hardly known. This is especially the case for the water-holding capacity (WHC) of meat, which is the ability of the muscle to store tissue fluids within its fibers after the cessation of normal physiological conditions. Low WHC leads to impaired meat-processing properties and does not meet consumers’ preferences. Several QTL and candidate genes for WHC, e.g., ryanodine receptor 2 (RYR2), integrins, μ-calpain, and desmin, have been found in pigs (Bee et al. 2007; Fujii et al. 1991; Malek et al. 2001; Ponsuksili et al. 2008; Wimmers et al. 2010; Zhang et al. 2006).
Because the extent and influence of genetic regulators on IMF and WHC are not yet fully understood, we performed a crossbred experiment between two phenotypically different Berlin Muscle Mouse inbred lines that have been long-term selected for high muscle mass before inbreeding. The study aimed to identify QTL that affect IMF and WHC. In addition, we analyzed traits like muscle glycogen and lactate contents, blood glucose levels, and body composition which might correlate with IMF and WHC.
Materials and methods
Animals
The Berlin Muscle Mouse population had been long-term selected for high body weight and high muscle mass to reflect the selective mechanisms in livestock breeding. Founder animals of the Berlin Muscle Mouse (BMM) population were originally purchased in several pet shops in Berlin, Germany. The selection process comprised several distinct phases. The beginning of the selection process constituted a phase of 23 generations of selection for high protein content of the carcass at the age of 60 days. Protein content was determined by chemical analyses. In a second phase, mice were selected for high body weight and low fat content at 42 days for 10 generations. Afterward, mice were monitored for high muscularity by palpation and mice with the highest muscularity on a scale of 1–5 were selected for the next generation. As a result of selection and likely natural mutation(s) over 25 generations, a mouse population with a high muscular phenotype had been generated. A high compact subline was perpetuated through random mating of selected animals (Varga et al. 1997). Sequencing of the myostatin gene in this line revealed a 12-bp deletion (Szabo et al. 1998) leading to a loss of function. Scale and weight-based selection continued for 28 generations. After 86 generations of selection, full sibs with distinct phenotypes were mated. These founder animals became the basis of seven Berlin Muscle Mouse inbred lines (BMMI), four of which carry the MstnCmpt-dl1Abc mutation and three are wild type. In this study we used the lines BMMI806 and BMMI816, which are hypermuscular but do not carry the myostatin mutation. At the time of setting up the crossbred experiment, the lines were in generation 21 of inbreeding.
Pedigree structure
Two pairs of full sibs of the Berlin Muscle Mouse inbred lines BMMI806 and BMMI816 were crossed reciprocally to generate F1, F2, and G3 intercross populations. For the latter, 93 F2 animals were randomly mated (Schmitt et al. 2009), while avoiding sibling mating, to generate 345 G3 animals.
Husbandry and feeding conditions
The mice were treated in accordance with and all experimental protocols were approved by the German Animal Welfare Authorities (approval No. G0405/08). The animals were maintained under conventional conditions at 22 ±2°C and controlled lighting, with a 12:12-h light:dark cycle. They were kept in groups of two to four animals of the same sex per Macrolon cage and had ad libitum access to food and water. Until the age of 70 days, the animals were fed a standard breeding diet (Altromin standard breeding diet No. 1314 TPF, Lage, Germany). This diet contained 27.0% crude protein, 5.0% crude fat, 4.5% crude fiber, 6.5% crude ash, 50.5% nitrogen-free extract (starch and sugar), vitamins, trace elements, and minerals (2988 kcal/kg metabolizable energy of which 27.0% energy was from proteins, 13.0% from fat, and 60.0% from carbohydrates).
Phenotypic measures
After a fasting period of 2 h, 71-day-old mice were anesthetized under isoflurane and decapitated. The Musculus longissimus (ML) and Musculus quadriceps (MQ) were dissected and weighed. The summed muscle weights of the left and right ML and the left and right MQ were recorded as muscle mass (MM). The right muscles were immediately frozen in liquid nitrogen and subsequently stored at −80°C. The left muscles were cooled down to 6°C for 1 h, subsequently frozen, and stored at −20°C until WHC and IMF were measured. Carcasses were stored at 6°C and pH values were taken within the M. biceps femoris at 1 and 24 h postmortem (ebro PHT 810, Ingolstadt, Germany). For the determination of WHC, frozen muscles were thawed and stored at 6°C for 24 h. Muscles were then gently centrifuged for 60 s at 604×g in Invitek 1.5-ml receiver tubes with filter inlays to collect the tissue fluid that was not held from the muscle (Eppendorf Minispin, Hamburg, Germany; Invitek, part of STRATEC Molecular GmbH, Berlin, Germany). The ratio of lost tissue fluid to tissue mass was designated as “drip loss.” IMF was measured as percentage of muscle weight using nuclear magnetic resonance technology (SMART Trac System, CEM, Kamp-Lintfort, Germany) (Kaerst et al. 2010). Body weights were recorded weekly. At 10 weeks, fat and lean masses were measured by quantitative magnetic resonance (QMR) analysis using the EchoMRI whole-body composition analyzer (Echo Medical Systems, Houston, TX, USA) (Neuschl et al. 2010; Tinsley et al. 2004). Blood glucose levels were measured after 2 h of fasting, prior to dissection at 10 weeks (Bayer “Contour,” Leverkusen, Germany). Muscle glycogen content was determined colorimetrically in the right ML (GOD/PAP method “Glucose liquicolor” by Human, Wiesbaden, Germany), as suggested by Barham and Trinder (1972). Lactate contents were determined colorimetrically in the right ML using the kit by Chung Lee (Lactate Assay Kit, SUNY, Buffalo, NY, USA).
Genotyping
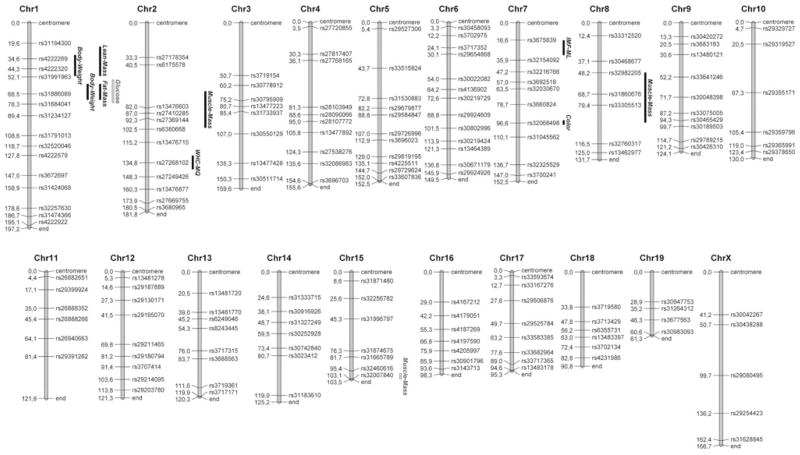
Parental BMMI lines were genotyped with the Mouse Diversity Array (Yang et al. 2009) comprising 623,124 single-nucleotide polymorphisms (SNPs). The SNP information provided evidence for allele fixation of 98.3 and 98.0% in lines BMMI806 and BMMI816, respectively, after 21 generations of inbreeding. Both lines differed by 8.8% at the SNP level. Using the information on diverse genomic regions between BMMI806 and BMMI816, 164 informative markers covering all chromosomes (except Y) in an average distance of 16.2 Mb were selected for genotyping the parents, the F2, and the G3 animals (Fig. 1). Regions larger than 10 Mb without informative markers did not differ in SNP alleles between parental lines and were not included in the linkage analysis. Genotyping was done at KBiosciences (Hoddesdon, UK). The genetic map was converted into the physical map using the Mouse Map Converter software from the Jackson Laboratory (Bar Harbor, ME, USA) (Cox et al. 2009). QTL refinement was done by analyzing informative SNPs from the Mouse Diversity Array as described before (Schmitt et al. 2007).
Fig. 1.
