Abstract
The ability of fungi to transition between unicellular and multicellular growth has a profound impact on our health and the economy. Many important fungal pathogens of humans, animals, and plants are dimorphic, and the ability to switch between morphological states has been associated with their virulence. Cryptococcus neoformans is a human fungal pathogen that causes life-threatening meningoencephalitis in immunocompromised and, in some cases, immunocompetent hosts. Cryptococcus neoformans grows vegetatively as a budding yeast and switches to hyphal growth during the sexual cycle, which is important in the study of cryptococcal pathogenicity because spores resulting from sexual development are infectious propagules and can colonize the lungs of a host. In addition, sexual reproduction contributes to the genotypic variability of Cryptococcus species, which may lead to increased fitness and virulence. Despite significant advances in our understanding of the mechanisms behind the development of C. neoformans, our knowledge is still incomplete. Recent studies have led to the emergence of many intriguing questions and hypotheses. In this review, we describe and discuss the most interesting aspects of C. neoformans development and address their impact on pathogenicity.
Keywords: morphogenesis, mating, sexual development, fungi, MAPK pathway, dimorphic fungi
Introduction
Some fungal species can shift between morphological forms during their life cycles. While mushrooms with elaborate and impressive fruiting bodies are considered monomorphic because their only morphological form is hyphae that develop directly from spores after germination, dimorphic fungi proliferate as unicellular organisms and can switch to multicellular growth under specific conditions. Dimorphic fungi are attractive models to study development due to their relatively simple morphological transition; for example a paradigm for morphological differentiation is the ascomycete Saccharomyces cerevisiae, whose yeast to pseudohyphal growth change has been studied extensively (Rua et al., 2001).
The ability of fungi to switch between unicellular and multicellular hyphal growth not only constitutes an important biological phenomenon, but also has a profound impact on our health and the economy. Many important fungal pathogens of humans, animals, and plants are dimorphic, and the ability to switch between morphological states has been associated with their virulence. Prominent examples of species with virulence-linked morphological transitions include the human opportunistic pathogen Candida albicans (Sudbery et al., 2004; Berman, 2006) and the maize pathogen Ustilago maydis (Nadal et al., 2008; Steinberg & Perez-Martin, 2008; Brefort et al., 2009). Several known systemic dimorphic fungi belonging to the ascomycete lineage grow as a spore-producing mold in the environment at ambient temperature and switch to yeast growth after being inhaled into the lungs of a human host as spores (Klein & Tebbets, 2007). Therefore, examining the mechanistic basis for morphological transitions in fungi may lead to the development of more effective antifungal therapies and strategies to prevent devastating crop diseases.
Cryptococcus neoformans is a human fungal pathogen that causes life-threatening meningoencephalitis in immunocompromised and, in some cases, immunocompetent hosts (Idnurm et al., 2005). Two C. neoformans serotypes are currently recognized: the most common causative agent of cryptococcosis, serotype A (C. neoformans var. grubii), and the relatively less virulent serotype D (C. neoformans var. neoformans) (Kwon-Chung & Varma, 2006; Lin & Heitman, 2006). Cryptococcus gattii has been classified into two additional serotypes, B and C, but these are currently considered to be sibling species (Kwon-Chung & Varma, 2006; Lin & Heitman, 2006).
Cryptococcus neoformans grows vegetatively as budding yeast and can be frequently found in tree hollows and pigeon guano. During the sexual cycle, Cryptococcus switches from yeast growth to hyphal growth. Despite this dramatic morphological transition, Cryptococcus is not considered by some to be a dimorphic fungus because yeast cells are the predominant form in the environment and in the human host, and it is likely that the morphological transition is not involved in infection. However, there are at least three important reasons why the development of Cryptococcus is relevant to its pathogenicity. First, spores that result from hyphal development during mating are infectious propagules. Upon inhalation, spores (in addition to desiccated yeast) can colonize the lungs of a host. Cryptococcus neoformans propagates to the bloodstream and crosses the blood–-brain barrier, ultimately colonizing brain tissue and leading to fatal consequences if not treated. Second, sexual reproduction contributes to the genotypic variability of Cryptococcus species, which may lead to increased fitness and virulence. Third, some genes located within the MAT locus are important during mating and during infection. Therefore, the development of Cryptococcus is not only an interesting paradigm for biologists, but is also important in the study of cryptococcal pathogenicity.
