Abstract
Infectious fungi are among a broad group of microbial pathogens that has and continues to emerge concomitantly due to the global AIDS pandemic as well as an overall increase of patients with compromised immune systems. In addition, many pathogens have been emerging and reemerging, causing disease in both individuals who have an identifiable immune defect and those who do not. The fungal pathogen Cryptococcus gattii can infect individuals with and without an identifiable immune defect, with a broad geographic range including both endemic areas and emerging outbreak regions. Infections in patients and animals can be severe and often fatal if untreated. We review the molecular epidemiology, population structure, clinical manifestations, and ecological niche of this emerging pathogen.
Keywords: Fungal infections, Cryptococcosis, Emerging outbreak, Pulmonary disease, Meningitis
1. Introduction
Infectious diseases cause a significant burden on human and animal health, plants and agriculture, and the global economy [1–3]. Among the microbial pathogens causing infectious diseases worldwide, fungi represent one major class. Advances in the healthcare of patients with noninfectious diseases coupled with the rise of HIV/AIDS globally, and continued outbreaks of fungal pathogens in otherwise healthy hosts, have led to an overall increase of fungi as etiologic agents producing human disease. Each of the four major fungal phyla has representatives that cause serious disease in both humans and a vast range of other animals. Although less prevalent than plant fungal pathogens, the animal fungal pathogens pose serious threats to entire animal populations (e.g., Batrachochytrium dendrobatidis in amphibians [4], Geomyces destructans in bats [5], and Nosema ceranae and/or Nosema-iridovirus co-infections in honey bees [6–9]) and continue to cause serious morbidity and mortality among immunocompromised humans and even individuals with no apparent immune defect worldwide.
In many cases, the incidence of fungal disease is increasing due to the rise in susceptible hosts. Furthermore, limited fungicidal treatment options exacerbate both morbidity and mortality. A major factor influencing the lack of effective and safe treatment is that, unlike bacteria and viruses, the fungi are eukaryotic siblings to the animals and share several conserved signaling cascades. These issues cause major obstacles in the search and development of new antimicrobials that target fungi without causing major toxic side effects in the host. Several fungal species frequently cause infections in immunocompromised human hosts (e.g., Candida albicans, Aspergillus fumigatus), while others are known to cause disease in otherwise healthy individuals (e.g., Coccidioides immitis, Coccidioides posadasii, Histoplasma capsulatum). The pathogenic Cryptococcus species complex, consisting of C. neoformans and C. gattii, causes disease in both of these populations [10,11]. This is particularly evident with C. gattii, which is endemic in many tropical and sub-tropical regions and has also been associated with outbreaks in humans and a wide range of mammals, making it of global public health interest [10–12].
C. gattii is a basidiomycetous yeast and is estimated to have diverged from C. neoformans ~37.5 million years ago [13]. In aggregate, pathogenic Cryptococcus species produce disease in almost one million individuals annually with over 620,000 attributable mortalities and account for ~1/3 of all HIV/AIDS associated deaths, surpassing tuberculosis mortality in Africa [14]. C. gattii has a distinct epidemiology from its sibling species C. neoformans, which more commonly causes disease in immunosuppressed hosts. Within the species, C. gattii can be further subdivided into two serotypes (B and C) [15], and four molecular types designated VGI, VGII, VGIII, and VGIV [16]. Based on phylogenetic analyses, each VG group appears to show no inter-molecular type nuclear genetic exchange, indicating that the four molecular types are cryptic species, which could be assigned their own species designations [17–23]. Molecular type discrimination is of principal importance clinically and epidemiologically, as types VGI and VGII have been associated with the majority of cases from otherwise healthy hosts with outbreaks in the North American Pacific Northwest, in the Northern Territory of Australia in Aboriginals, and in the central province of Papua New Guinea. On the other hand, molecular types VGIII and VGIV appear to more commonly produce disease in immunocompromised patients including those with HIV/AIDS with documented case series in Africa and the United States. In fact, molecular types VGIII and VGIV appear to be similar to the epidemiological profiles observed for the sibling species, C. neoformans (Table 1) [10,11,14,24–28].
Table 1.
Descriptions of the four C. gattii molecular types.
| Molecular type |
Clinical features |
Environmental features | Distribution |
|---|---|---|---|
| VGI | Most common molecular type in humans and animals and highly clonal | Commonly associated with Eucalyptus trees, particularly in Australia [17,58] | Global with high distribution in Australia |
| VGII | Responsible for Pacific Northwest outbreak, clonal in outbreak region but diverse globally, highly virulent genotypes (VGIIa, VGIIc) identified | Associated with native tree species, with common isolation from Douglas-fir and Alder trees in British Columbia [59] | Global, also the cause of the first outbreak in a temperate climate on Vancouver Island |
| VGIII | Frequently associated with infection of HIV/AIDS patients | Isolates are highly fertile, and have been found in Corymbia ficifolia (Red Flowering Gum, Colombia) [124] and Eucalyptus (California) [22] | Global, high levels observed in Southern California, Mexico, and South America |
| VGIV | Frequently associated with infection of HIV/AIDS patients | Largely unknown, but one positive isolate from an Almond tree [43,125,126] | Rare, reported in Africa, India, and South America |
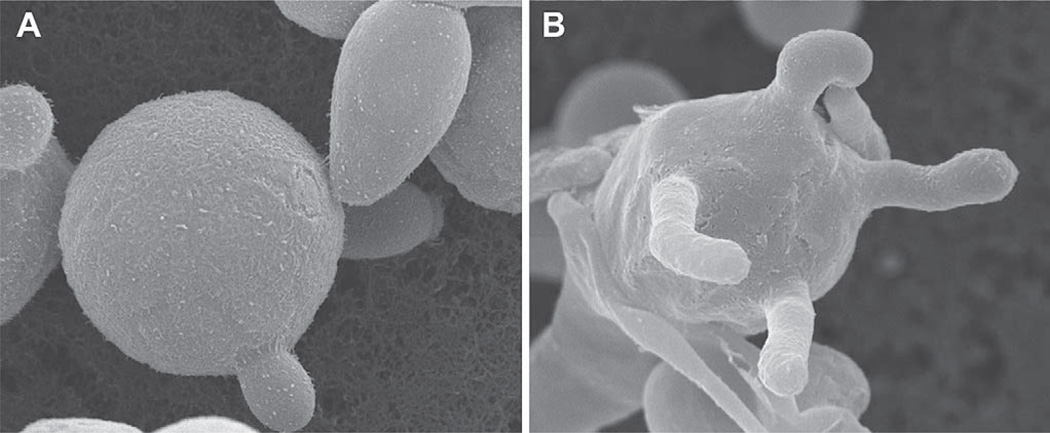
Overall, recent epidemiological studies of C. gattii suggest that this species is an emerging fungal pathogen with an expanding geographic range and environmental niche. In nature, C. gattii reproduces asexually as a budding yeast (Fig. 1A) and sexually to produce infectious spores (Fig. 1B); it is likely that the inhalation of both desiccated yeast cells and spores cause infections. While infections remain relatively rare, the level of mortality, percentage of patients with prolonged hospital visits, lengthy treatment regimens, and long-term sequelae remain common among infected patients [11,29,30]. Furthermore, this fungal disease can occur in a range of domestic, agricultural, wild terrestrial, and marine mammals [20,23,31–40]. To better understand the dynamics of these infections from a global perspective, a multidisciplinary approach is necessary. For these reasons, aspects of molecular biology, ecology, molecular epidemiology, traditional epidemiology, clinical practices, veterinary treatments, and therapeutics must be bridged together with each respective specialty aware of the basic tenets and principles of the current research in each respective field. In this review, sections are divided into specific areas of focus, with the aim of broadly summarizing important findings and future directions for this emerging/re-emerging infectious disease. Additionally, one section exclusively covers key aspects of the North American Pacific Northwest outbreak, as it represents the largest primary outbreak of C. gattii reported, and has emerged in an environmental climate that had not been previously associated with this disease and supports new features to the epidemiology of this encapsulated yeast [17,19,20,23,24,29,32,41].
Fig. 1.
Morphology of C. gattii. Scanning electron microscopy illustrates C. gattii asexually producing as a budding yeast (A) and sexually reproducing to form a basidium with four emerging basidiospores (B).
2. Molecular typing methods
As previously mentioned, C. gattii can be divided into four discrete lineages (i.e., molecular types) termed VGI–VGIV [17,18,42–44]. This is significant as each molecular type has distinct characteristics. Molecular types VGI and VGII are the two types most frequently associated with illness in otherwise healthy individuals [24]. Infections due to VGI have been reported to occur at high rates among certain populations in Australia such as Aboriginal groups but there have also been numerous cases elsewhere globally. In contrast, the numbers of VGII infection are high in the Pacific Northwest, in which over 95% of all cases are attributable to this molecular type [11,17,18,24,45]. In comparison, molecular types VGIII and VGIV have largely been associated with illness in HIV/AIDS patients, which is similar to the epidemiological profile observed for C. neoformans [10,11,14,24,25]. Molecular type VGIII has been isolated from a number of regions worldwide [17,18], and was found to be the predominant molecular type (>93%) in a cohort of HIV/AIDS patients infected with C. gattii from Southern California [46] (Byrnes et al., PLoS Pathogens, in revision). Molecular type VGIV appears to be rare in the global population but has been reported among HIV/AIDS patients in sub-Saharan Africa [25]. Overall, these distinctions are significant as there are clear associations with host preferences and some geographical restriction of disease.