MapChart plot of 164 reference single-nucleotide polymorphisms used in this study. Positions are given in Mb (Ensembl release 37). Bars indicate 1-LOD support intervals of identified significant QTL
QTL and statistical analyses
For QTL analysis, 332 G3 animals were used. Analyses for single-QTL detection and detection of interacting QTL were performed using QTLRel (http://cran.r-project.org/web/packages/QTLRel/index.html). Unlike the F2 animals, the G3 animals may be unequally related to each other. Ignoring the unequal relatedness may result in a serious inflation of false-positive QTL rates. Thus, we applied the mixed model described by Cheng et al. (2010) to analyze our data. All phenotypes were log-transformed to obtain normal distribution. Direction-of-cross and sex were included as additive covariates in the model and used as interactive covariates to test their effects on QTL. Genome-wide significant QTL (P < 0.05) were included in a mixed model for each trait to calculate the respective trait variance in the G3 population. Trait-specific significance thresholds were estimated from 5,000 permutations (Churchill and Doerge 1994). Additional analyses were performed using R/qtl (Broman et al. 2003). Basic statistics were performed using the SAS software package (SAS System for Windows, Release 9.2, SAS Institute, Cary, NC, USA).
Results
Phenotypes
Compared to male BMMI806, male BMMI816 showed 46% less total fat mass along with 36 and 22% less IMF in the M. longissimus and M. quadriceps, respectively. The mean fasting blood glucose levels were also significantly lower in BMMI816 (by 22% in males and 19% in females) than in BMMI806. On average, female BMMI816 mice reached 8% higher body weights and 11% lower muscle masses than BMMI806 females. The analysis of WHC revealed that the MQ of female BMMI816 also showed 51% more drip loss, which indicates a lower WHC than female BMMI806. Sex differences were also distinct between lines. While in line BMMI816 females showed lower WHC and higher fat mass and IMF than males, in line BMMI806 males and females did not differ significantly with respect to WHC and IMF.
When we compared the mean values of the crossbred populations with the parental lines, a dominance effect was observed in BMMI816 F1 males for lower lean mass, fat mass, and IMF, while blood glucose levels, WHC of the ML, and lactate levels were intermediate or resembled the BMMI806 phenotype. In female F1 animals, even over-dominance effects were found for body weight and lean mass, which were higher, and for IMF of the MQ, which was lower than in the parental lines. The BMMI816 phenotype dominated IMF of the ML, WHC, and blood glucose levels, while the BMMI806 phenotype prevailed for muscle mass, fat mass, and lactate levels in the F1 females.
In the males of the G3 population, we found overdominance effects for IMF of the MQ, WHC, muscle mass, and muscle lactate levels with respect to the respective values of the parental lines. The lower BMMI816 phenotypes prevailed for fat mass, IMF of the ML, and blood glucose levels. In female G3 overdominance was found for body weight and lean mass on the one hand, which were significantly higher, and for IMF and lactate on the other hand, which were lower as compared to both parental lines. The BMMI816 phenotype prevailed by low WHC and blood glucose levels, whereas the BMMI806 phenotype dominated in the higher muscle mass and the lower fat mass in females (Table 1).
Table 1.
Body traits of parental, F1, and G3 animals
| Trait | P
|
F1 |
G3 |
|||||
|---|---|---|---|---|---|---|---|---|
| BMMI816
|
BMMI806
|
Males | Females | Males | Females | |||
| Males | Females | Males | Females | |||||
| Body weight (g) | 36.88 (3.68) | 29.02 (2.77)** | 36.43 (4.69) | 26.77 (2.16) | 35.70 (4.21) | 30.73 (2.69)a,bbb | 38.99 (3.18) | 31.60 (2.72)aa,bbb |
| Lean mass (g) | 31.09 (2.19) | 23.04 (2.02) | 29.31 (2.29) | 23.87 (1.99) | 30.84 (3.55)bb | 26.00 (2.74)aa,b | 32.56 (2.58)bbb | 25.94 (2.16)aa,bb |
| Muscle mass (g) | 1.14 (0.10) | 0.78 (0.09)* | 1.12 (0.12) | 0.88 (0.07) | 1.14 (0.22) | 0.93 (0.12)aaa | 1.25 (0.14)aa,bbb | 0.96 (0.10)aaa |
| Fat mass (g) | 2.37 (0.65)*** | 4.26 (1.42) | 4.41 (1.46) | 3.13 (1.19) | 2.00 (0.91)bbb | 2.28 (0.63)a | 2.50 (1.32)bbb | 2.42 (1.03)aa |
| IMF-ML (%) | 1.68 (0.30)*** | 2.07 (0.42) | 2.64 (0.34) | 2.45 (0.49) | 1.69 (0.37)bbb | 1.84 (0.57)bbb | 1.62 (0.39)bbb | 1.58 (0.39)aaa,bbb |
| IMF-MQ (%) | 1.41 (0.20)*** | 1.94 (0.17) | 1.81 (0.30) | 1.80 (0.29) | 1.41 (0.23)bbb | 1.58 (0.32)aaa,bb | 1.28 (0.19)a,bbb | 1.32 (0.19)aaa,bbb |
| WHC-ML (%) | 0.92 (0.25) | 1.29 (0.35) | 1.09 (0.37) | 1.01 (0.38) | 1.29 (0.64)a | 1.36 (0.51)b | 1.38 (0.47)aaa,bb | 1.48 (0.43)bbb |
| WHC-MQ (%) | 0.89 (0.27) | 1.22 (0.34)* | 0.85 (0.33) | 0.81 (0.26) | 1.15 (0.65) | 1.18 (0.51)bb | 1.19 (0.42)aa,bbb | 1.16 (0.35)bbb |
| Glucose (mg/dl) | 91 (17)*** | 84 (13)*** | 120 (26) | 104 (17) | 106 (26)a,bb | 86 (18)bbb | 96 (17)bbb | 83 (12)bbb |
| Glycogen (mg/g) | 2.78 (0.89) | 2.83 (1.68) | 2.47 (0.76) | 2.45 (1.38) | 1.56 (0.87)a | 1.74 (1.62) | 1.82 (0.80) | 1.81 (0.73) |
| Lactate (mg/g) | 0.08 (0.01) | 0.08 (0.02) | 0.06 (0) | 0.07 (0.03) | 0.06 (0.01)a | 0.05 (0.01)a | 0.05 (0.03)aaa,bbb | 0.04 (0.02)a,bbb |
Values are mean (standard deviation)
IMF intramuscular fat content, WHC water-holding capacity (measured as drip loss in percent of muscle weight), ML Musculus longissimus, MQ Musculus quadriceps, Glucose blood glucose level
Phenotypes of the parental lines are compared with each other and the F1 and G3, separated for sex. Asterisks indicate significant differences between the parental lines; letters indicate significant differences between the F1 or G3 and BMMI816
or BMMI806
lines. Number of asterisks and letters show levels of significance
P < 0.001,
P < 0.01,
P < 0.05)
In both sexes of the G3 population, body weight was positively correlated with fat, lean mass, and muscle mass. Significant positive correlations were also found between the IMF of the ML and MQ, but also with total fat mass. Fasting glucose levels were highly dependent on not only body composition but also on the IMF in ML and to a lesser extent in MQ. The higher the IMF was the higher the blood glucose level, which might indicate reduced glucose uptake by the muscle cells and low insulin sensitivity. High fat mass contributed to increased IMF, while high muscle mass (and total lean mass) had positive effects on lowered IMF in both sexes. Interestingly, an opposite correlation between muscle mass and fat mass was observed for both sexes: high muscle mass was negatively correlated with fat mass in males but positively correlated in females. In both sexes, high glycogen content was associated with high drip loss in the ML and MQ, high pH of muscle 1 h postmortem in females, and low pH after 24 h in males (Table 2).
Table 2.