Despite significant advances in our understanding of the mechanisms behind the Cryptococcus development, our knowledge is still limited. Recent studies have led to the emergence of intriguing questions and hypotheses, many of which are discussed below. Several excellent reviews describe the sexual reproduction of Cryptococcus, and the species’ mating-type locus structure and the signal transduction involved in mating (Wang & Heitman, 1999; Idnurm et al., 2005; Nielsen & Heitman, 2007; Hsueh & Heitman, 2008; Lin, 2009; Morrow & Fraser, 2009; Kruzel & Hull, 2010; Raudaskoski & Kothe, 2010; Wang, 2011), or discuss the unique properties of the Cryptococcus mating types (McClelland et al., 2004). In this review, we describe development in a broad sense as the events leading to all morphological transitions that contribute to the proliferation and increased fitness of C. neoformans both in the environment and within a human host. While we discuss the most salient aspects and questions of cryptococcal development, we refer the reader to a recently published work describing Cryptococcus biology for more detailed information (Heitman et al., 2011).
Morphological forms of C. neoformans
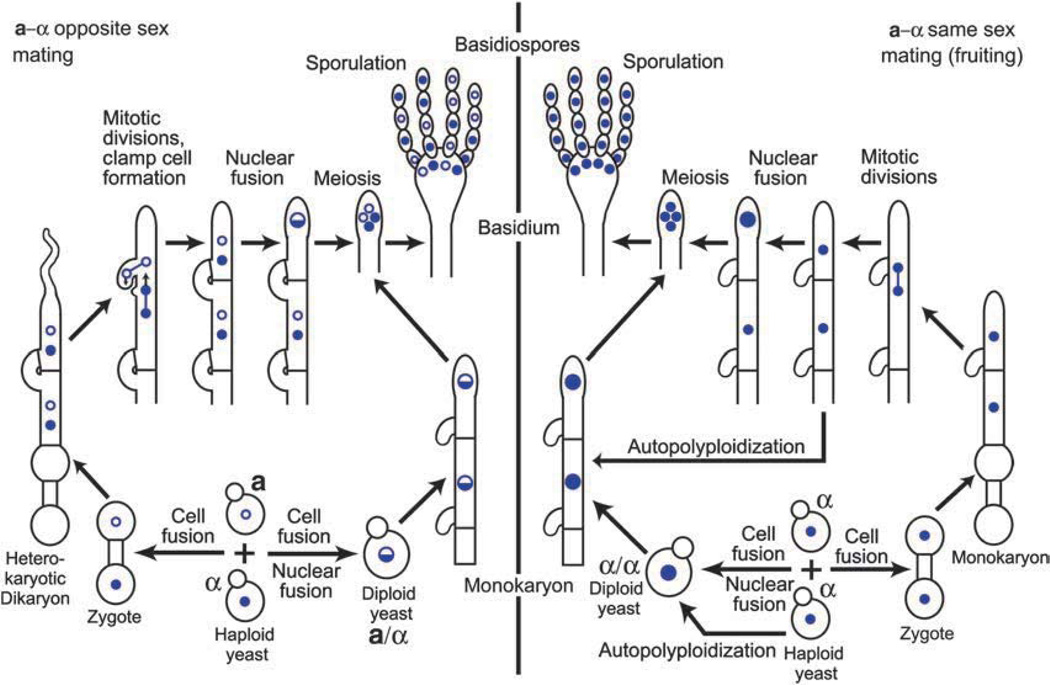
The Cryptococcus life cycle consists of vegetative and sexual growth phases (Fig. 1). Cryptococcus neoformans exists in at least two morphological states during vegetative growth. The most prevalent form in its natural habitat and in clinical samples is unicellular budding yeast, which reproduce by mitotic division. Alternative vegetative forms are pseudohyphae. Pseudohyphae are linked yeast cells that do not completely separate after mitotic divisions and serve as an intermediate form between yeast and true hyphae. Cryptococcus neoformans pseudohyphae have only occasionally been reported in clinical samples (Shadomy & Utz, 1966; Freed et al., 1971; Lurie & Shadomy, 1971; Williamson et al., 1996; Gazzoni et al., 2009) and in the environment and may represent a strategy to avoid natural predators (Neilson et al., 1978). Similar to S. cerevisiae, C. neoformans utilizes the RAM pathway to control the switch from yeast to pseudohyphae, but mechanistic details of this transition are lacking at present (Walton et al., 2006; Verma-Gaur et al., 2008).