The discrimination of molecular types is useful, but in some cases too limited for one to gain a more accurate understanding of the molecular epidemiology in any given region. To fully appreciate and track outbreaks and disparate cases in regions, a more rigorous molecular examination of isolates is necessary. This type of comprehensive analysis often reveals specific genotypes that are associated with clinical and veterinary cases. Several genotypes have been identified through molecular approaches including Amplified Fragment Length Polymorphism analysis (AFLP), Random Amplification of Polymorphic DNA (RAPD), and Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) analysis [16,41]. These molecular approaches are a rapid way to identify both the molecular type and common genotypes, but with the increasing discriminatory power of direct DNA sequence analysis and the continued reduction in costs, sequence analysis for molecular epidemiology is becoming increasingly applied to the analysis of C. neoformans and C. gattii strains [17,20,23,47,48].
The most widely utilized sequence-based typing method for the analysis of C. gattii molecular epidemiology is multilocus sequence typing (MLST, i.e., molecular bar-coding) [49]. This method is robust as it is portable between labs and allows for a simple deposition of sequence alleles into public sequence-repository databases. Most often, discriminatory sequences based on unlinked genomic loci are employed for the analysis of isolates [17,47,48].MLST revealed that the four molecular types exhibit no examples of nuclear allelic exchange in >300 global isolates, supporting the hypothesis that they are cryptic species [17,20]. Furthermore, MLST analysis of a large number of global isolates showed genotypic diversity within each molecular type, often associated with specific geographic regions, such as a high prevalence of VGI in Australia and common VGIV genotypes restricted to sub-Saharan Africa [17,20,25].
One important application of molecular epidemiology was applied with the emergence of the Vancouver Island outbreak in 1999 and its subsequent expansion throughout the North American Pacific Northwest. First AFLP, then MLST, confirmed that there are two genotypes largely responsible for the outbreak, VGIIa/major and VGIIb/minor [22,41]. Subsequent studies on the outbreak were then conducted to confirm these genotypes in the region and also to document that the outbreak had expanded into the United States [17,31,50]. Additionally, sequence-based analysis revealed that a third outbreak genotype, unique to Oregon (VGIIc/novel), is also contributing to this ongoing outbreak [19,20,32]. The use of sequence typing and its role in understanding the Pacific Northwest outbreak will be detailed further in the section focused on this outbreak.
MLST analysis is also useful in identifying fungal infection cases as a result of travel. A significant finding of the Pacific Northwest outbreak has been that the VGIIa/major genotype has yet to be found elsewhere globally in the environment or clinically. This observation was then useful when cases from patients with a travel history to the endemic Vancouver Island region began to appear with travel-associated VGIIa/major genotype infections now documented in patients from Alberta (Canada), Denmark, the Netherlands, and Switzerland [51–53]. There has also recently been a report of a patient in Japan infected with the VGIIa/major genotype (identical at 11/11 MLST markers examined), but further analysis will be necessary to determine if this is a true non travel-associated VGIIa/major infection or a related but distinguishable genotype as is known to occur elsewhere globally [20,54]. Unrelated to the outbreak was also the first reported case of C. gattii infection from the Southeastern United States [21]. In this case, the isolate was determined to be molecular type VGI (and distinct from VGII and rare VGI isolates in the Pacific Northwest) based on MLST analysis. This patient had traveled to central California, in which the VGI molecular type is known to be endemic.
While the analysis of coding regions is highly useful for discriminating genotypes of pathogenic fungi, in some cases molecular epidemiology aspects cannot be resolved in these highly clonal populations. This is especially evident from studies of the VGI and VGII molecular types of C. gattii, in which isolates are often indistinguishable based on MLST analysis alone. To increase the resolution of typing, several approaches have been employed to generate microsatellite-based markers, including Variable Number of Tandem Repeat (VNTR) markers. The initial study to employ this approach in C. gattii was focused on largely clonal C. gattii isolates from Australia and the United States that were VGI molecular type [21]. In this case, MLST analysis of an isolate from a North Carolina patient provided initial evidence that it was indistinguishable from clinical isolates from Australia and California and environmental isolates from Australia; however, after the addition of microsatellite markers, the environmental isolates were distinguishable from other clinical isolates based on both size variation in microsatellite PCR products and sequence differences in the VNTR markers [21].
The large clonal outbreak of C. gattii continues in the North American Pacific Northwest. As the isolates from the three outbreak genotypes (VGIIa/major, VGIIb/minor, VGIIc/novel) are highly clonal, the sequence-based VNTR approach was applied so that a combination of repeat variations and single nucleotide substitutions, insertions, or deletions could be assessed [20]. Overall, this approach demonstrated that there was some underlying diversity in the VGIIa/major genotype, particularly among three isolates from Oregon and several associated isolates from California that are closely related but not indistinguishable from the VGIIa major genotype based on SNPs in variable regions of the genome. Several of the distinguishing SNPs are related to similar alleles seen elsewhere globally in divergent VGII strains. However, these differences occur in non-coding regions and are therefore likely related due to homoplasy (i.e., similarity in isolates of different ancestry and therefore of independent origin and not identical by descent) and not shared ancestry. The results also showed that the groups of VGIIb/minor and VGIIc isolates examined were indistinguishable from each other (all VGIIc originated from Oregon), although a report of a VGIIc isolate from a patient in Idaho with travel history to Oregon showed a single base pair difference at an MLST locus, indicating a likely recent divergence/drift due to expansion of this genotype [32]. Overall, the application of the VNTR sequence-based method proved highly discriminatory to differentiate C. gattii isolates that otherwise appeared indistinguishable at the previous loci examined. These studies also aided in expanding our knowledge of the overall structure of global C. gattii genotypes.
3. Global overview of clinical and environmental populations and insights into the origins of the pathogenic Cryptococcus species complex
3.1. Global overview of clinical and environmental populations
Molecular typing is a method often utilized to study pathogenic microbes for purposes of outbreak analyses. In addition, the analyses of conserved non-repetitive sequences are the fundamental basis for population studies. Unlike SNPs, repetitive sequences are often subject to homoplasy and thus avoided, although these markers can be of use for non-phylogenetic clustering analysis, which is often insightful for studies of global population structures (see Section 2). For these reasons, studies often include only unlinked conserved unique sequences (predominantly coding) for phylogenetic approaches and population genetic analysis and in some cases combine non-coding variable markers for large clustering approaches. In this section we review approaches that have been employed to analyze the population genetics of C. gattii, including approaches to identify evidence for same- and opposite-sex mating, global and regional population studies, and overall trends in global populations, both environmentally and clinically.
While C. gattii is often studied due to its importance as a mammalian pathogen, its common ecological niche is in the environment. For this reason, and to also understand the roles of mating and recombination (both of which can lead to novel and pathogenic genotypes), studies of naturally occurring isolates and their population structures are of fundamental importance. Several landmark studies of C. gattii in the environment have been conducted in Australia. Studies focused on environmental, clinical, and veterinary isolates have been approached from a population perspective. Studies during 2003 of Eucalyptus tree isolates yielded results consistent with only clonality and no signs of sexual recombination [55]. However, more recent studies analyzing populations collected from individual Eucalyptus trees showed evidence for both same- and opposite-sex mating [56]. Taken together, this suggests that while clonal reproduction may be a common means of reproduction for this fungus, both same- and opposite-sex mating are occurring in the environment, perhaps in highly restricted local environments that in some cases may even be as restricted as a single colonized tree. Furthermore, studies analyzing the population structures of clinical and veterinary isolates in Australia found evidence for clonality in some populations, and a combination of clonality and sexual reproduction in others [57]. Supporting studies showed that these isolates retain their ability to sexually reproduce in laboratory-based mating assays [58]. Population-based studies in Australia provide a foundation for studies in other regions, which also often yielded results suggesting both clonality and recombination in local and global based analyses, with further additional evidence for laboratory-based mating supporting these results [17,20,22,45,59]. Studies examining both small and geographically dispersed isolates from the environment and infected hosts are fundamental to elucidating population structures, speciation dynamics, and evidence for same- and opposite-sex mating in naturally occurring populations.
3.2. Insights into the origins of the pathogenic Cryptococcus species complex
In addition to population studies, comparative genetic and ecological studies of both the pathogenic species of focus and related non-pathogenic species can yield insights into pathogen emergence. The origin and evolution of pathogens remain central questions in studies of both plant and animal diseases. One method to examine the likely origins of pathogens is to phylogenetically place the species into the context of closely related relatives, which in the case of C. neoformans and C. gattii are saprobic. The closest related species to the pathogenic Cryptococcus species are associated with insects or insect frass. Although these sibling species are often less studied than their medically relevant counterparts, they can offer significant insights into the evolution of the animal pathogens and how these pathogenic species might have arisen from insect-associated saprophytes.