Spearman’s correlation coefficients between different traits in the G3 population
| Males
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Body weight (g) | Lean mass (g) | Fat mass (g) | Muscle mass (g) | IMF-ML (%) | IMF-MQ (%) | WHC-ML (%) | WHC-MQ (%) | Glucose (mg/dl) | pH1 | pH24 | Glycogen (mg/g) | Lactate (mg/g) | |
| Body weight (g) | 0.91*** | 0.34*** | 0.64*** | 0.09 | 0.06 | 0.02 | −0.05 | 0.24*** | −0.01 | 0.03 | −0.10 | 0.13 | |
| Lean mass (g) | 0.90*** | 0.01 | 0.78*** | −0.15• | −0.18* | 0.05 | 0.00 | 0.22** | −0.08 | 0.04 | −0.03 | 0.11 | |
| Fat mass (g) | 0.42*** | 0.11 | −0.18* | 0.71*** | 0.70*** | −0.07 | −0.08 | 0.17* | 0.20** | −0.07 | −0.15* | 0.05 | |
| Muscle mass (g) | 0.68*** | 0.71*** | 0.17* | −0.24** | −0.26*** | 0.03 | −0.05 | 0.13 | −0.05 | 0.07 | −0.14 | 0.11 | |
| IMF-ML (%) | 0.02 | −0.12 | 0.50*** | −0.15 | 0.67*** | −0.03 | −0.09 | 0.10 | 0.16* | −0.15* | −0.15• | −0.04 | |
| IMF-MQ (%) | −0.10 | −0.25*** | 0.45*** | −0.23** | 0.49*** | −0.12 | −0.03 | 0.05 | 0.02 | −0.26*** | −0.13 | 0.03 | |
| WHC-ML (%) | −0.03 | −0.03 | 0.07 | 0.04 | 0.11 | 0.01 | 0.41*** | −0.02 | 0.10 | −0.15* | 0.36*** | 0.16* | |
| WHC-MQ (%) | −0.02 | −0.03 | 0.00 | −0.07 | 0.03 | 0.03 | 0.14 | −0.06 | −0.02 | −0.24*** | 0.37*** | 0.04 | |
| Glucose (mg/dl) | 0.15 | 0.17* | 0.10 | 0.04 | 0.17* | 0.08 | 0.13 | 0.10 | 0.10 | 0.17* | 0.12 | 0.00 | |
| pH1 | 0.17* | 0.11 | 0.10 | 0.23** | −0.06 | −0.06 | −0.01 | 0.02 | 0.08 | 0.34*** | 0.02 | 0.07 | |
| pH24 | 0.16 | 0.08 | 0.07 | 0.14 | −0.06 | −0.06 | −0.06 | −0.21** | 0.01 | 0.25** | −0.26*** | 0.01 | |
| Glycogen (mg/g) | −0.01 | 0.01 | −0.02 | 0.14 | −0.05 | −0.04 | 0.17* | 0.22** | 0.03 | 0.21** | −0.18* | 0.01 | |
| Lactate (mg/g) | −0.01 | −0.04 | 0.00 | −0.03 | −0.04 | −0.14 | 0.08 | −0.04 | 0.01 | −0.01 | 0.16* | −0.02 | |
| Females | |||||||||||||
IMF intramuscular fat content, WHC water-holding capacity, measured as drip loss in percent of muscle weight, ML M. longissimus, MQ M. quadriceps, Glucose blood glucose level
Asterisks indicate the significance of correlations:
P < 0.001,
P < 0.01,
P < 0.05,
P < 0.10
QTL effects
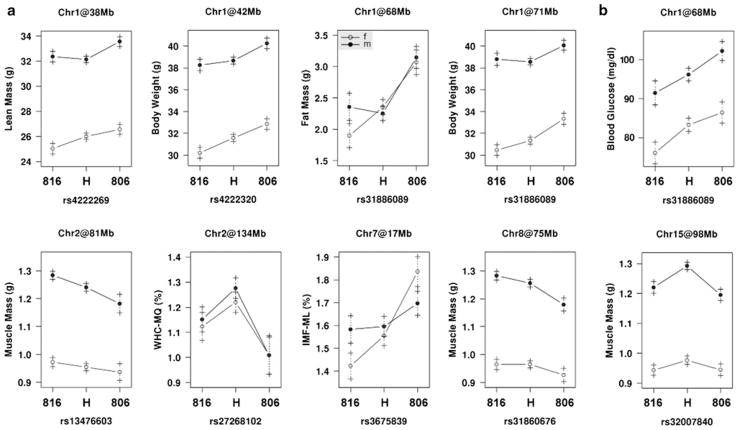
Despite the small difference between the parental lines, two QTL for body weight were identified on chromosome (Chr) 1 at 42 Mb (with a LOD support interval between 31 and 51 Mb) and at 71 Mb (59–74 Mb). Animals carrying the BMMI816 alleles showed lower values for this trait. These two QTL for body weight overlapped with QTL for fat and lean mass, since a QTL affecting fat mass was also mapped on Chr 1 at 68 Mb (59–74 Mb) and another QTL for lean mass was mapped on Chr 1 at 38 Mb (23–51 Mb). The alleles for increasing fat mass and lean mass were both inherited from the BMMI806 line. These results are shown in Table 3 and Fig. 2. Furthermore, we identified a QTL for IMF of the ML on Chr 7 at 17 Mb (17–31 Mb), which explains 5.70% of the phenotypic variance in the G3 population; BMMI806 alleles increased IMF in an additive manner.
Table 3.
QTL significant at the genome-wide level of 5% for various traits in the G3 population at the age of 10 weeks
| Trait | Chr | Mba | SIb | Markerc | LODd | a (SE)e | d (SE)e | % G3 varf | iSNP%g | #genesh |
|---|---|---|---|---|---|---|---|---|---|---|
| Lean mass (g) | 1 | 38 | 23–51 | rs4222269 | 3.66 | −0.83 (0.22) | −0.36 (0.29) | 2.22 | 20.1 | 71 |
| Body weight (g) | 1 | 42 | 31–51 | rs4222320 | 6.46 | −1.41 (0.31) | −0.27 (0.36) | 4.21 | 19.1 | 65 |
| Fat mass (g) | 1 | 68 | 59–74 | rs31886089 | 7.02 | −0.53 (0.11) | −0.27 (0.13) | 9.84 | 14.6 | 35 |
| Body weight (g) | 1 | 71 | 59–74 | rs31886089 | 7.99 | −1.54 (0.34) | −0.69 (0.39) | 5.32 | 14.6 | 35 |
| Muscle mass (g) | 2 | 81 | 66–101 | rs13476603 | 4.29 | 0.06 (0.01) | 0.03 (0.02) | 3.11 | 16.1 | 92 |
| WHC-MQ (%) | 2 | 134 | 128–141 | rs27268102 | 3.93 | 0.06 (0.03) | 0.17 (0.04) | 4.60 | 20.4 | 30 |
| IMF-ML (%) | 7 | 17 | 17–31 | rs3675839 | 3.72 | −0.13 (0.03) | −0.06 (0.04) | 5.70 | 4.4 | 37 |
| Muscle mass (g) | 8 | 75 | 48–96 | rs31860676 | 3.63 | 0.05 (0.01) | 0.02 (0.01) | 2.59 | 6.3 | 51 |
Most likely chromosomal location given as Mb position
1-LOD support interval (SI) in Mb
Marker closest to the chromosomal position with the highest LOD score
LOD scores from the full model estimations
Additive (a) and dominance (d) effect and their standard errors (SE) determined with the nontransformed raw trait values, therefore given in the respective unit; the direction of a and d, respectively given as the effect of the BMMI816 allele
G3 phenotypic variance (%) explained by the QTL; QTL effect given as the reduction of the residual sum of squares fitting 1 vs. 0 QTL
Percentage of informative SNP markers in the LOD support interval
Number of genes that are located in informative loci within the SI
Fig. 2.
Effect plots showing means and standard errors of the three genotype classes at all significant QTL peak positions for males (filled circles) and females (open circles); QTL significant under a P < 0.05 and b P < 0.10 genome-wide threshold; 816 homozygous BMMI816 allele, 806 homozygous BMMI806 allele, H heterozygous animals
Another significant QTL was found for WHC of the MQ on Chr 2 at 134 Mb (128–141 Mb). This QTL accounted for 4.60% of the G3 variance. The locus showed over-dominance, with heterozygous animals having the highest drip loss (i.e., lowest WHC). For muscle mass, significant QTL were located on Chr 2 at 81 Mb (66–101 Mb) and on Chr 8 at 75 Mb (48–96 Mb). For both QTL, the BMMI816 alleles increased the muscle mass and explained 3.11 and 2.59% of the total phenotypic variance, respectively. QTL on Chr 2 that regulate muscle mass were described before. Their peak positions were located at 76 Mb (64–127 Mb) and 125 Mb (16–161 Mb) (Lionikas et al. 2005). On Chr 8, QTL for muscle mass were detected at 81 Mb (46–86 Mb) (Lionikas et al. 2006) and 110 Mb (27–145 Mb) (Brockmann et al. 2004). An additional QTL for muscle mass was located on Chr 15 at 98 Mb (98–103 Mb). It was found under the 10% genome-wide significance threshold (LOD = 3.45) with additive and dominance effects of 0.01 and 0.05 g, respectively, and had not described before. No significant QTL for glycogen, lactate, and the variation of pH values were identified.