Fig. 1.
Sexual cycle of Cryptococcus neoformans. During the opposite-sex mating, a and α haploid yeast cells secrete peptide pheromones that stimulate cell–cell fusion. The resulting zygote develops first as hyphae, which are dikaryotic. After the first clamp cell is formed, a narrow ‘pioneer’ hypha is present at the apex. During hyphal growth, a basidium develops from the apical cell. Nuclear fusion and meiosis take place most likely concomitant with the formation of the basidium. Postmeiotic nuclei undergo rounds of mitotic divisions and four chains of spores are formed by subsequent budding from the surface of the basidium. Alternatively, a and α haploid yeast cells can form an a/α diploid, which grows as yeast at 37 °C in a rich medium and forms hyphae at 24 °C on mating-inducing medium. These diploid-derived hyphae are monokaryotic, have unfused clamp cells, and produce a and α spores. During same-sex mating, two yeast cells of the same mating type (α is depicted) undergo fusion and form monokaryotic hyphae with haploid nuclei. Concomitant with the formation of the basidium, nuclear fusion and meiosis occur. The resulting recombinant spores are of a single mating type. A single yeast cell can undergo autopolyploidization (i.e. endoreplication), resulting in a diploid, which develops into monokaryotic hyphae with diploid nuclei and unfused clamp cells. Autopolyploidization can also occur in the hyphae. Both opposite- and same-sex mating hyphae can develop chlamydospores and blastospores (not depicted) (see the text for a more detailed description).
Mating of C. neoformans results in a third growth form: hyphal growth. During mating, two compatible yeast cells fuse, but the two parental nuclei remain separate, leading to the formation of dikaryotic hyphae, a hallmark feature of basidiomycetes (Fig. 1). Another basidiomycete-specific characteristic is the specialized clamp cells that form between each cellular compartment in the hyphae to maintain the dikaryotic state. The ultimate developmental stage in the sexual cycle of C. neoformans occurs in a terminal, specialized cell called the basidium, in which the fusion of the parental nuclei and meiosis take place. Nuclear meiotic products undergo rounds of mitotic division, and the mitotic nuclei are packaged into spores that bud from the apical surface of the basidium to form four spore chains, a feature that distinguishes the genus Filobasidiella from other members of the Basidiomycota. Upon germination, spores develop into yeast, which concludes the sexual cycle. Opposite- sex mating can also produce diploids that grow as yeast at 37 °C and filament under inducing conditions, resulting in monokaryotic hyphae, which are decorated with unfused clamp cells and capable of producing basidia and spores (Sia et al., 2000).
In addition to classical opposite-sex mating, C. neoformans can also undergo same-sex mating, which is referred to as monokaryotic or haploid fruiting (Wickes et al., 1996; Lin et al., 2005) and was originally reported by Erke (1976) as homothallism. Similar to opposite-sex mating, monokaryotic fruiting also results in the formation of hyphae and leads to the formation of basidia and spores. However, the filaments are monokaryotic and the clamp cells are not fused, similar to hyphae produced by a/α diploids. Although this unusual form of sexual reproduction is not α-mating type specific (Tscharke et al., 2003), it is strongly associated with the α-mating type (Wickes et al., 1996; Lin et al., 2006).
Mating is a possible source of hybrid C. neoformans species. Hybrids are most frequently formed between the A and the D serotypes of either the same or opposite mating types (αADa, aADα, αADα) and are of significant clinical importance (Litvintseva et al., 2005a; Lin et al., 2007). Diploids derived from genetically distinct strains of the same serotype (αAAα) have also been reported (Lin et al., 2009). Interestingly, even hybrids between C. neoformans and C. gattii have been described from the environment (Bovers et al., 2006) and in clinical samples (Bovers et al., 2008).