Phylogenetic analyses indicate that the Cryptococcus species complex likely arose from the Tremella lineage, clustered closely with the Tremellales, Trichosporonales, Filobasidiales, and the Cystofilobasidiales [60–62]. Several of the species within these lineages are saprophytes that are commonly associated with insect debris, leading to the hypothesis that the pathogens emerged from an association within this environmental niche [63]. In support of this hypothesis, C. gattii has been isolated from both insect frass and wasp nests and C. neoformans has been isolated from honeybee hives, indicating that these animal pathogens may still in some cases act as an insect-associated saprophyte or pathogen in the environment [61,64,65]. While the evolutionary factors influencing the manifestations of mammalian pathogens from saprobes are unclear, the support for the hypothesis of emergence based on phylogenetic and ecological studies provides insights into the evolutionary history of the pathogenic Cryptococcus species complex.
In addition to the ecologic studies mentioned above, interactions between Cryptococcus and insects have been further studied by the development of a well-validated insect model of pathogenesis in the heterologous insect host Galleria mellonella [66–68]. Outcomes from this yeast–insect system have been shown to correlate with the murine model of infection. In fact, studies examining both C. neoformans and C. gattii wild-type and mutant isolates show similar mortality/attenuation properties compared to the mammalian infection model [21,63,69]. Additionally, the pathogenic Cryptococcus species were shown to be more virulent than their non-pathogenic insect-associated relatives [63]. This invertebrate model of infection is inexpensive and poses fewer ethical issues, allowing for more facile, high throughput analyses of the cryptococcal virulence composite.
4. Environmental aspects of C. gattii
The ubiquity of Cryptococcus neoformans in the environment has been described as the inevitable consequence of the association with pigeons and the admixture of bird guano, cryptococci, and soil [70]. In the case of C. neoformans, it was the increase in the number of susceptible hosts with the advent of the HIV/AIDS epidemic that resulted in the clinical appreciation of the relationship of environmental exposure to acquisition of disease. Compare this then to the relative obscurity of disease caused by C. gattii, seemingly geographically restricted based on early studies of stored clinical isolates [12]. In 1987, David Ellis, challenged by the lack of an identified environmental source of C. neoformans var. gattii to account for the excess cases of gattii-associated disease in Northern Australia, began to search for the ecological niche of this organism [71]. In succeeding years, Ellis and Pfeiffer described the successful isolation of C. gattii from Eucalyptus trees in Australia and California [72–74]. The rarity of gattii-associated disease in geographic locations that did not support the importation or growth of cold-intolerant Eucalypts fit this segregation theory, despite the lack of evidence of an environmental source in areas with both Eucalypts and gattii-associated disease such as Papua New Guinea [27,75,76]. In North America, it was unusual for clinical laboratories to differentiate pathogenic cryptococci and all were assumed to be C. neoformans.
In 2001, however, a veterinary pathologist was the first to notice an increasing number of cases of cryptococcosis, all originating from a large island west of mainland British Columbia. Vancouver Island has a population representing only approximately 17% of the entire province. Human cases were also starting to appear, also disproportionately represented from the smaller population pool. The cases were characterized as not associated with HIV/AIDS and the ensuing emergence, described in animals by Stephen et al. [33] and reviewed in humans by Galanis and MacDougall [29], continues to the present day. The subsequent discovery of cryptococcal disease in Washington and Oregon are described in detail in other sections of this review [19,20,23,24,31,50,51,53,77,78].
Investigation of trees, soil, air, and water in the vicinity of animal and human cases resident on Vancouver Island resulted in the isolation of C. gattii major (VGIIa) and minor (VGIIb) genotypes co-existing within the same ecologic niche [17,22,41]. In contrast to the Australian experience, where the majority of cases of gattii-cryptococcosis were in aboriginal people living in sparsely populated areas, the British Columbia cases were predominantly from urban (e.g., Victoria is the provincial capital of BC) or scenic suburban communities popular with retirees due to the mild climate [71]. Airborne C. gattii propagules are present in endemic areas of BC in highest concentration during the dry summer months (April through October, peaking in July) [59]. The seasonality of airborne propagules is not associated with the flowering or production of pollen of any of the trees found colonized with the pathogen, nor in the diagnosis of the disease as the time of onset of symptoms varies considerably as described by MacDougall and Fyfe [79]. Duncan et al. [80] found an epidemiological association between logging activities or soil disturbance within 10 km of the residences of veterinary cases of gattii-cryptococcosis. Environmental investigation within the 10 km radius of cases has been fruitful in locating trees and soil colonized with C. gattii. These investigations have further elucidated areas that are permanently colonized, yielding positive samples over the last decade of repeated sampling (53).
C. gattii colonization is not homogeneous throughout the eastern coast of Vancouver Island, nor is the relative proportion of the major to minor genotypes the same between permanently colonized areas. Recent analysis stratified by GIS location identified several sites where 100% of isolates belonged to the major genotype (VGIIa), compared to sites where three genotypes (VGIIa, 37%, VGIIb 23% and VGI (14%) are routinely co-isolated along with C. neoformans molecular types VNI and VNIII (Bartlett and Kidd, unpublished data). However, to date, VGIIc has not been identified on Vancouver Island (Byrnes, Hagen, Bartlett, Heitman, unpublished data).
Air and soil samples have yielded some interesting new information regarding the emergent C. gattii. Concentrations of 105 per gram soil are not unusual at some of the permanently colonized sites, with the highest concentrations often found outside the drip line of the tree. Of the Vancouver Island samples, C. gattii concentrations are negatively correlated with organic carbon content and soil moisture [59]. Using Andersen six-stage air impaction sampling, the size range of the airborne propagules was determined, with approximately 88% being in the size range 3.3 to >7 µm, and 12% smaller than 3.3 µm, which is the range considered to be small enough to be inhaled deep into the lung and the alveoli ([59]; Bartlett, unpublished data). Not surprisingly, disturbance of soil or the aerosolization of wood particulate during logging or limbing of infected trees increases the airborne concentration of C. gattii 10–100 fold ([59]; Bartlett and Hingston, unpublished data).
Early in the investigation of the environmental niche of C. gattii, speculation arose regarding the eventual spread of the pathogen. Using databases of the addresses of veterinary or human cases, and the GIS locations of positive environmental samples, ecological niche modeling was used to predict the eventual range of potential colonization to aid in planning the public health response to the emergence. The environmental predictors tested in the model included topographic, climatic, biogeoclimatic and soil data. The significant predictors were low lying elevations, daily January average temperatures above freezing, and presence within the Coastal Douglas-fir and Coastal Western Hemlock xeric maritime biogeoclimatic zones [81]. These same conditions are present on the southwestern coast of British Columbia and further south into Puget Sound. It was not a surprise, therefore, when the first mainland cases without a travel history to Vancouver Island were identified, first in veterinary cases, subsequently in humans in British Columbia [23], Washington [50], and Oregon [19,20,23,31,78]. In corroboration, positive environmental samples have been collected from air, soil and wood from mainland BC and Washington.
Much discussion has resulted from theories regarding how and when C. gattii came to BC. Of interest is a clinical culture isolated in Seattle (NIH444) and deposited in the American Type Culture Collection by June Kwon-Chung in the early 1970s that is indistinguishable from the major genotype, VGIIa. Duncan et al. [34] looked for potential wildlife reservoirs, but isolation of C. gattii from feral birds and mammals has been sporadic and not predictive. In the case of marine mammals, the travel path of the host was unknown prior to washing ashore with fatal cryptococcosis [33,37]. Anecdotally, C. gattii was recovered from the feces of two psittacine birds being treated for cryptococcomas, but the role that the fecal material of caged birds has in spreading C. gattii into the environment is unknown (Bartlett, unpublished data). To date, none of the cold-tolerant eucalyptus tree species surveyed in BC or Washington have been positive for C. gattii.
In summary, the emergence of C. gattii in the Pacific Northwest was unexpected, but the knowledge gained has been a unique example of the need for an interdisciplinary approach to craft the public health response. Early in the emergence, information was rapidly disseminated to primary care physicians and veterinarians in British Columbia. Public media coverage was less helpful in conveying accurate information [82]. The economic consequences have yet to be tallied, and include the cost of hospitalizations, antifungal medications, and loss of tourism to the area [83]. The value to understanding this novel pathogen, however, has been enormous, as evidenced by the speed with which the genome sequencing has been undertaken by an international consortium of researchers [84]. There is still much to be learned about the complex interactions between the environment, microbial ecology, and the ramifications for human health from present and future fungal pathogens.
5. Clinical aspects: human and veterinary infections
Cryptococcosis, the manifestation of infections caused by the pathogenic Cryptococcus species, is a severe and often life-threatening disease in humans and myriad wild, agrarian, and domestic mammals. Furthermore, infections can occur in both terrestrial and marine mammals, highlighting the ubiquitous nature of this pathogen in a wide range of terrestrial and aquatic environments. These infections are also complex clinically, as they commonly result in pneumonia, meningitis, and cryptococcoma formations in humans, and additionally may present as skin infections, particularly in feline and canine cases but also in humans [11,21,34–37,80,85–91]. The clinical course of infections are complex, and treatments are challenging, often long-term, and costly. These issues become particularly apparent in developing regions of the world where access to more effective therapeutic strategies is frequently limited and maintaining aggressive treatments for extended periods of time is not feasible.