In addition, we identified two suggestive QTL under a somewhat more liberal 10% genome-wide significance threshold (P < 0.1). For fasting blood glucose levels, we found a suggestive QTL on Chr 1 at 68 Mb (55–77 Mb, LOD = 3.40). Consistent with the parental phenotypes, the additive effect of the BMMI806 allele for blood glucose levels was 4.7 mg/dl (dominance effect = 0.8 mg/dl). This QTL coincided with the QTL interval for fat mass and body weight.
Moreover, a significant QTL for the pigmentation of the BMMI816 was identified on Chr 7 at 97 Mb (94–98 Mb). The BMMI816 line showed black pigmentation except for the ventral side of the body, while the BMMI806 animals were not pigmented. Those G3 animals that were homozygous or heterozygous for the BMMI816 allele at this locus exhibited the specified pigmentation.
Informative loci
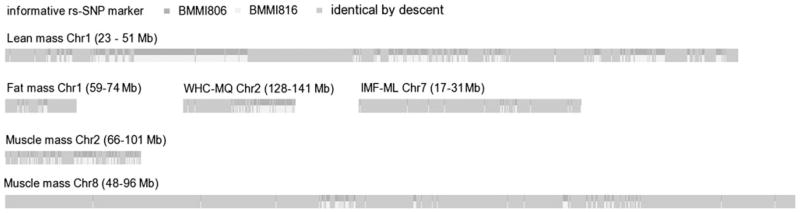
Additional QTL refinement was accomplished when we analyzed the degree of divergence between the parental BMMI lines. The extent of divergence and the number of genes in these informative loci are given in Table 3, according to the data of all SNP markers from the Mouse Diversity Array. A list of putative candidate genes, which are located in the divergent regions, is given in Table 4A and B for the respective examined traits. However, albeit a high SNP density on the array, it is very likely that not all divergent loci were detected. Therefore, it cannot be excluded that genetic determinants underlying certain QTL effects are located between the visible divergent regions. Additional data, e.g., sequencing and RNA expression, are needed to verify candidate genes that were obtained from this analysis. Using rs-SNP marker information of the Mouse Diversity Array and a perl script, we generated Fig. 3, which illustrates the ratio between divergent regions and regions identical by descent (IBD) between the two parental lines for the detected QTL (Mott 2005). The regions that are divergent between the two parental strains most likely contain the genes responsible for the detected QTL effects. The close relatedness of the parental lines explains the high amount of IBD haplotypes.
Table 4.
List of candidate genes for different traits in informative regions of the QTL
| A
| |||
|---|---|---|---|
| Lean mass (Chr1:23–51 Mb) | |||
| 1110058L19Rik | Cnga3 | Inpp4a | Tesp1 |
| 1700024G10Rik | Cnnm3 | Khdrbs2 | Tesp2 |
| 2010300C02Rik | Cnnm4 | Lincr | Tmem131 |
| 2300002O18Rik | Col19a1 | Lman2l | Tpp2 |
| 4832428D23Rik | Col5a2 | Lmbrd1 | Tsga10 |
| 4921511C04Rik | Creg2 | Lonrf2 | Txndc9 |
| 4921533L14Rik | D1Bwg0212e | Map4k4 | Ugcgl1 |
| 6330578E17Rik | Dnahc7 | Mgat4a | Unc50 |
| 9430069J07Rik | Dnahc7b | Mitd1 | Uxs1 |
| A230074B11Rik | Dst | Mrpl30 | Wdr75 |
| Actr1b | EG212225 | Nms | Zap70 |
| Aff3 | Eif5b | Npas2 | |
| Arid5a | ENSMUSG00000053185 | Pdcl3 | |
| Bai3 | Ercc5 | Phf3 | |
| BC043098 | Fhl2 | Plekhb2 | |
| BC050210 | Gm776 | Prim2 | |
| Bivm | Gulp1 | Rev1 | |
| C230029F24Rik | Il18r1 | Slc39a10 | |
| Cfc1 | Il1r2 | Slc40a1 | |
| Chst10 | Il1rl2 | Tbc1d8 | |
|
| |||
| Fat mass (Chr1:59–74 Mb) | IMF-ML (Chr7:17–31 Mb) | ||
|
| |||
| 1110028C15Rik | Klf7 | 1700049G17Rik | Pvrl2 |
| 9430067K14Rik | March4 | 4933426I21Rik | Ryr1 |
| A830006F12Rik | Mpp4 | Actn4 | Samd4b |
| Abi2 | Mreg | Akt2 | Sertad3 |
| Acadl | Mtap2 | BC057627 | Sfrs16 |
| Als2cr13 | Nbeal1 | Bckdha | Shkbp1 |
| BC042720 | Pard3b | Blvrb | Sipa1l3 |
| Bmpr2 | Pecr | Ceacam13 | Spnb4 |
| C030018G13Rik | Pip5k3 | Ckm | Sympk |
| Carf | Pthr2 | Cyp2b10 | Tex101 |
| Cd28 | Spag16 | Cyp2s1 | Trappc6a |
| Cps1 | Tmem169 | Ercc2 | Xrcc1 |
| Cyp20a1 | Wdr12 | Gemin7 | Zfp30 |
| D630023F18Rik | Xrcc5 | Grik5 | Zfp568 |
| Erbb4 | Zdbf2 | Nlrp4e | Zfp60 |
| Gm973 | Nlrp5 | Zfp607 | |
| Ica1l | Numbl | Zfp84 | |
| Icos | Pou2f2 | ||
| Idh1 | Prx | ||
| Ikzf2 | Ptgir | ||
|
| |||
| WHC-MQ (Chr2:128–141 Mb)
| |||
| 1110034G24Rik | Plcb1 | ||
| 2210009G21Rik | Prei4 | ||
| 2310003L22Rik | Sel1l2 | ||
| 2900006F19Rik | Smox | ||
| 4921508D12Rik | Snap25 | ||
| 4930473A02Rik | Snrpb | ||
| Anapc1 | Sptlc3 | ||
| Atrn | Tasp1 | ||
| Btbd3 | Txndc13 | ||
| Cdc25b | Zc3h8 | ||
| Cds2 | |||
| Ebf4 | |||
| Esf1 | |||
| F830045P16Rik | |||
| Flrt3 | |||
| Hao1 | |||
| Jag1 | |||
| Mertk | |||
| Mkks | |||
| Pdyn | |||
| B
| ||||
|---|---|---|---|---|
| Muscle mass (Chr2:66–101 Mb) | ||||
| 1110051M20Rik | Cugbp1 | Madd | Olfr1258 | Syt13 |
| 2700094K13Rik | D030051N19Rik | Metapl1 | Olfr1271 | Tfpi |
| 4833418A01Rik | D230010M03Rik | Mtch2 | Osbpl6 | Tnks1bp1 |
| 4833423E24Rik | D2Ertd391e | Myo3b | P2rx3 | Ttc21b |
| 4932414N04Rik | D430039N05Rik | Nckap1 | Pde1a | Ube2e3 |
| Abcb11 | Ddb2 | Ndufs3 | Pex16 | Ube2l6 |
| Acp2 | Dnajc10 | Nup160 | Phf21a | Xirp2 |
| Agbl2 | Dusp19 | Nup35 | Ppp1r1c | Ypel4 |
| Agtrl1 | ENSMUSG00000075313 | Olfr1032 | Prdx6_rs1 | Zc3h15 |
| Arhgap1 | Ext2 | Olfr1033 | Prg2 | Zdhhc5 |
| B3galt1 | F2 | Olfr1107 | Psmc3 | Zfp533 |
| BC003993 | Hsd17b12 | Olfr1115 | Ptprj | Zfp804a |
| Calcrl | Itga4 | Olfr1182 | Rapgef4 | |
| Chn1 | Itgav | Olfr1193 | Scn1a | |
| Chst1 | Kbtbd4 | Olfr1206 | Scn7a | |
| Ckap5 | Lass6 | Olfr1213 | Slc43a1 | |
| Clp1 | Lnp | Olfr1215 | Smtnl1 | |
| Creb3l1 | Lrp2 | Olfr1219 | Ssfa2 | |
| Cry2 | Lrp4 | Olfr1221 | Ssrp1 | |
| Ctnnd1 | Lrrc55 | Olfr1245 | Stk39 | |
|
| ||||
| Muscle Mass (Chr8:48–96 Mb) | Muscle Mass (Chr15:98–103 Mb) | |||
|
| ||||
| 1700007B14Rik | Es22 | Sh2d4a | Zfp641 | |
| 1810029B16Rik | Frem3 | Sh3rf1 | Pou6f1 | |
| 2810422J05Rik | Fto | Slc10a7 | Adcy6 | |
| 4732435N03Rik | Gab1 | Stox2 | Slc4a8 | |
| Abcc12 | Gatad2a | Tktl2 | Cacnb3 | |
| Adcy7 | Inpp4b | Tmem188 | Scn8a | |
| Aga | Large | Tox3 | Ccdc65 | |
| AI931714 | March1 | Ttc29 | Espl1 | |
| Arhgap10 | Nanos3 | Vegfc | Atp5g2 | |
| Atp6v1b2 | Nat2 | Wdr17 | Mll2 | |
| BC033932 | Nat3 | Zfp423 | Spats2 | |
| BC053440 | Nek1 | Lima1 | ||
| BC057552 | Nkd1 | |||
| Brd7 | Nod2 | |||
| Cyp4f18 | Nr3c2 | |||
| D630040G17Rik | Odz3 | |||
| Ednra | Otud4 | |||
| Ell | Palld | |||
| EG330776 | Psd3 | |||
| EG70793 | Rab8a | |||
|
| ||||
| Pigmentation (Chr7:94–98 Mb)
| ||||
| Nox4 | ||||
| Tyr | ||||
| Grm5 | ||||
| Ctsc | ||||
| Rab38 | ||||
| Me3 | ||||
| Sytl2 | ||||
Fig. 3.
Illustration of divergent genomic regions and regions being identical by descent between BMMI806 and BMMI816 mice for different quantitative traits, based on significant QTL and respective rs-SNP marker information
Discussion