Two groups recently described a peculiar morphological form of C. neoformans that occurs during infection (Okagaki et al., 2010; Zaragoza et al., 2010). These studies found unusually large yeast-like Cryptococcus cells in clinical samples, now known as giant cells, and it remains to be elucidated how this morphological change is triggered.
Cryptococcus neoformans mating
Cryptococcus neoformans is a heterothallic fungus with a bipolar mating system in which a single mating locus defines each mating type (a and α) (Kwon-Chung, 1976b). The mating-type locus of C. neoformans is significantly larger than the MAT loci of most other fungi, spanning over 100 kb with > 20 genes. Several MAT locus genes function in mating, including genes encoding the Ste20 p21-activated kinase (PAK), the MAPKKK Ste11, and the transcription factor Ste12 (Lengeler et al., 2002; Fraser et al., 2004). The bipolar MAT locus of Cryptococcus may have evolved from an ancestral tetrapolar MAT system through a series of genomic rearrangements (Fraser et al., 2004; Hsueh et al., 2008). Similar to other fungi, the cell-type identity of Cryptococcus is specified by pheromone and receptor genes and homeodomain transcription factor genes within the MAT locus that serve as master regulators of sexual reproduction (Lengeler et al., 2002).
Cryptococcus species and serotypes vary in their ability to undergo the sexual cycle. Most serotype D strains mate, while the mating ability of serotype A and C. gattii is strain specific (Fraser et al., 2003; Halliday & Carter, 2003; Nielsen et al., 2003). However, the genomes of all strains suggest that the ability to undergo sexual reproduction is conserved.
Mating of C. neoformans has never been directly observed in nature or within the host, and only specific conditions in the laboratory can trigger sexual reproduction between compatible yeast cells (Kwon-Chung, 1976a; Heitman, 2006). However, both mating and sporulation have been observed on media containing pigeon guano and on live plants under laboratory conditions, suggesting that the sexual cycle occurs occasionally in its natural environment (Nielsen et al., 2007; Xue et al., 2007). This is further supported by recent population genetics studies that provide compelling evidence for sexual reproduction in both C. neoformans and C. gattii in nature (Campbell et al., 2005; Bui et al., 2008; Hiremath et al., 2008). Currently, the filamentous form (teleomorph) of C. neoformans is classified under the genus Filobasidiella, which also contains the closely related species Filobasidiella depauperata (Heitman et al., 2011). Interestingly, F. depauperata grows only as hyphae and has no yeast state (Rodriguez-Carres et al., 2010). It will be interesting to uncover the underlying genetic and/or epigenetic factors that govern yeast and hyphal growth in these sister species.
One of the interesting, but unexplained aspects of C. neoformans biology is the overwhelming predominance of the α-mating type in the environment and clinical isolates. This phenomenon could explain why mating in nature is rare (Idnurm et al., 2005). One exception is the recently documented population of Cryptococcus in sub-Saharan Africa, which contains an equal ratio of a- and α-mating-type cells and has a relatively high recombination rate (Litvintseva et al., 2003, 2005b). In serotype D, the α-mating type is more virulent than the a-mating type (Kwon-Chung et al., 1992), which led to the hypothesis that α cells have a higher fitness than the a cells (McClelland et al., 2004). However, in serotype A, both mating types have similar virulence (Nielsen et al., 2003). Therefore, the mechanism underlying the skewed mating-type proportion in natural populations is still unknown.
What signals trigger mating in C. neoformans?
In contrast to S. cerevisiae, which prefers the diploid state in nature and for which pheromone stimulation is sufficient to initiate mating, C. neoformans proliferates as a haploid yeast and specific environmental conditions in addition to pheromone signals are necessary to initiate the sexual cycle. Several external cues can promote or inhibit mating in C. neoformans. Nutritional signals that promote mating have been identified by generating defined media and analyzing media of unknown compositions. Among these factors, myo-inositol, copper ions, and nitrogen starvation were most significant (Xue et al., 2007; Kent et al., 2008). These results were also supported by genetic studies (Torres-Guererro & Edman, 1994; Walton et al., 2005; Rutherford et al., 2008). Xue et al. (2007) established that mating can occur on plant surfaces using Arabidopsis thaliana and Eucalyptus camaldulensis under laboratory conditions. Furthermore, myo-inositol and the plant hormone indole acetic acid were identified as two major plant-derived stimulators of mating (Xue et al., 2007). Cryptococcus neoformans var. grubii and var. neoformans, but not C. gattii grow and mate robustly on media prepared with pigeon guano (Staib, 1981; Staib & Blisse, 1982), indicating that the nutritional composition of pigeon guano provides a highly favorable environment for C. neoformans growth and mating. The actual components in pigeon guano that stimulate mating are unknown, but these studies provide an insight into why avian excreta are a common ecological niche for C. neoformans.