Along with the differences in ecological and epidemiological features between C. gatti and C. neoformans, there has been a history of attempts to distinguish their clinical presentations and outcomes [92]. There are two large reviews of C. gattii compared to C. neoformans prior to the recent Vancouver Island outbreak and generally these comparisons came to the same clinical conclusions [93,94]. First, C. gattii occurs by percentage significantly more common in immunocompetent patients than C. neoformans. Second, C. gattii frequently produces disease with focal cryptococcomas in the brain or lung. These mass lesions are associated with either CNS complications such as hydrocephalus, cranial nerve palsies, and seizures or large inflammatory lesions in the lung that either require prolonged therapy and/or surgical intervention. Third, there are some suggestions by clinical experience that C. gattii requires more prolonged antifungal therapy and more commonly surgical removal of cryptococcomas. In fact, there remain inconsistent data on the in vitro susceptibility of C. gattii strains. For instance, there are both studies that support a similar MIC range to azoles as C. neoformans [95,96] and some recent data from the Vancouver Island outbreak strains that suggest some of the strains/genotypes possess increased resistance (West and Marr, unpublished observations). It may be that genetic subtypes vary in their susceptibility [97]. It is also likely that the majority of the clinical resistance for C. gattii disease is influenced by host factors such as immune reconstitution syndrome (IRIS) and with primarily immunocompetent patients the actual outcome of these difficult to manage infections is better than C. neoformans. In HIV-infected patients, it appears that C. neoformans and C. gattii have the same outcome in one center [98]. Specifically, in the Pacific NW outbreak with these new C. gattii strains there has been less mortality in Canada compared to United States [29,99]. The reason for this is unclear and some of it may still be related to a smaller sample size within the United States. After review of present data on outcomes, in vitro drug susceptibility and clinical experience, the 2010 IDSA guidelines have attempted to reflect the present knowledge base and that is to treat C. gattii similar to C. neoformans [30]. However, there was mention of how C. gattii may challenge the clinician in its management of cryptococcomas (brain and lung), chronic conditions of hydrocephalus, and at times slow improvement in symptoms on standard antifungal regimens.
Although the clinical manifestations may overlap between C. gattii and C. neoformans, it has been proposed that C. gattii infections and disease may more commonly present during the initial (acute) infection rather than present with reactivation disease [100]. This pathogenetic understanding is based on two indirect observations. First, serological studies in children show that they commonly have antibodies to C. neoformans but rarely C. gattii and second, C. neoformans commonly presents as the immunosuppressed patient’s immunity wanes and much less of C. gattii disease occurs in immunosuppressed patients. However, it is important to emphasize that this impression is not absolute. It is also noted that serologies may be influenced by epidemiologic exposures and specific risk factors to the fungus. Furthermore, reactivations in some cases of C. gattii are supported by transplant infections (occurrence in non-endemic locations after living in an endemic area) [101] (although the patient in this case did also have travel to Africa 6–7 months prior to disease presentation) or natural animal infections that appear to be recurrent with the same C. gattii strain [102]. Even the use of the reduced numbers of immunosuppressed patients with C. gattii is not a solid insight into pathogenesis since we are now observing more cases during HIV infection and even with the Northwest United States outbreak the number of C. gattii infections in immunosuppressed individuals is substantial. Therefore, although there is some suggestion that the pathophysiology of C. gattii and C. neoformans might be different this is uncertain to what extent and to its clinical relevance. A final point to the pathophysiology and clinical presentation between C. gattii and C. neoformans is that the species can be configured differently for virulence factors. For example, in the trehalose pathway that is essential for disease production, the pathway’s genetic influence on virulence factors such as capsule formation and melanin production differ between C. gattii and C. neoformans [103,104]. Therefore, our present knowledge base has documented differences in virulence factors between species and strains and this may be important to outcome in individual patients. Furthermore, the Pacific NW outbreak has taught us that with its massive human exposure to C. gattii propagules, only a small fraction of exposed humans came down with disease and this demonstrates that the genetic susceptibility of the host remains a primary factor in what direction this infection will travel.
In the clinical setting, the current IDSA cryptococcal treatment guidelines outline similar treatment schedules for C. gattii and C. neformans because there are no definitive trials that prospectively compare regimens and outcomes between the two species [30]. However, the guidelines do acknowledge that special care should be used to examine for the more likely possibility of cryptococcoma formation in C. gattii cases. Also, care should be given to appreciating the immune status of the patient, as there are a higher percentage of immunocompetent patients with cryptococcal infections attributable to C. gattii [30]. Therefore, these patients may present with impressive inflammatory masses and immune reconstitution inflammatory syndrome may become a prominent feature during treatment course and thus require clinical attention. In another clinical concern, several studies have recently documented some azole resistance in C. gattii, particularly among some of the highly virulent molecular type VGII genotypes responsible for the Pacific Northwest outbreak [32,105]. Although whether C. gattii strains in general have higher minimum inhibitory concentrations (MICs) than C. neoformans strains remains uncertain, clearly the apparently clinical response to treatment suggests that C. gattii infections may appear somewhat resistant to many treatment strategies. The CDC recommends that amphotericin B or its lipid products should always be used as an induction therapy followed by fluconazole maintenance therapy for C. gattii for the Pacific Northwest outbreak strains, whereas in non-CNS C. neoformans cases fluconazole can still be used successfully (Harris et al. 48th IDSA poster presentation #642, [30]). When the patient’s immune status, with its increased predilection for cryptococcoma formation and higher rates of azole resistance are considered together, it suggests that C. gattii infections may need to be treated more aggressively than other cryptococcal infections and this may be particularly true in the Pacific Northwest outbreak setting. Another clinical factor is that C. gattii infections without risk factor appearance (i.e., normal host) may often go undiagnosed for longer periods of time and not be appreciated by clinical providers. There is generally a lack of speciation between C. gattii and C. neoformans in clinical labs. Overall, C. gattii infections in humans are complex and clinical studies specifically addressing C. gattii prognosis and treatment in a prospective manner need to be conducted.
While a large focus of cryptococcal disease is on human infections, veterinary infections are also a significant cause of animal morbidity and mortality worldwide. Infections have been documented in a broad range of domestic, agrarian, and wild mammals, indicating that many mammals may be subject to disease and that cases in wild animals may frequently go unrecognized [19,31,33–37,80,85–87,89,91,106–108]. Infections in animals are of concern for economic factors (agrarian), quality of life for owners (domestic), and for wildlife concern particularly in the cases of koalas and marine mammals [19,20,33,37,77,78,91,109]. Furthermore, in cases of novel outbreaks (Vancouver Island outbreak and United States expansion), animal infections served as sentinel cases and helped highlight specific geographic expansion and emergence because many of the animals were non-migratory [21,33,34,78].
Cryptococcal infections in humans and other animals, particularly those caused by C. gattii, are not difficult to diagnose but the clinician must consider it in their differential diagnosis. The disease presentation can be diverse in humans and animals. Advances in early diagnosis and access to effecting treatments in humans and animals can decrease attributable morbidity and mortality; study of yeast strains with clinical information can help understand the evolution of virulence. In fact, the Vancouver Island outbreak represents an ideal opportunity to study the genetic host susceptibility to cryptococcal disease. Clearly, the island inhabitants (>700,000 individuals) had substantial exposure to the fungus but only a small fraction came down with disease. Genetic studies may identify find polymorphisms in innate immune genes in those with disease. Clinical and veterinary aspects also highlight the multidisciplinary and collaborative approach needed to address the many issues surrounding C. gattii infections.
6. The Pacific NW outbreak: review and outlook
As of 1999, C. gattii has emerged as a primary pathogen in North America, including both Canada and the United States [17,19,20,22–24,29,41,77]. This outbreak now spans a large geographic range, with levels of infection as high or higher than anywhere else globally. For instance, the annual incidence on Vancouver Island is approximately 25 cases/million people [29]. The only two reports with higher overall incidences are one examination of native Aboriginals in the Northern Territory of Australia, and a study conducted in the central province of Papua New Guinea [26,27,29]. The VGI and VGII molecular types are the primary ones associated with illness in otherwise healthy individuals [24]. Infections due to VGI have been reported at high rates among populations in Australia, while the levels of VGII infection are higher in the Pacific NW, where >95% of all cases are attributable to this molecular type [11,17,18,24,45]. The appearance of C. gattii in North America is startling because this is the first major emergence of this species in a temperate climate [59,79], and unlike C. neoformans, this pathogen had previously been geographically restricted to tropical and sub-tropical regions throughout the world [11,12,110,111]. To examine the evolutionary aspects of this unprecedented outbreak, efforts were undertaken to study the molecular epidemiology and characteristics of isolates collected from humans, animals, and the environment. These efforts have and will continue to shed light onto several key features of this outbreak, while other contributing factors remain elusive.