The present study was performed to identify QTL for muscle traits, with IMF and WHC being the main traits of interest. It was unclear whether IMF-influencing genes would be different from genes responsible for the distribution and quantity of normal adipose tissue (Tanomura et al. 2002). From our results that showed different QTL for IMF and total fat mass, we conclude that there are indeed different genetic determinants that control total fat mass and IMF. The highly significant QTL for total fat mass on Chr 1 in our cross was also seen in the scans for IMF of the M. longissimus but had no significant effects (P < 0.37) and did not affect IMF in the M. quadriceps. This QTL likely acts predominantly on fat accumulation in the adipose but not in the muscle tissue. The total fat mass QTL on Chr 1 (Fig. 1) overlapped with a suggestive QTL (P < 0.10) for fasting blood glucose levels. It cannot be excluded that this chromosomal region is a major regulator of both glucose utilization and fat metabolism in our cross. It has been shown repeatedly that increased adiposity is linked to insulin resistance and higher blood glucose levels as a result of impaired glucose clearance (Kahn et al. 2006). In addition, high intramuscular triglyceride levels, which are highly correlated with high body fat mass in our cross, inhibit the insulin-signaling cascade and lower glucose uptake (Chadt et al. 2008; Powell et al. 2004; Schmitz-Peiffer 2000; Tanomura et al. 2002). Both high fasting blood glucose levels and increased IMF were observed in BMMI806 mice. Different genes in the QTL region could contribute to the QTL effect. One interesting candidate gene in the QTL region on Chr 1 at 67 Mb is the long-chain acyl-CoA-dehydrogenase gene (Acadl), which is directly involved in the first step of β oxidation by converting acyl-CoA from fatty acids into Δ2-trans-enoyl-CoA. Decreased oxidation of mitochondrial fatty acid was shown in Acadl-deficient mice (Zhang et al. 2007). It seems possible that impaired fatty acid utilization could result in increased fat mass but also in high IMF, especially since skeleton muscles lack de novo lipogenesis (Eaton 2002). Interestingly, data from the Mouse Diversity Array showed three SNPs to be different in this gene between BMMI806 and BMMI816 mice. The first SNP is located in an intron and the second and the third SNPs are located within exons and code synonymously. However, since it cannot be ruled out that adjacent base pairs are also different leading to a nonsynonymous coding, Acadl seems to be a good candidate gene that deserves further attention.
As mentioned, it is known from studies in humans and mice that muscular triglycerides and certain lipid species inside the muscle fibers have similar metabolic effects, e.g., on the glucose uptake (Ebeling et al. 1998; Pan et al. 1997; Phillips et al. 1996; Yu et al. 2002). To some extent, the biological function of muscle lipid stores seems to be similar in different species in terms of lipids as an energy fuel and their role in homeostasis. However, the respective patterns of fatty acids might be slightly different in different species and can even be manipulated (Ludden et al. 2009; Wood et al. 2004). In our study we measured the total fat content within the muscle and found differences between parental mouse lines.
Several fat metabolism genes are located in the QTL region on Chr 7 that affects IMF, e.g., the lipase gene (Lipe) and the apolipo-protein E gene (Apoe) (Hansson et al. 2005; Hunt et al. 2006), which might be responsible for the high IMF phenotype of the BMMI806. However, both genes are located in regions that are noninformative on the measured SNP level.
Still, there might be sequence alterations in these regions that remained undetected by the Mouse Diversity Array. Another interesting candidate gene in this region is thymoma viral proto-oncogene 2 (Akt2 or Pkb) at 28 Mb. Akt2 is involved in the insulin-dependent regulation of lipid metabolism and triglyceride storage (Leavens et al. 2009). There is a SNP marker in this gene that differs between the parental lines, and the BMMI806 animals have high fasting blood glucose levels that might also indicate a certain insulin resistance. This shows that including information about regions that are identical by descent between the parental lines can help to narrow down the list of candidate genes in a QTL, but additional data, such as from RNA expression or sequencing, are needed to finally confirm candidate genes (Table 4A, B).
The comparison of syntenic regions between mice and other mammals via the Ensembl database revealed supporting evidence for the presented data. The murine QTL region for IMF on Chr 7 is syntenic with regions on the Sus scrofa chromosomes (SSC) SSC6 (25–43 Mb) and SSC14 (138–140 Mb) (Birney et al. 2004; Rohrer et al. 1996). In the syntenic regions on SSC6, one QTL for IMF and five additional fat-related QTL have been identified in pigs (de Koning et al. 2000; Liu et al. 2008; Paszek et al. 2001), while the small region on SSC14 is located in a region where two fat-related QTL have been found (Dragos-Wendrich et al. 2003; Knott et al. 1998). Therefore, it seems possible that genes in these syntenic regions have similar functions in both species. The presented mouse model, which is simplified in its genomic structure, could support the identification of genes that might affect IMF in pigs.
The second objective of this study was the identification of QTL for WHC. One QTL for this trait that was specific for the M. quadriceps was identified on Chr 2. The M. longissimus did not show differences in WHC between the parental lines. The particular fiber type composition of the two different muscles could be responsible for differences in WHC and IMF. Depending on the ratio of oxidative and glycogenic fiber types, the metabolism of the tissue can be different (Armstrong and Phelps 1984; Hamalainen and Pette 1993; Rehfeldt et al. 2010). Moreover, a correlation existed between glycogen and WHC, since higher glycogen content was also associated with higher drip loss. It is known that high glycogen storage impairs the WHC of the tissue. With postmortem hypoxia, glycogen degradation results in an increased accumulation of lactate, which among other factors lowers cellular pH values. The timing and extent of this lactate accumulation depend on the amount of stored glycogen. The concomitant pH drop might alter the structural integrity of the muscle fibers’ cellular scaffold and membrane proteins, which results in an increased loss of cell form and cell fluid (Bee et al. 2007; Choe et al. 2008). The parental BMMI816 line showed higher glycogen content and inferior WHC.