Three environmental factors found in the human host inhibit the mating of C. neoformans: a temperature of 37 °C, high humidity, and 5% carbon dioxide (CO2). This is consistent with the hypothesis that mating does not occur during infection. Similar to some thermodimorphic fungal pathogens such as Histoplasma capsulatum, a temperature of 37 °C promotes yeast growth of C. neoformans and blocks hyphal growth (Sia et al., 2000; Klein & Tebbets, 2007; Nguyen & Sil, 2008). Mechanisms of temperature-controlled morphogenesis in Cryptococcus are unknown, but the regulatory pathways may involve calcineurin and Ras1, as these components are essential for mating and growth at a high temperature (Odom et al., 1997; Alspaugh et al., 2000; Nichols et al., 2007; Kozubowski et al., 2009). Mating of C. neoformans is more efficient under dry compared with humid conditions and has never been observed in liquid, but the mechanism of this regulation is unknown.
Cell fusion during mating is inhibited by 5% CO2, likely because of decreased pheromone production (Bahn et al., 2005). Bahn et al. (2005) proposed that CO2-mediated mating inhibition results from increased intracellular levels of produced by the carbonic anhydrase Can2 at high CO2 levels. In the can2Δ mutant, the small amount of bicarbonate generated through nonenzymatic spontaneous hydration of CO2 is sufficient to support vegetative growth, but not to inhibit mating. However, proper development of basidia and sporulation requires CAN2; while sporulation is dependent on the correct nuclear distribution within hyphae, CAN2 was not associated with nuclear migration (Bahn et al., 2005).
Mating of C. neoformans on a medium supplemented with V8 juice is inhibited by continuous exposure to white light, whereas mating on plants is not significantly inhibited by light (Xue et al., 2007). Two conserved photoreceptor genes, BWC1 and BWC2, mediate mating inhibition by light (Idnurm & Heitman, 2005; Lu et al., 2005; Yeh et al., 2009). Idnurm & Heitman (2005) demonstrated that Bwc1 and Bwc2 interact physically in a two-hybrid assay and function either directly or indirectly to repress the transcription of MFα1 and SXI1α, two key genes that regulate mating and completion of the sexual cycle. Accordingly, BWC1 and BWC2 were required for efficient light-driven inhibition of cell fusion (dependent on MFα1) and subsequent filamentation (dependent on SXI1α). The inhibition of mating by light, specifically UV irradiation, which induces DNA damage, may have evolved to avoid the DNA damage-sensitive process of meiosis (Idnurm & Heitman, 2005; Heitman et al., 2011). Interestingly, bwc1Δ and bwc2Δ mutants were significantly less virulent in a murine model of infection, suggesting an intriguing connection between the ability to sense light and pathogenicity (Idnurm & Heitman, 2005).