A central question from the analysis of the Vancouver Island outbreak relates to the origin of the first novel genotype observed as part of the outbreak, VGIIa/major. This genotype has been responsible for the vast majority of all infections reported in British Columbia [22,41]. As in the sibling species C. neoformans, many C. gattii populations are predominantly comprised of a mating type isolates, and to date all isolates related to the outbreak have been exclusively α. A seminal finding by Lin and colleagues in 2005 was the discovery that monokaryotic fruiting represents a novel mode of α–α unisexual reproduction (including meiosis) [112–114]. This finding, in combination with the discovery of an α/α VGIIa/major diploid isolate from Vancouver Island (RB59), and molecular comparisons between the VGIIa/major genotype and the less prevalent VGIIb/minor genotype that is also found in Australia led to the hypothesis that same-sex mating may have produced the virulent VGIIa genotype and is responsible for ongoing production of infectious spores. An alternative hypothesis is that opposite-sex mating, possibly in South America where MATa isolates similar in genotype have been discovered, contributing to the origin of the outbreak isolate genotype [17,115]. In fact, the VGIIa/major subgroup has been shown to be more fertile in comparison to the VGIIb/minor subgroup [20,116]. Further studies of the global VGII population also suggest that recombination is still occurring [20]. While the origins of C. gattii VGIIa/major in North America remain uncertain, it is clear that this emerging pathogen has recently invaded the United States, and that in addition a new unique and highly virulent genotype in Oregon, VGIIc, has also arisen [19,20,31].
In response to this outbreak, a multidisciplinary C. gattii working group was established to address the epidemiology, clinical features, and basic science questions surrounding this outbreak [78]. The overall incidence remains low even in endemic areas and very little is currently known about how or why specific humans and animals develop disease. It probably involves unique host factors, including possible genetic predisposition in addition to steroid treatment and other identified predisposing risk factors [117]. In addition, the precise origins of the VGIIa/major and VGIIc/novel genotypes remain elusive: the VGIIa outbreak isolates are indistinguishable from isolate NIH444, indicating it has been circulating in the region for at least ~40 years; the VGIIb outbreak isolates are indistinguishable from fertile recombining Australian populations and therefore likely originated there; and VGIIc outbreak isolates are unique to Oregon so likely originate from an under-sampled global niche or arose locally. Substantial progress has been achieved in addressing the molecular epidemiology and expansion of the outbreak, and also the phenotypic characteristics that make these genotypes unique. However, many critical questions remain to be addressed in the future to understand the evolutionary dynamics of this unprecedented C. gattii emergence in North America. Expanded environmental sampling, further phenotypic characterization of associations with host animals and plants, and genome sequencing of C. gattii mitochondrial and nuclear genomes in addition to those available for the VGI isolate WM276 and the VGIIa/major outbreak isolate R265 [84] should be conducted. Furthermore, host genetic profiling is needed.
7. Conclusions
Until 1999, the vast majority of studies surrounding Cryptococcus pathogenesis focused on the globally distributed sibling species to C. gattii, C. neoformans. While the North American Pacific Northwest C. gattii outbreak has increased the profile of infections caused by the less-common species, we now also appreciate that C. gattii may be more frequently found in HIV/AIDS patients than previously estimated [46] (Byrnes et al, PLoS Pathogens, in revision), and that outside of the Pacific Northwest outbreak there may also be a number of attributable cases from any of the four molecular types in both otherwise healthy and immunosuppressed hosts [21,51–54,118].
Examination of isolates by high-resolution molecular and phenotypic analyses can facilitate in determining and understanding the dynamics of expansions into new regions, help to reveal the emergence of novel genotypes, and allows discrimination of cases not related to specific outbreak genotypes. Furthermore, molecular typing can support whether or not specific cases are likely travel-associated. Characterizing genotypes and molecular types also allows for the classification and phenotypic comparisons of virulence, highlighting as in the case of the Pacific Northwest that specific genotypes causing disease are highly virulent in animal models of infection. In-depth analysis of populations also allows for recombination studies, leading to a greater understanding of the roles that both same- and opposite-sex mating may play in the formation of infectious spores and the generation of genetic diversity and thus novel genotypes via meiotic events.
Recent lines of evidence suggest that mitochondrial inheritance and recombination could also play significant roles in the evolution and virulence composite of C. gattii [119–121]. Mitochondrial recombination or exchange requires cell–cell fusion, and it is hypothesized that this type of event could also lead to other nuclear or plasmid genetic exchange [122,123]. Future studies should also expand environmental sampling, apply further phenotypic characterizations of associations with host animals and plants, and utilize next generation sequencing to analyze more representative C. gattii mitochondrial and nuclear genomes. Additionally, epidemiological, clinical, and veterinary studies need to be continued, with an active multidisciplinary collaboration between all of those with vested interests in this area of study.
Why C. gattii emerged in a temperate climate for the first time remains unclear, and importantly it is not known how far the outbreak will expand and if new risk areas may occur in other temperate regions. Furthermore, if the outbreak will inexorably expand down the coast or alternatively discontinuously jump to new regions remains unclear. This punctuates the Pacific Northwest outbreak as a focal point for studies and a paradigm for novel emergent fungal diseases. To this end, it is critical that clinical laboratories and veterinary diagnostic laboratories begin to consider assigning species identity to clinical isolates, thereby distinguishing C. neoformans and C. gattii. In addition to advancing the field of C. gattii research, the reviewed studies also contribute to the ongoing research surrounding the dynamics of emerging and reemerging diseases and outbreak examinations. In host-microbe interactions one tenant is clear and well-described by Lewis Carroll’s Red Queen adage: “It takes all the running you can do, to keep in the same place.” As advances in treatment continue, microbes gain drug resistance/virulence attributes and new pathogens emerge, tipping the balance between the host and pathogen populations toward disease emergence. This co-evolution will undoubtedly continue, and further understanding of both the host and its pathogens are required so that advances in the prognosis, treatment, prevention, and eradication of infectious diseases can be achieved.
Acknowledgments
We thank Wenjun Li, Deborah Springer, and Sheng Sun for reading the manuscript and providing comments. These studies were supported by NIH/NIAID R37 grant AI39115 and R01 grant AI50113 to JH; NIH/NIAID R01 grant AI73896 to JP; and funding from the British Columbia Lung Association, WorkSafe BC, and the Michael Smith Foundation for Health Research to KB.
References
- 1.Fauci ML. Cohen, Changing patterns of infectious disease. Nature. 2000;406:762–767. doi: 10.1038/35021206. [DOI] [PubMed] [Google Scholar]