For the lactate content of the M. longissimus, a significant, positive correlation was found with the WHC in the M. longissimus in males, and with the pH 24 h after dissection in females. Since the M. longissimus samples, in which the glycogen and lactate contents were measured, were taken immediately after exsanguinations, it is possible that hypoxia did not last long enough to observe stronger correlations of lactate and glycogen with carcass pH. However, the results yet support the model of postmortem scaffold destruction and concomitant loss of tissue fluid due to increased tissue acidity that is influenced by the levels of muscular glycogen and lactate at the time of death. For the QTL for WHC on Chr 2 there was a syntenic region on SSC17 (13–37 Mb) in which no WHC-related QTL was found.
Comparative genomics and interspecies research are used to transfer the information obtained from our mouse study into the field of livestock research to which it is supposed to contribute. Using syntenic regions in the context of QTL studies can thereby provide additional information about the plausibility of identified QTL. However, it must be considered that the chance of finding syntenic regions between mouse and pig on a chromosome that contains any QTL is high, depending on the number of QTL and the size of the respective QTL intervals in both species. This is the case, e.g., for IMF and WHC (as drip loss), for which 15 of the 19 porcine chromosomes contain several significant QTL. Therefore, chances are relatively high to find a QTL for IMF on any of the pig chromosomes. However, for the murine IMF QTL on Chr 7 (17–31 Mb), only two syntenic regions exist, one on SSC6 (25–43 Mb) and one on SSC14 (138–140 Mb), but only the region on SSC6 contains a QTL for IMF. More than 50% of all murine genes are identified as having orthologs in pigs, which does not prove but indicate many putative similar gene functions (BioMart, Ensembl). Comparing mouse data to the results from pigs contributes additional information not only about orthology but also about the actual functions of conserved genes in both species.
Both mouse lines used in this study are hypermuscular and share large portions of their genomes. Nevertheless, they show large differences in their phenotypes. The increased body weights of the BMMI lines reflect the selection response during the historic breeding process. It appeared reasonable to examine these lines as models for livestock, since commercially used races of pigs or cattle are generally larger than their wild ancestors, as are the BMMI lines compared to wild-type mice. By generating a G3 population, we expected narrower QTL intervals but had to use a relatively more complicated analysis. Breeding additional generations will probably enable us to fine map these regions better. The QTL identified in the G3 in this study provide evidence to look for interesting alterations in the BMMI genome that explain the different phenotypes. Since the lines are related and have most haplotypes in common, we also expect fewer differences in the DNA sequence between the lines as compared to unrelated lines. This circumstance also could decrease the number of putative candidate genes in subsequent sequencing and expression studies.
Conclusion
The Animal Genome database (animalgenome.org) reports 54 and 37 significant QTL for IMF and WHC, respectively, but only a few candidate genes have been identified in pigs so far. Use of our inbred mouse lines in a controlled environment with the whole range of tools that are available for mice (with their long history of being genetic models) and their short generation interval might contribute to increasing the knowledge about livestock genetics for the examined traits. However, the success of cloning QTL in mice was quite limited in the past. The use of outbred populations seems to be more effective for identifying causative mutations (Brockmann and Bevova 2002; Valdar et al. 2003). Using advanced intercross lines (AIL) seems to be beneficial in this regard compared to the classic F2 approach (Darvasi and Soller 1995). The AIL approach leads to more recombination events and smaller confidence intervals, while the close relatedness between the inbred parental lines initially decreases the number of regions that probably harbor phenotype-affecting alterations in the genome. We took this into account by using a G3 instead of an F2 population. Analyses of RNA expression and sequencing based on the identified QTL region should allow the identification of the causative mutation(s) for the examined traits in the near future.
To our knowledge this is the first report on QTL mapping for IMF, WHC, and associated traits in mice. The localization of different genomic regions affecting IMF and WHC yielded information for fine mapping these regions and identifying putative candidate genes. The identified loci partly coincide with QTL for these traits in other species, especially pig. Therefore, the BMMI model is an interesting genetic resource not only to identify genes, which could also contribute to QTL effects in other species, but also to test identified candidate genes for their functions in genetically modified mice.
Acknowledgments
This project was supported by the Deutsche Forschungsgemeinschaft (Project BR1285/8-1) and National Institutes of Health grant R21DA024845. We thank Gary Churchill for the support of the Center for Genome Dynamics for high density SNP genotyping at The Jackson Laboratory. Furthermore, we thank Carsten Berndt, Alexandra Weyrich, Melanie Riedel, and Annett Kannegiesser for technical assistance.
Contributor Information
Stefan Kärst, Email: stefan.kaerst@agrar.hu-berlin.de, Department for Crop and Animal Sciences, Humboldt-Universität zu Berlin, Invalidenstraße 42, 10115 Berlin, Germany.
Riyan Cheng, Department of Human Genetics, University of Chicago, 920 East 58th Street, Chicago, IL 60637, USA.
Armin O. Schmitt, Department for Crop and Animal Sciences, Humboldt-Universität zu Berlin, Invalidenstraße 42, 10115 Berlin, Germany. Faculty of Science and Technology, Universitätsplatz 5—piazza Universitä, 539100 Bozen-Bolzano, Italy
Hyuna Yang, Department of Genetics, School of Medicine, University of North Carolina, 120 Mason Farm Road, Chapel Hill, NC 27599-7264, USA.
Fernando Pardo Manuel de Villena, Department of Genetics, School of Medicine, University of North Carolina, 120 Mason Farm Road, Chapel Hill, NC 27599-7264, USA.