Cell fusion, the initial step during sexual development
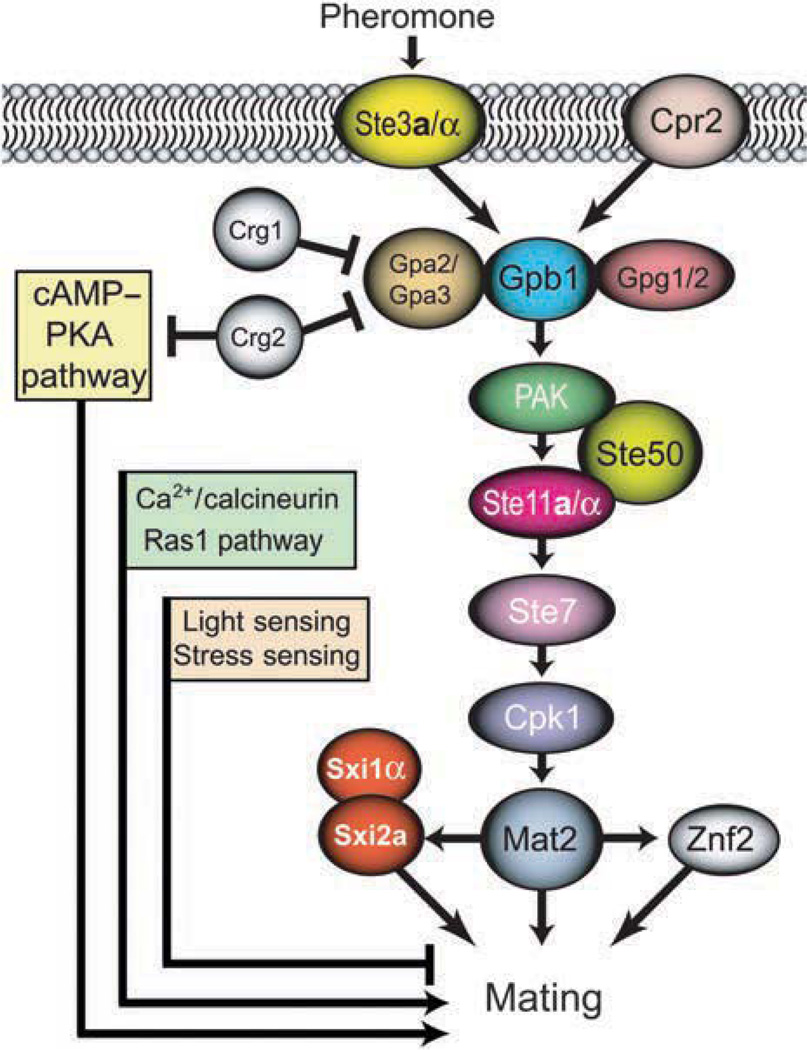
For cell fusion to occur, at least one mating cell usually produces a conjugation tube directed towards the mating partner. This early mating response is orchestrated by a dedicated mating signaling pathway. Pheromone stimulation in C. neoformans is an essential, although not sufficient, mating signal necessary for fusion to occur between opposite mating-type cells. The mating-type locus of each mating type encodes specific pheromones and receptors that determine the compatibility of mating partners (Stanton et al., 2010). Similar to S. cerevisiae, in C. neoformans, pheromone stimulation of the pheromone receptor transduces the signal through heterotrimeric G proteins and triggers signaling via the mitogen-activated protein kinase (MAPK) signaling pathway (Fig. 2) (Alspaugh et al., 1998; Wang & Heitman, 1999).
Fig. 2.
The pheromone response pathway in Cryptococcus neoformans. Other pathways that either positively regulate sexual development (cAMP-PKA pathway, Ca2+-calcineurin pathway) or inhibit mating (light sensing, stress sensing) are also indicated (see the text for details).