- 2.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morens DM, Folkers GK, Fauci AS. Emerging infections: a perpetual challenge. Lancet Infect. Dis. 2008;8:710–719. doi: 10.1016/S1473-3099(08)70256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisher MC, Garner TW, Walker SF. Global emergence of Batrachochytrium dendrobatidis and amphibian chytridiomycosis in space, time, and host. Annu. Rev. Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- 5.Blehert DS, Hicks AC, Behr M, Meteyer CU, Berlowski-Zier BM, Buckles EL, Coleman JT, Darling SR, Gargas A, Niver R, Okoniewski JC, Rudd RJ, Stone WB. Bat white-nose syndrome: an emerging fungal pathogen? Science. 2009;323:227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- 6.Bromenshenk JJ, Henderson CB, Wick CH, Stanford MF, Zulich AW, Jabbour RE, Deshpande SV, McCubbin PE, Seccomb RA, Welch PM, Williams T, Firth DR, Skowronski E, Lehmann MM, Bilimoria SL, Gress J, Wanner KW, Cramer RA., Jr Iridovirus and microsporidian linked to honey bee colony decline. PLoS One. 2010;5:e13181. doi: 10.1371/journal.pone.0013181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Evans JD, Smith IB, Pettis JS. Nosema ceranae is a long-present and wide-spread microsporidian infection of the European honey bee (Apis mellifera) in the United States. J. Invertebr. Pathol. 2008;97:186–188. doi: 10.1016/j.jip.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Cornman RS, Chen YP, Schatz MC, Street C, Zhao Y, Desany B, Egholm M, Hutchison S, Pettis JS, Lipkin WI, Evans JD. Genomic analyses of the microsporidian Nosema ceranae, an emergent pathogen of honey bees. PLoS Pathog. 2009;5:e1000466. doi: 10.1371/journal.ppat.1000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, Hornig M, Geiser DM, Martinson V, vanEngelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin WI. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 10.Perfect JR. Cryptococcosis. Infect. Dis. Clin. North Am. 1989;3:77–102. [PubMed] [Google Scholar]
- 11.Sorrell TC. Cryptococcus neoformans variety gattii. Med. Mycol. 2001;39:155–168. [PubMed] [Google Scholar]
- 12.Kwon-Chung KJ, Bennett JE. High prevalence of Cryptococcus neoformans var. gattii in tropical and subtropical regions. Zentralbl. Bakteriol. Mikrobiol. Hyg. [A] 1984;257:213–218. [PubMed] [Google Scholar]
- 13.Kwon-Chung KJ, Boekhout T, Fell JW, Diaz M. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae) Taxon. 2002;51:804–806. [Google Scholar]
- 14.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 15.Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–587. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- 16.Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, Abeln EC, Dromer F, Meyer W. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 17.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 18.Bovers M, Hagen F, Kuramae EE, Boekhout T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet. Biol. 2008;45:400–421. doi: 10.1016/j.fgb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Byrnes EJ, 3rd, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the Pacific Northwest in the United States. J. Infect. Dis. 2009;199:1081–1086. doi: 10.1086/597306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrnes EJ, 3rd, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrnes EJ, 3rd, Li W, Lewit Y, Perfect JR, Carter DA, Cox GM, Heitman J. First reported case of Cryptococcus gattii in the Southeastern USA: implications for travel-associated acquisition of an emerging pathogen. PLoS One. 2009;4:e5851. doi: 10.1371/journal.pone.0005851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kidd SE, Guo H, Bartlett KH, Xu J, Kronstad JW. Comparative gene genealogies indicate that two clonal lineages of Cryptococcus gattii in British Columbia resemble strains from other geographical areas. Eukaryot. Cell. 2005;4:1629–1638. doi: 10.1128/EC.4.10.1629-1638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacDougall L, Kidd SE, Galanis E, Mak S, Leslie MJ, Cieslak PR, Kronstad JW, Morshed MG, Bartlett KH. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg. Infect. Dis. 2007;13:42–50. doi: 10.3201/eid1301.060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrnes EJ, 3rd, Heitman J. Cryptococcus gattii outbreak expands into the Northwestern United States with fatal consequences. F1000 Biology Reports. 2009;1:62. doi: 10.3410/B1-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J. Infect. Dis. 2005;192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- 26.Fisher D, Burrow J, Lo D, Currie B. Cryptococcus neoformans in tropical northern Australia: predominantly variant gattii with good outcomes. Aust. N.Z.J. Med. 1993;23:678–682. doi: 10.1111/j.1445-5994.1993.tb04726.x. [DOI] [PubMed] [Google Scholar]
- 27.Seaton RA. The management of cryptococcal meningitis in Papua New Guinea. P.N.G. Med. J. 1996;39:67–73. [PubMed] [Google Scholar]
- 28.Seaton RA, Wembri JP, Armstrong P, Ombiga J, Naraqi S, Kevau I. Symptomatic human immunodeficiency virus (HIV) infection in Papua New Guinea. Aust. N.Z.J. Med. 1996;26:783–788. doi: 10.1111/j.1445-5994.1996.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 29.Galanis E, MacDougall L. Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999–2007. Emerg. Infect. Dis. 2010;16:251–257. doi: 10.3201/eid1602.090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byrnes EJ, 3rd, Bildfell RJ, Dearing PL, Valentine BA, Heitman J. Cryptococcus gattii with bimorphic colony types in a dog in western Oregon: additional evidence for expansion of the Vancouver Island outbreak. J. Vet. Diagn. Invest. 2009;21:133–136. doi: 10.1177/104063870902100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iqbal N, Debess EE, Wohrle R, Sun B, Nett RJ, Ahlquist AM, Chiller T. Correlation of genotype and in vitro susceptibilities of Cryptococcus gattii from the Pacific Northwest of the United States. J. Clin. Microbiol. 2009;48:539–544. doi: 10.1128/JCM.01505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephen C, Lester S, Black W, Fyfe M, Raverty S. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 2002;43:792–794. [PMC free article] [PubMed] [Google Scholar]
- 34.Duncan C, Schwantje H, Stephen C, Campbell J, Bartlett K. Cryptococcus gattii in wildlife of Vancouver Island, British Columbia, Canada. J. Wildl. Dis. 2006;42:175–178. doi: 10.7589/0090-3558-42.1.175. [DOI] [PubMed] [Google Scholar]
- 35.Duncan C, Stephen C, Lester S, Bartlett KH. Sub-clinical infection and asymptomatic carriage of Cryptococcus gattii in dogs and cats during an outbreak of cryptococcosis. Med. Mycol. 2005;43:511–516. doi: 10.1080/13693780500036019. [DOI] [PubMed] [Google Scholar]
- 36.Malik R, Alderton B, Finlaison D, Krockenberger MB, Karaoglu H, Meyer W, Martin P, France MP, McGill J, Lester SJ, O’Brien CR, Love DN. Cryptococcosis in ferrets: a diverse spectrum of clinical disease. Aust. Vet. J. 2002;80:749–755. doi: 10.1111/j.1751-0813.2002.tb11343.x. [DOI] [PubMed] [Google Scholar]
- 37.Miller WG, Padhye AA, van Bonn W, Jensen E, Brandt ME, Ridgway SH. Cryptococcosis in a bottlenose dolphin (Tursiops truncatus) caused by Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 2002;40:721–724. doi: 10.1128/JCM.40.2.721-724.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duncan C, Bartlett KH, Lester S, Bobsien B, Campbell J, Stephen C, Raverty S. Surveillance for Cryptococcus gattii in horses of Vancouver Island, British Columbia, Canada. Med. Mycol. 2011;7:1–5. doi: 10.3109/13693786.2011.560196. [DOI] [PubMed] [Google Scholar]
- 39.Lester SJ, Kowalewich NJ, Bartlett KH, Krockenberger MB, Fairfax TM, Malik R. Clinicopathologic features of an unusual outbreak of cryptococcosis in dogs, cats, ferrets, and a bird: 38 cases (January to July 2003) J. Am. Vet. Med. Assoc. 2004;225:1716–1722. doi: 10.2460/javma.2004.225.1716. [DOI] [PubMed] [Google Scholar]
- 40.Lester SJ, Malik R, Bartlett KH, Duncan CG. Cryptococcosis: update and emergence of Cryptococcus gattii. Vet. Clin. Pathol. 2011;40:4–17. doi: 10.1111/j.1939-165X.2010.00281.x. [DOI] [PubMed] [Google Scholar]
- 41.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada) Proc. Natl. Acad. Sci. U. S.A. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, Methling K, Viviani MA, Chindamporn A, Sukroongreung S, John MA, Ellis DH, Sorrell TC. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–1799. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 43.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellis D, Marriott D, Hajjeh RA, Warnock D, Meyer W, Barton R. Epidemiology: surveillance of fungal infections. Med. Mycol. 2000;38 Suppl. 1:173–182. [PubMed] [Google Scholar]
- 45.Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot. Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaturvedi S, Dyavaiah M, Larsen RA, Chaturvedi V. Cryptococcus gattii in AIDS patients, southern California. Emerg. Infect. Dis. 2005;11:1686–1692. doi: 10.3201/eid1111.040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Litvintseva AP, Thakur R, Vilgalys R, Mitchell TG. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics. 2006;172:2223–2238. doi: 10.1534/genetics.105.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, Esposto MC, Fisher M, Gilgado F, Hagen F, Kaocharoen S, Litvintseva AP, Mitchell TG, Simwami SP, Trilles L, Viviani MA, Kwon-Chung J. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 2009;47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Upton A, Fraser JA, Kidd SE, Bretz C, Bartlett KH, Heitman J, Marr KA. First contemporary case of human infection with Cryptococcus gattii in Puget Sound: evidence for spread of the Vancouver Island outbreak. J. Clin. Microbiol. 2007;45:3086–3088. doi: 10.1128/JCM.00593-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindberg J, Hagen F, Laursen A, Stenderup J, Boekhout T. Cryptococcus gattii risk for tourists visiting Vancouver Island, Canada. Emerg. Infect. Dis. 2007;13:178–179. doi: 10.3201/eid1301.060945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Georgi A, Schneemann M, Tintelnot K, Calligaris-Maibach RC, Meyer S, Weber R, Bosshard PP. Cryptococcus gattii meningoencephalitis in an immunocompetent person 13 months after exposure. Infection. 2009;37:370–373. doi: 10.1007/s15010-008-8211-z. [DOI] [PubMed] [Google Scholar]