Abraham A. Palmer, Department of Human Genetics, University of Chicago, 920 East 58th Street, Chicago, IL 60637, USA
Gudrun A. Brockmann, Department for Crop and Animal Sciences, Humboldt-Universität zu Berlin, Invalidenstraße 42, 10115 Berlin, Germany
References
- Armstrong RB, Phelps RO. Muscle fiber type composition of the rat hindlimb. Am J Anat. 1984;171:259–272. doi: 10.1002/aja.1001710303. [DOI] [PubMed] [Google Scholar]
- Barham D, Trinder P. An improved colour reagent for the determination of blood glucose by the oxidase system. Analyst. 1972;97:142–145. doi: 10.1039/an9729700142. [DOI] [PubMed] [Google Scholar]
- Bee G, Anderson AL, Lonergan SM, Huff-Lonergan E. Rate and extent of pH decline affect proteolysis of cytoskeletal proteins and water-holding capacity in pork. Meat Sci. 2007;76:359–365. doi: 10.1016/j.meatsci.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T, Down T, Eyras E, Fernandez-Suarez XM, Gane P, Gibbins B, Gilbert J, Hammond M, Hotz HR, Iyer V, Jekosch K, Kahari A, Kasprzyk A, Keefe D, Keenan S, Lehvaslaiho H, McVicker G, Melsopp C, Meidl P, Mongin E, Pettett R, Potter S, Proctor G, Rae M, Searle S, Slater G, Smedley D, Smith J, Spooner W, Stabenau A, Stalker J, Storey R, Ureta-Vidal A, Woodwark KC, Cameron G, Durbin R, Cox A, Hubbard T, Clamp M. An overview of Ensembl. Genome Res. 2004;14:925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann GA, Bevova MR. Using mouse models to dissect the genetics of obesity. Trends Genet. 2002;18:367–376. doi: 10.1016/s0168-9525(02)02703-8. [DOI] [PubMed] [Google Scholar]
- Brockmann GA, Karatayli E, Haley CS, Renne U, Rottmann OJ, Karle S. QTLs for pre- and postweaning body weight and body composition in selected mice. Mamm Genome. 2004;15:593–609. doi: 10.1007/s00335-004-3026-4. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Cannata S, Engle TE, Moeller SJ, Zerby HN, Radunz AE, Green MD, Bass PD, Belk KE. Effect of visual marbling on sensory properties and quality traits of pork loin. Meat Sci. 2010;85(3):428–434. doi: 10.1016/j.meatsci.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Chadt A, Leicht K, Deshmukh A, Jiang LQ, Scherneck S, Bernhardt U, Dreja T, Vogel H, Schmolz K, Kluge R, Zierath JR, Hultschig C, Hoeben RC, Schurmann A, Joost HG, Al-Hasani H. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat Genet. 2008;40:1354–1359. doi: 10.1038/ng.244. [DOI] [PubMed] [Google Scholar]
- Cheng R, Lim JE, Samocha KE, Sokoloff G, Abney M, Skol AD, Palmer AA. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics. 2010;185:1033–1044. doi: 10.1534/genetics.110.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe JH, Choi YM, Lee SH, Shin HG, Ryu YC, Hong KC, Kim BC. The relation between glycogen, lactate content and muscle fiber type composition, and their influence on postmortem glycolytic rate and pork quality. Meat Sci. 2008;80:355–362. doi: 10.1016/j.meatsci.2007.12.019. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE, Bult C, Paigen B, Flint J, Tsaih SW, Churchill GA, Broman KW. A new standard genetic map for the laboratory mouse. Genetics. 2009;182:1335–1344. doi: 10.1534/genetics.109.105486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141:1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning DJ, Janss LL, Rattink AP, van Oers PA, de Vries BJ, Groenen MA, van der Poel JJ, de Groot PN, Brascamp EW, van Arendonk JA. Detection of quantitative trait loci for backfat thickness and intramuscular fat content in pigs (Sus scrofa) Genetics. 1999;152:1679–1690. doi: 10.1093/genetics/152.4.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning DJ, Rattink AP, Harlizius B, van Arendonk JA, Brascamp EW, Groenen MA. Genome-wide scan for body composition in pigs reveals important role of imprinting. Proc Natl Acad Sci USA. 2000;97:7947–7950. doi: 10.1073/pnas.140216397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragos-Wendrich M, Sternstein I, Brunsch C, Moser G, Bartensch-lager H, Reiner G, Geldermann H. Linkage and QTL mapping for Sus scrofa chromosome 14. J Anim Breed Genet. 2003;120:111–118. [Google Scholar]
- Eaton S. Control of mitochondrial [beta]-oxidation flux. Progr Lipid Res. 2002;41:197–239. doi: 10.1016/s0163-7827(01)00024-8. [DOI] [PubMed] [Google Scholar]
- Ebeling P, Essen-Gustavsson B, Tuominen JA, Koivisto VA. Intramuscular triglyceride content is increased in IDDM. Dia-betologia. 1998;41:111–115. doi: 10.1007/s001250050875. [DOI] [PubMed] [Google Scholar]
- Fernandez X, Monin G, Talmant A, Mourot J, Lebret B. Influence of intramuscular fat content on the quality of pig meat-1. Composition of the lipid fraction and sensory characteristics of m. longissimus lumborum. Meat Sci. 1999;53:59–65. doi: 10.1016/s0309-1740(99)00037-6. [DOI] [PubMed] [Google Scholar]
- Fujii J, Otsu K, Zorzato F, de Leon S, Khanna VK, Weiler JE, O’Brien PJ, MacLennan DH. Identification of a mutation in porcine ryanodine receptor associated with malignant hyperthermia. Science. 1991;253:448–451. doi: 10.1126/science.1862346. [DOI] [PubMed] [Google Scholar]
- Hamalainen N, Pette D. The histochemical profiles of fast fiber types IIB, IID, and IIA in skeletal muscles of mouse, rat, and rabbit. J Histochem Cytochem. 1993;41:733–743. doi: 10.1177/41.5.8468455. [DOI] [PubMed] [Google Scholar]
- Hansson O, Donsmark M, Ling C, Nevsten P, Danfelter M, Andersen JL, Galbo H, Holm C. Transcriptome and proteome analysis of soleus muscle of hormone-sensitive lipase-null mice. J Lipid Res. 2005;46:2614–2623. doi: 10.1194/jlr.M500028-JLR200. [DOI] [PubMed] [Google Scholar]
- Hunt MC, Rautanen A, Westin MA, Svensson LT, Alexson SE. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. FASEB J. 2006;20:1855–1864. doi: 10.1096/fj.06-6042com. [DOI] [PubMed] [Google Scholar]
- Kaerst S, Schmitt A, Brockmann G. A novel method for measuring of fat content in low-weight tissue: a NMR study. WebmedCentral OBESITY 2010. 2010;1(12):WMC001368. http://www.webmedcentral.com/article_view/1368.
- Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- Knott SA, Marklund L, Haley CS, Andersson K, Davies W, Ellegren H, Fredholm M, Hansson I, Hoyheim B, Lundstrom K, Moller M, Andersson L. Multiple marker mapping of quantitative trait loci in a cross between outbred wild boar and large white pigs. Genetics. 1998;149:1069–1080. doi: 10.1093/genetics/149.2.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambe NR, Macfarlane JM, Richardson RI, Matika O, Haresign W, Bünger L. The effect of the Texel muscling QTL (TM-QTL) on meat quality traits in crossbred lambs. Meat Sci. 2010;85:684–690. doi: 10.1016/j.meatsci.2010.03.025. [DOI] [PubMed] [Google Scholar]
- Leavens KF, Easton RM, Shulman GI, Previs SF, Birnbaum MJ. Akt2 is required for hepatic lipid accumulation in models of insulin resistance. Cell Metab. 2009;10:405–418. doi: 10.1016/j.cmet.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionikas A, Blizard DA, Gerhard GS, Vandenbergh DJ, Stout JT, Vogler GP, McClearn GE, Larsson L. Genetic determinants of weight of fast- and slow-twitch skeletal muscle in 500-day-old mice of the C57BL/6J and DBA/2J lineage. Physiol Genom. 2005;21:184–192. doi: 10.1152/physiolgenomics.00209.2004. [DOI] [PubMed] [Google Scholar]
- Lionikas A, Blizard D, Vandenbergh D, Stout J, Vogler G, McClearn G, Larsson L. Genetic determinants of weight of fast- and slow-twitch skeletal muscles in old mice. Mamm Genome. 2006;17:615–628. doi: 10.1007/s00335-005-0177-x. [DOI] [PubMed] [Google Scholar]
- Liu G, Kim JJ, Jonas E, Wimmers K, Ponsuksili S, Murani E, Phatsara C, Tholen E, Juengst H, Tesfaye D, Chen JL, Schellander K. Combined line-cross and half-sib QTL analysis in Duroc-Pietrain population. Mamm Genome. 2008;19:429–438. doi: 10.1007/s00335-008-9132-y. [DOI] [PubMed] [Google Scholar]
- Ludden PA, Kucuk O, Rule DC, Hess BW. Growth and carcass fatty acid composition of beef steers fed soybean oil for increasing duration before slaughter. Meat Sci. 2009;82:185–192. doi: 10.1016/j.meatsci.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Malek M, Dekkers JC, Lee HK, Baas TJ, Prusa K, Huff-Lonergan E, Rothschild MF. A molecular genome scan analysis to identify chromosomal regions influencing economic traits in the pig. II. Meat and muscle composition. Mamm Genome. 2001;12:637–645. doi: 10.1007/s003350020019. [DOI] [PubMed] [Google Scholar]
- Mott R. Perl script “ril.pl”. 2005 http://mus.well.ox.ac.uk/mouse/INBREDS/RIL/index.shtml.