Two G protein-dependent signaling pathways that influence mating in C. neoformans have been characterized. One pathway responds to pheromone while the other is stimulated by nutritional signals (Fig. 2) (Alspaugh et al., 1998; Wang & Heitman, 1999). Similar to S. cerevisiae, in C. neoformans, GTP binding activates the G protein α subunit when pheromone binds to the pheromone receptor, releasing the Gβγ complex to activate downstream effectors. Despite similarities, G protein signaling in C. neoformans is more complex than in S. cerevisiae due to the more complex nature of its development. For example, among the three G protein α subunits expressed in C. neoformans (Gpa1, Gpa2, and Gpa3), Gpa1 functions upstream of adenylyl cyclase and protein kinase A (PKA) and is required for the pathogenesis and development of C. neoformans, while Gpa2 and Gpa3 are involved in mating (Alspaugh et al., 1997, 2002; D’Souza et al., 2001). Similarly, GIP2 and GPB1 encode β-subunits that function in nutrient sensing and mating, respectively (Wang et al., 2000; Hsueh et al., 2007; Li et al., 2007), and γ- subunits are encoded by GPG1 and GPG2 (Li et al., 2007). Both the Gpa2 and the Gpa3 α subunits interact physically with the pheromone receptor Ste3. However, only Gpa2 was shown to interact with the β-subunit Gpb1 (Li et al., 2007). Moreover, while the expression of GPA2 is induced during mating, GPA3 is induced by nutrient limitation (Hsueh et al., 2007) and the two genes seem to have both overlapping and opposing functions in pheromone sensing (Hsueh et al., 2007; Li et al., 2007). The α subunits are negatively regulated by the RGS domain proteins Crg1 and Crg2, which stimulate GTP hydrolysis. Similar to what was described in S. cerevisiae, Crg1 is in a complex with the pheromone receptor Ste3, and Crg2, which is membrane bound, also interacts with the pheromone receptor, possibly with a lower affinity (Hsueh et al., 2007). While Crg1 is induced by pheromone and acts exclusively in pheromone sensing, Crg2 is constitutively expressed and can inhibit both the mating and the Gpa1-cAMP pathways (Hsueh et al., 2007; Shen et al., 2008; Xue et al., 2008). The cAMP/PKA pathway regulates responses to several nutritional starvation signals, including nitrogen, iron, and glucose, and may be responsible for the nutritional regulation of the MFα1 gene (Alspaugh et al., 1997). In addition, mutants lacking the α subunit Gpa1 exhibit mating defects, further suggesting cross-talk between the pheromone signaling and the cAMP/PKA pathway.
Similar to other fungi, the MAPK cascade transduces the signal triggered by pheromone in C. neoformans (Fig. 2) (Davidson et al., 2003). Upon pheromone stimulation, the Gβγ subunit heterodimer activates a member of the PAK family. Cryptococcus neoformans genome encodes two PAK homologues, Ste20 and Pak1, which together are essential for cell viability (Wang et al., 2002) and involved in mating (Wang et al., 2002; Nichols et al., 2004). PAK transduces the signal to the MAPK pathway components Ste11 (MAPKKK), Ste7 (MAPKK), and Cpk1 (MAPK), resulting in the activation of the mating response. Another conserved component of the mating pathway was described recently: the adaptor protein Ste50 (Jung et al., 2011). Unlike in S. cerevisiae, where Ste50 acts in multiple MAPK pathways, C. neoformans Ste50 appears to be mating specific. Further, it does not appear to be important for virulence, in contrast to the U. maydis homologue. Fu et al. (2011) have shown that the STE50 gene is required for response to pheromone, cell fusion, and the production of dikaryotic hyphae. Interestingly, Ste50 is also required for the production of monokaryotic hyphae during monokaryotic fruiting (Fu et al., 2011). The main role of Ste50 appears to be similar to the S. cerevisiae homologue in that it serves as an adapter protein for bringing Ste20 and Ste11 together to activate Ste11 (Fu et al., 2011).
Another unique characteristic of the C. neoformans MAPK pathway is that STE20 and STE11 are represented by mating-type-specific alleles (Clarke et al., 2001; Davidson et al., 2003). Initial cell fusion is blocked in ste11a/α, ste7, and cpk1 mutants (Davidson et al., 2003), but cell fusion is not dependent on Ste20 (Nichols et al., 2004), indicating that Pak1 may share this role.
A homologue of the S. cerevisiae pheromone-responsive transcription factor Ste12 has been cloned from C. neoformans (Wickes et al., 1997). STE12α plays only a minor role in mating, but may be one of several transcription factors regulating the response to pheromone (Yue et al., 1999; Davidson et al., 2003). Overexpression of STE12α restores mating and fruiting in MAPK mutants (Davidson et al., 2003) and dramatically induces the expression of the pheromone MFα1 gene, but the deletion of STE12α has no effect on MFα1 expression, suggesting that Ste12α does not function downstream of the MAPK cascade in a simple linear pathway (Davidson et al., 2003).
In S. cerevisiae, the response to pheromone involves two major events: cell cycle arrest and conjugation between two cells of opposite mating type. Cell cycle arrest is mediated by Fus3 (a member of the MAPK family) and its substrate Far1 by inhibiting G1 cyclins (Peter et al., 1993). It is unclear whether cell cycle arrest occurs in response to a pheromone signal in C. neoformans, and if it does, the stage of the cell cycle at which the arrest takes place (Kruzel & Hull, 2010).