- 53.Hagen F, Assen SV, Luijckx GJ, Boekhout T, Kampinga GA. Activated dormant Cryptococcus gattii infection in a Dutch tourist who visited Vancouver Island (Canada): a molecular epidemiological approach. Med. Mycol. 2009;48:528–531. doi: 10.3109/13693780903300319. [DOI] [PubMed] [Google Scholar]
- 54.Okamoto K, Hatakeyama S, Itoyama S, Nukui Y, Yoshino Y, Kitazawa T, Yotsuyanagi H, Ikeda R, Sugita T, Koike K. Cryptococcus gattii genotype VGIIa infection in man, Japan, 2007. Emerg. Infect. Dis. 2010;16:1155–1157. doi: 10.3201/eid1607.100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halliday CL, Carter DA. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var gattii isolates from Australia. J. Clin. Microbiol. 2003;41:703–711. doi: 10.1128/JCM.41.2.703-711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saul N, Krockenberger M, Carter D. Evidence of recombination in mixed-mating-type and alpha-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot. Cell. 2008;7:727–734. doi: 10.1128/EC.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot. Cell. 2005;4:1403–1409. doi: 10.1128/EC.4.8.1403-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell LT, Fraser JA, Nichols CB, Dietrich FS, Carter D, Heitman J. Clinical and environmental isolates of Cryptococcus gattii from Australia that retain sexual fecundity. Eukaryot. Cell. 2005;4:1410–1419. doi: 10.1128/EC.4.8.1410-1419.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, Hingston AO, Kronstad JW, Bartlett KH. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl. Environ. Microbiol. 2007;73:1433–1443. doi: 10.1128/AEM.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sampaio JP, Inacio J, Fonseca A, Gadanho M, Spencer-Martins I, Scorzetti G, Fell JW. Auriculibuller fuscus gen. nov., sp. nov. and Bullera japonica sp. nov., novel taxa in the Tremellales. Int. J. Syst. Evol. Microbiol. 2004;54:987–993. doi: 10.1099/ijs.0.02970-0. [DOI] [PubMed] [Google Scholar]
- 61.Ergin C, Ilkit M, Kaftanoglu O. Detection of Cryptococcus neoformans var. grubii in honeybee (Apis mellifera) colonies. Mycoses. 2004;47:431–434. doi: 10.1111/j.1439-0507.2004.01018.x. [DOI] [PubMed] [Google Scholar]
- 62.Rimek D, Haase G, Luck A, Casper J, Podbielski A. First report of a case of meningitis caused by Cryptococcus adeliensis in a patient with acute myeloid leukemia. J. Clin. Microbiol. 2004;42:481–483. doi: 10.1128/JCM.42.1.481-483.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Findley K, Rodriguez-Carres M, Metin B, Kroiss J, Fonseca A, Vilgalys R, Heitman J. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryot. Cell. 2009;8:353–361. doi: 10.1128/EC.00373-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gezuele E, Calegari L, Sanabŕıa D, Davel G, Civila E. Isolation in Uruguay of Cryptococcus neoformans var. gattii from nest of the wasp Polybia occidentalis. Revista Iberoamericana de Micoloǵıa. 1993;10:5–6. [Google Scholar]
- 65.Kidd SE, Sorrell TC, Meyer W. Isolation of two molecular types of Cryptococcus neoformans var. gattii from insect frass. Med. Mycol. 2003;41:171–176. doi: 10.1080/mmy.41.2.171.176. [DOI] [PubMed] [Google Scholar]
- 66.Fuchs BB, Mylonakis E. Using non-mammalian hosts to study fungal virulence and host defense. Curr. Opin. Microbiol. 2006;9:346–351. doi: 10.1016/j.mib.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 67.London R, Orozco BS, Mylonakis E. The pursuit of cryptococcal pathogenesis: heterologous hosts and the study of cryptococcal hostpathogen interactions. FEMS Yeast Res. 2006;6:567–573. doi: 10.1111/j.1567-1364.2006.00056.x. [DOI] [PubMed] [Google Scholar]
- 68.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 2005;73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, Niang R, Casadevall A. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics. 2001;107:E66. doi: 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- 71.Ellis DH. Cryptococcus neoformans var. gattii in Australia. J. Clin. Microbiol. 1987;25:430–431. doi: 10.1128/jcm.25.2.430-431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pfeiffer T, Ellis D. Environmental isolation of Cryptococcus neoformans var. gattii from California. J. Infect. Dis. 1991;163:929–930. doi: 10.1093/infdis/163.4.929. [DOI] [PubMed] [Google Scholar]
- 73.Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J. Clin. Microbiol. 1990;28:1642–1644. doi: 10.1128/jcm.28.7.1642-1644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pfeiffer TJ, Ellis DH. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus tereticornis. J. Med. Vet. Mycol. 1992;30:407–408. [PubMed] [Google Scholar]
- 75.Seaton RA, Hamilton AJ, Hay RJ, Warrell DA. Exposure to Cryptococcus neoformans var. gattii—a seroepidemiological study. Trans. R. Soc. Trop. Med. Hyg. 1996;90:508–512. doi: 10.1016/s0035-9203(96)90297-7. [DOI] [PubMed] [Google Scholar]
- 76.Laurenson IF, Lalloo DG, Naraqi S, Seaton RA, Trevett AJ, Matuka A, Kevau IH. Cryptococcus neoformans in Papua New Guinea: a common pathogen but an elusive source. J. Med. Vet. Mycol. 1997;35:437–440. doi: 10.1080/02681219780001561. [DOI] [PubMed] [Google Scholar]
- 77.Bartlett KH, Kidd SE, Kronstad JW. The emergence of Cryptococcus gattii in British Columbia and the Pacific Northwest. Curr. Infect. Dis. Rep. 2008;10:58–65. doi: 10.1007/s11908-008-0011-1. [DOI] [PubMed] [Google Scholar]
- 78.Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, Hoang L, Leslie MJ, MacDougall L, Magill SS, Morshed MG, Marr KA. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg. Infect. Dis. 2009;15:1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.MacDougall L, Fyfe M. Emergence of Cryptococcus gattii in a novel environment provides clues to its incubation period. J. Clin. Microbiol. 2006;44:1851–1852. doi: 10.1128/JCM.44.5.1851-1852.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Duncan CG, Stephen C, Campbell J. Evaluation of risk factors for Cryptococcus gattii infection in dogs and cats. J. Am. Vet. Med. Assoc. 2006;228:377–382. doi: 10.2460/javma.228.3.377. [DOI] [PubMed] [Google Scholar]
- 81.Mak S, Klinkenberg B, Bartlett K, Fyfe M. Ecological niche modeling of Cryptococcus gattii in British Columbia, Canada. Environ. Health. Perspect. 2010;118:653–658. doi: 10.1289/ehp.0901448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nicol AM, Hurrell C, McDowall W, Bartlett K, Elmieh N. Communicating the risks of a new, emerging pathogen: the case of Cryptococcus gattii. Risk. Anal. 2008;28:373–386. doi: 10.1111/j.1539-6924.2008.01024.x. [DOI] [PubMed] [Google Scholar]
- 83.Chambers C, MacDougall L, Li M, Galanis E. Tourism and specific risk areas for Cryptococcus gattii, Vancouver Island, Canada. Emerg. Infect. Dis. 2008;14:1781–1783. doi: 10.3201/eid1411.080532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.D’Souza CA, Kronstad JW, Taylor G, Warren R, Yuen M, Hu G, Jung WH, Sham A, Kidd SE, Tangen K, Lee N, Zeilmaker T, Sawkins J, McVicker G, Shah S, Gnerre S, Griggs A, Zeng Q, Bartlett K, Li W, Wang X, Heitman J, Stajich JE, Fraser JA, Meyer W, Carter D, Schein J, Krzywinski M, Kwon-Chung KJ, Varma A, Wang J, Brunham R, Fyfe M, Ouellette BF, Siddiqui A, Marra M, Jones S, Holt R, Birren BW, Galagan JE, Cuomo CA. Genome variation in Cryptococcus gattii, an emerging pathogen of Immunocompetent hosts. MBio. 2011;2:e00342–e00310. doi: 10.1128/mBio.00342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Beatty JA, Barrs VR, Swinney GR, Martin PA, Malik R. Peripheral vestibular disease associated with cryptococcosis in three cats. J. Feline. Med. Surg. 2000;2:29–34. doi: 10.1053/jfms.2000.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Begg LM, Hughes KJ, Kessell A, Krockenberger MB, Wigney DI, Malik R. Successful treatment of cryptococcal pneumonia in a pony mare. Aust. Vet. J. 2004;82:686–692. doi: 10.1111/j.1751-0813.2004.tb12155.x. [DOI] [PubMed] [Google Scholar]
- 87.Duncan C, Stephen C, Campbell J. Clinical characteristics and predictors of mortality for Cryptococcus gattii infection in dogs and cats of southwestern British Columbia. Can. Vet. J. 2006;47:993–998. [PMC free article] [PubMed] [Google Scholar]
- 88.Duncan C, Stephen C, Lester S, Bartlett KH. Follow-up study of dogs and cats with asymptomatic Cryptococcus gattii infection or nasal colonization. Med. Mycol. 2005;43:663–666. doi: 10.1080/13693780500220076. [DOI] [PubMed] [Google Scholar]
- 89.Malik R, Martin P, McGill J, Martin A, Love DN. Successful treatment of invasive nasal cryptococcosis in a ferret. Aust. Vet. J. 2000;78:158–159. doi: 10.1111/j.1751-0813.2000.tb10582.x. [DOI] [PubMed] [Google Scholar]