- Neuschl C, Hantschel C, Wagener A, Schmitt AO, Illig T, Brockmann GA. A unique genetic defect on chromosome 3 is responsible for juvenile obesity in the Berlin Fat Mouse. Int J Obes (Lond) 2010;34(12):1706–1714. doi: 10.1038/ijo.2010.97. [DOI] [PubMed] [Google Scholar]
- Ochoa O, Shireman PK, McManus LM. Altered inflammation increases intramuscular fat accumulation and impairs skeletal muscle regeneration following ischemic injury in CCR2−/−mice. J Am Coll Surg. 2006;203:S101. doi: 10.1152/ajpcell.00154.2006. [DOI] [PubMed] [Google Scholar]
- Pan DA, Lillioja S, Kriketos AD, Milner MR, Baur LA, Bogardus C, Jenkins AB, Storlien LH. Skeletal muscle triglyceride levels are inversely related to insulin action. Diabetes. 1997;46:983–988. doi: 10.2337/diab.46.6.983. [DOI] [PubMed] [Google Scholar]
- Paszek AA, Wilkie PJ, Flickinger GH, Miller LM, Louis CF, Rohrer GA, Alexander LJ, Beattie CW, Schook LB. Interval mapping of carcass and meat quality traits in a divergent swine cross. Anim Biotechnol. 2001;12:155–165. doi: 10.1081/ABIO-100108342. [DOI] [PubMed] [Google Scholar]
- Phillips DIW, Caddy S, Ilic V, Fielding BA, Frayn KN, Borthwick AC, Taylor R. Intramuscular triglyceride and muscle insulin sensitivity: evidence for a relationship in nondiabetic subjects. Metabolism. 1996;45:947–950. doi: 10.1016/s0026-0495(96)90260-7. [DOI] [PubMed] [Google Scholar]
- Ponsuksili S, Jonas E, Murani E, Phatsara C, Srikanchai T, Walz C, Schwerin M, Schellander K, Wimmers K. Trait correlated expression combined with expression QTL analysis reveals biological pathways and candidate genes affecting water holding capacity of muscle. BMC Genom. 2008;9:367. doi: 10.1186/1471-2164-9-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DJ, Turban S, Gray A, Hajduch E, Hundal HS. Intracellular ceramide synthesis and protein kinase Czeta activation play an essential role in palmitate-induced insulin resistance in rat L6 skeletal muscle cells. Biochem J. 2004;382:619–629. doi: 10.1042/BJ20040139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeldt C, Renne U, Sawitzky M, Binder G, Hoeflich A. Increased fat mass, decreased myofiber size, and a shift to glycolytic muscle metabolism in adolescent male transgenic mice overexpressing IGFBP-2. Am J Physiol Endocrinol Metab. 2010;299:E287–E298. doi: 10.1152/ajpendo.00492.2009. [DOI] [PubMed] [Google Scholar]
- Rohrer GA, Alexander LJ, Hu Z, Smith TP, Keele JW, Beattie CW. A comprehensive map of the porcine genome. Genome Res. 1996;6:371–391. doi: 10.1101/gr.6.5.371. [DOI] [PubMed] [Google Scholar]
- Schmitt AO, Al-Hasani H, Cheverud JM, Pomp D, Bunger L, Brockmann GA. Fine mapping of mouse QTLs for fatness using SNP data. OMICS. 2007;11:341–350. doi: 10.1089/omi.2007.0015. [DOI] [PubMed] [Google Scholar]
- Schmitt A, Bortfeldt R, Neuschl C, Brockmann G. RandoMate: a program for the generation of random mating schemes for small laboratory animals. Mamm Genome. 2009;20:321–325. doi: 10.1007/s00335-009-9185-6. [DOI] [PubMed] [Google Scholar]
- Schmitz-Peiffer C. Signalling aspects of insulin resistance in skeletal muscle: mechanisms induced by lipid oversupply. Cell Signal. 2000;12:583–594. doi: 10.1016/s0898-6568(00)00110-8. [DOI] [PubMed] [Google Scholar]
- Schwab CR, Mote BE, Du ZQ, Amoako R, Baas TJ, Rothschild MF. An evaluation of four candidate genes for use in selection programmes aimed at increased intramuscular fat in Duroc swine. J Anim Breed Genet. 2009;126:228–236. doi: 10.1111/j.1439-0388.2008.00770.x. [DOI] [PubMed] [Google Scholar]
- Szabo G, Dallmann G, Muller G, Patthy L, Soller M, Varga L. A deletion in the myostatin gene causes the compact (Cmpt) hypermuscular mutation in mice. Mamm Genome. 1998;9:671–672. doi: 10.1007/s003359900843. [DOI] [PubMed] [Google Scholar]
- Tanomura H, Miyake T, Taniguchi Y, Manabe N, Kose H, Matsumoto K, Yamada T, Sasaki Y. Detection of a quantitative trait locus for intramuscular fat accumulation using the OLETF rat. J Vet Med Sci. 2002;64:45–50. doi: 10.1292/jvms.64.45. [DOI] [PubMed] [Google Scholar]
- Tinsley FC, Taicher GZ, Heiman ML. Evaluation of a quantitative magnetic resonance method for mouse whole body composition analysis. Obesity. 2004;12:150–160. doi: 10.1038/oby.2004.20. [DOI] [PubMed] [Google Scholar]
- Tyra M, Ropka-Molik K, Eckert R, Piórkowska K, Oczkowicz M. H-FABP and LEPR gene expression profile in skeletal muscles and liver during ontogenesis in various breeds of pigs. Domest Anim Endocrinol. 2010;40(3):147–154. doi: 10.1016/j.domaniend.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Underwood KR, Tong J, Zhu MJ, Shen QW, Means WJ, Ford SP, Paisley SI, Hess BW, Du M. Relationship between kinase phosphorylation, muscle fiber typing, and glycogen accumulation in longissimus muscle of beef cattle with high and low intramuscular fat. J Agric Food Chem. 2007;55:9698–9703. doi: 10.1021/jf071573z. [DOI] [PubMed] [Google Scholar]
- Valdar WJ, Flint J, Mott R. QTL fine-mapping with recombinant-inbred heterogeneous stocks and in vitro heterogeneous stocks. Mamm Genome. 2003;14:830–838. doi: 10.1007/s00335-003-3021-1. [DOI] [PubMed] [Google Scholar]
- Varga L, Szabo G, Darvasi A, Muller G, Sass M, Soller M. Inheritance and mapping of Compact (Cmpt), a new mutation causing hypermuscularity in mice. Genetics. 1997;147:755–764. doi: 10.1093/genetics/147.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers K, Murani E, Ponsuksili S. Functional genomics and genetical genomics approaches towards elucidating networks of genes affecting meat performance in pigs. Brief Funct Genom. 2010;9:251–258. doi: 10.1093/bfgp/elq003. [DOI] [PubMed] [Google Scholar]
- Wood JD, Richardson RI, Nute GR, Fisher AV, Campo MM, Kasapidou E, Sheard PR, Enser M. Effects of fatty acids on meat quality: a review. Meat Sci. 2004;66:21–32. doi: 10.1016/S0309-1740(03)00022-6. [DOI] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, de Villena FP, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6:663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaspelkis BB, Singh MK, Krisan AD, Collins DE, Kwong CC, Bernard JR, Crain AM. Chronic leptin treatment enhances insulin-stimulated glucose disposal in skeletal muscle of high-fat fed rodents. Life Sci. 2004;74:1801–1816. doi: 10.1016/j.lfs.2003.08.037. [DOI] [PubMed] [Google Scholar]
- Yu C, Chen Y, Cline GW, Zhang D, Zong H, Wang Y, Bergeron R, Kim JK, Cushman SW, Cooney GJ, Atcheson B, White MF, Kraegen EW, Shulman GI. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- Zhang WG, Lonergan SM, Gardner MA, Huff-Lonergan E. Contribution of postmortem changes of integrin, desmin and [mu]-calpain to variation in water holding capacity of pork. Meat Sci. 2006;74:578–585. doi: 10.1016/j.meatsci.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Zhang D, Liu ZX, Choi CS, Tian L, Kibbey R, Dong J, Cline GW, Wood PA, Shulman GI. Mitochondrial dysfunction due to long-chain Acyl-CoA dehydrogenase deficiency causes hepatic steatosis and hepatic insulin resistance. Proc Natl Acad Sci USA. 2007;104:17075–17080. doi: 10.1073/pnas.0707060104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SM, Ren LJ, Guo L, Cheng ML, Zhang X, Ge CR, Gao SZ. Muscle lipid metabolism gene expression in pigs with different H-FABP genotypes. Livestock Sci. 2010;128:101–107. [Google Scholar]