- 90.Morgan J, McCarthy KM, Gould S, Fan K, Arthington-Skaggs B, Iqbal N, Stamey K, Hajjeh RA, Brandt ME. Cryptococcus gattii infection: characteristics and epidemiology of cases identified in a South African province with high HIV seroprevalence, 2002–2004. Clin. Infect. Dis. 2006;43:1077–1080. doi: 10.1086/507897. [DOI] [PubMed] [Google Scholar]
- 91.Krockenberger MB, Canfield PJ, Malik R. Cryptococcus neoformans var. gattii in the koala (Phascolarctos cinereus): a review of 43 cases of cryptococcosis. Med. Mycol. 2003;41:225–234. doi: 10.1080/369378031000137242. [DOI] [PubMed] [Google Scholar]
- 92.Chen S, Sorrell T, Nimmo G, Speed B, Currie B, Ellis D, Marriott D, Pfeiffer T, Parr D, Byth K. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin. Infect. Dis. 2000;31:499–508. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 93.Mitchell DH, Sorrell TC, Allworth AM, Heath CH, McGregor AR, Papanaoum K, Richards MJ, Gottlieb T. Cryptococcal disease of the CNS in immunocompetent hosts: influence of cryptococcal variety on clinical manifestations and outcome. Clin. Infect. Dis. 1995;20:611–616. doi: 10.1093/clinids/20.3.611. [DOI] [PubMed] [Google Scholar]
- 94.Speed B, Dunt D. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 1995;21:28–34. doi: 10.1093/clinids/21.1.28. discussion 35–26. [DOI] [PubMed] [Google Scholar]
- 95.Thompson GR, 3rd, Wiederhold NP, Fothergill AW, Vallor AC, Wickes BL, Patterson TF. Antifungal susceptibilities among different serotypes of Cryptococcus gattii and Cryptococcus neoformans. Antimicrob. Agents. Chemother. 2009;53:309–311. doi: 10.1128/AAC.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Torres-Rodriguez JM, Alvarado-Ramirez E, Murciano F, Sellart M. MICs and minimum fungicidal concentrations of posaconazole, voriconazole and fluconazole for Cryptococcus neoformans and Cryptococcus gattii. J. Antimicrob. Chemother. 2008;62:205–206. doi: 10.1093/jac/dkn132. [DOI] [PubMed] [Google Scholar]
- 97.Cheng PY, Sham A, Kronstad JW. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect. Immun. 2009;77:4284–4294. doi: 10.1128/IAI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Steele KT, Thakur R, Nthobatsang R, Steenhoff AP, Bisson GP. In-hospital mortality of HIV-infected cryptococcal meningitis patients with C. gattii and infection in Gaborone, Botswana. Med. Mycol. 2010;48:1112–1115. doi: 10.3109/13693781003774689. [DOI] [PubMed] [Google Scholar]
- 99.CDC. Emergence of Cryptococcus gattii– Pacific Northwest, 2004–2010. Morb. Mortal. Wkly. Rep. 2010;59:865–868. [PubMed] [Google Scholar]
- 100.Sorrell TC, Chen SCA, Phillips P, Marr KA. Clinical perspectives on Cryptococcus neoformans and Cryptococcus gattii: implications for diagnosis and management. In: Heitman J, Kozel T, Kwon-Chung J, Perfect JR, Casadevall A, editors. Cryptococcus: From Human Pathogen to Model Yeast. Washington, DC: ASM Press; 2011. pp. 595–606. [Google Scholar]
- 101.Dromer F, Ronin O, Dupont B. Isolation of Cryptococcus neoformans var. gattii from an Asian patient in France: evidence for dormant infection in healthy subjects. J. Med. Vet. Mycol. 1992;30:395–397. [PubMed] [Google Scholar]
- 102.Kluger EK, Karaoglu HK, Krockenberger MB, Della Torre PK, Meyer W, Malik R. Recrudescent cryptococcosis, caused by Cryptococcus gattii (molecular type VGII), over a 13-year period in a Birman cat. Med. Mycol. 2006;44:561–566. doi: 10.1080/13693780600582847. [DOI] [PubMed] [Google Scholar]
- 103.Ngamskulrungroj P, Himmelreich U, Breger JA, Wilson C, Chayakulkeeree M, Krockenberger MB, Malik R, Daniel HM, Toffaletti D, Djordjevic JT, Mylonakis E, Meyer W, Perfect JR. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii. Infect. Immun. 2009;77:4584–4596. doi: 10.1128/IAI.00565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Petzold EW, Himmelreich U, Mylonakis E, Rude T, Toffaletti D, Cox GM, Miller JL, Perfect JR. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect. Immun. 2006;74:5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hagen F, Illnait-Zaragozi MT, Bartlett KH, Swinne D, Geertsen E, Klaassen CH, Boekhout T, Meis JF. In vitro antifungal susceptibilities and AFLP genotyping of a worldwide collection of 350 clinical, veterinary and environmental Cryptococcus gattii isolates. Antimicrob. Agents. Chemother. 2010;54:5139–5145. doi: 10.1128/AAC.00746-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McGill S, Malik R, Saul N, Beetson S, Secombe C, Robertson I, Irwin P. Cryptococcosis in domestic animals in Western Australia: a retrospective study from 1995–2006. Med. Mycol. 2009;47:625–639. doi: 10.1080/13693780802512519. [DOI] [PubMed] [Google Scholar]
- 107.Barrs VR, Allan GS, Martin P, Beatty JA, Malik R. Feline pyothorax: a retrospective study of 27 cases in Australia. J. Feline. Med. Surg. 2005;7:211–222. doi: 10.1016/j.jfms.2004.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bui T, Lin X, Malik R, Heitman J, Carter D. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, alpha mating type populations. Eukaryot. Cell. 2008;7:1771–1780. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rotstein DS, West K, Levine G, Lockhart SR, Raverty S, Morshed MG, Rowles T. Cryptococcus gattii VGI in a spinner dolphin (Stenella longirostris) from Hawaii. J. Zoo Wildl. Med. 2010;41:181–183. doi: 10.1638/2009-0145.1. [DOI] [PubMed] [Google Scholar]
- 110.Casadevall A, Perfect JR, editors. Cryptococcus neoformans. Washington, DC: ASM Press; 1998. [Google Scholar]
- 111.Kwon-Chung KJ, Bennett JE. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 1984;120:123–130. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 112.Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 113.Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. alphaADalpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–1990. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. Diploids in the Cryptococcus neoformans serotype A population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Escandon P, Sanchez A, Martinez M, Meyer W, Castaneda E. Molecular epidemiology of clinical and environmental isolates of the Cryptococcus neoformans species complex reveals a high genetic diversity and the presence of the molecular type VGII mating type a in Colombia. FEMS Yeast Res. 2006;6:625–635. doi: 10.1111/j.1567-1364.2006.00055.x. [DOI] [PubMed] [Google Scholar]
- 116.Ngamskulrungroj P, Sorrell TC, Chindamporn A, Chaiprasert A, Poonwan N, Meyer W. Association between fertility and molecular subtype of global isolates of Cryptococcus gattii molecular type VGII. Med. Mycol. 2008:1–9. doi: 10.1080/13693780802210734. [DOI] [PubMed] [Google Scholar]
- 117.Macdougall L, Fyfe M, Romney M, Starr M, Galanis E. Risk Factors for Cryptococcus gattii Infection, British Columbia, Canada. Emerg. Infect. Dis. 2011;17:193–199. doi: 10.3201/eid1702.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bodasing N, Seaton RA, Shankland GS, Kennedy D. Cryptococcus neoformans var. gattii meningitis in an HIV-positive patient: first observation in the United Kingdom. J. Infect. 2004;49:253–255. doi: 10.1016/j.jinf.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 119.Bovers M, Hagen F, Kuramae EE, Boekhout T. Promiscuous mitochondria in Cryptococcus gattii. FEMS Yeast Res. 2009;9:489–503. doi: 10.1111/j.1567-1364.2009.00494.x. [DOI] [PubMed] [Google Scholar]
- 120.Xu J, Yan Z, Guo H. Divergence, hybridization, and recombination in the mitochondrial genome of the human pathogenic yeast Cryptococcus gattii. Mol. Ecol. 2009;18:2628–2642. doi: 10.1111/j.1365-294X.2009.04227.x. [DOI] [PubMed] [Google Scholar]
- 121.Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, Polacheck I, Boekhout T, May RC. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12980–12985. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu J, Ali RY, Gregory DA, Amick D, Lambert SE, Yoell HJ, Vilgalys RJ, Mitchell TG. Uniparental mitochondrial transmission in sexual crosses in Cryptococcus neoformans. Curr.Microbiol. 2000;40:269–273. doi: 10.1007/s002849910053. [DOI] [PubMed] [Google Scholar]
- 123.Yan Z, Hull CM, Sun S, Heitman J, Xu J. The mating type-specific homeodomain genes SXI1alpha and SXI2 a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr. Genet. 2007;51:187–195. doi: 10.1007/s00294-006-0115-9. [DOI] [PubMed] [Google Scholar]
- 124.Escandon P, Sanchez A, Firacative C, Castaneda E. Isolation of Cryptococcus gattii molecular type VGIII, from Corymbia ficifolia detritus in Colombia. Med. Mycol. 2010;48:675–678. doi: 10.3109/13693780903420633. [DOI] [PubMed] [Google Scholar]
- 125.Callejas A, Ordonez N, Rodriguez MC, Castaneda E. First isolation of Cryptococcus neoformans var. gattii, serotype C, from the environment in Colombia. Med. Mycol. 1998;36:341–344. [PubMed] [Google Scholar]
- 126.Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerg. Infect. Dis. 2010;16:14–20. doi: 10.3201/eid1601.090369. [DOI] [PMC free article] [PubMed] [Google Scholar]