Unlike in S. cerevisiae, where both mating partners extend a projection during mating, only one of the cells typically generates a projection in Cryptococcus. Some reports indicate that only cells of the α-mating type generate conjugation tubes, and cells of the a-mating type swell (Davidson et al., 2000; Wang et al., 2000; McClelland et al., 2004). This divergent response of cells of opposite mating type may be characteristic only of serotype D, because in serotype A, the formation of conjugation tubes by either mating partner has been observed and swelling of a cells does not seem to occur (Nichols et al., 2004; L. Kozubowski & J. Heitman, unpublished data). Future studies should determine whether conjugation tube formation is serotype specific.
Mating projections are visible by ~4 h, which coincides with an increase of pheromone transcript levels (Shen et al., 2002; Chang et al., 2003), suggesting that these phenomena are related. It is expected that polarisome components analogous to those in S. cerevisiae govern the formation of conjugation tubes in C. neoformans, but these mechanisms have not yet been explored in detail. A PAK homologue, Pak1, is necessary for the establishment of polarized protrusions during mating (Nichols et al., 2004). Surprisingly, Pak1 is not involved in pheromone induction during mating, in contrast to the role of its homologue in S. cerevisiae, Ste20 (Nichols et al., 2004).
After one or both cells of opposite mating type produce a conjugation tube, fusion between the two cells occurs. Under laboratory conditions, only a relatively small percentage of the cell population exhibits a mating response and undergoes fusion events. Less than four fusion events per 50 cells were reported in serotype A after 24 h (Nichols et al., 2004). The exact mechanism of cell fusion has not been studied in Cryptococcus. In S. cerevisiae, the initial cell wall attachment between two cells is mediated by GPI-anchored agglutinin proteins (Chen et al., 2007). This is followed by cell wall remodeling and the formation of a pore through which fusion of the plasma membrane takes place (White & Rose, 2001). Only two proteins are known to participate in cell fusion during mating in S. cerevisiae: Prm1 and Fig1. Only a Prm1 homologue is encoded in the C. neoformans genome, but its role has not been investigated (Heiman & Walter, 2000).
Development of postmating hyphae
A common feature of fungi belonging to the basidiomycete phylum is the formation of dikaryotic hyphae, which ultimately give rise to spore-producing structures. For example, U. maydis develops dikaryotic filaments upon mating that penetrate host tissue during plant infection (Banuett, 1992). The initial step in the regulation of the dikaryotic growth phase is conserved and involves homeodomain mating-type-specific transcription factors that heterodimerize (Kues & Casselton, 1992). Cell fusion in C. neoformans is followed by the development of hyphae that grow as a dikaryon until sporulation occurs (Figs 1 and 3). Then, the two homeodomain transcription factors Sxi1α and Sxi2a, which are derived from opposite mating partners, dimerize and trigger hyphal transition and development (Hull et al., 2002, 2005). The Sxi1α/Sxi2a heterodimer binds to DNA sequences that differ from sequences described in other fungi (Stanton et al., 2009). In the absence of one or the other homeodomain protein, cells can fuse, but subsequent hyphal development fails to progress. Hence, the Sxi1α/Sxi2a heterodimer plays a central role in establishing the zygotic developmental fate that follows cell fusion during mating. SXI1α and SXI2a expression is elevated upon cell fusion and does not require the presence of the Sxi1α/Sxi2a-regulatory heterodimer or pheromone, indicating that other mating-specific factors are responsible for this gene expression upregulation. However, the expression of SXI1α and SXI2a results in the repression of pheromone gene expression, underscoring the importance of such repression in sexual development following cell fusion. When Sxi1α was expressed in a cells or Sxi2a in α cells, complete sexual development with sporulation occurred, even in the absence of the opposite mating-type partner (Hull et al., 2002, 2005). In this artificially induced sexual cycle, the hyphae produced were monokaryotic, and the clamp cells did not fuse, reminiscent of same-sex mating or self-filamentation of a diploid (Sia et al., 2000).
Fig. 3.
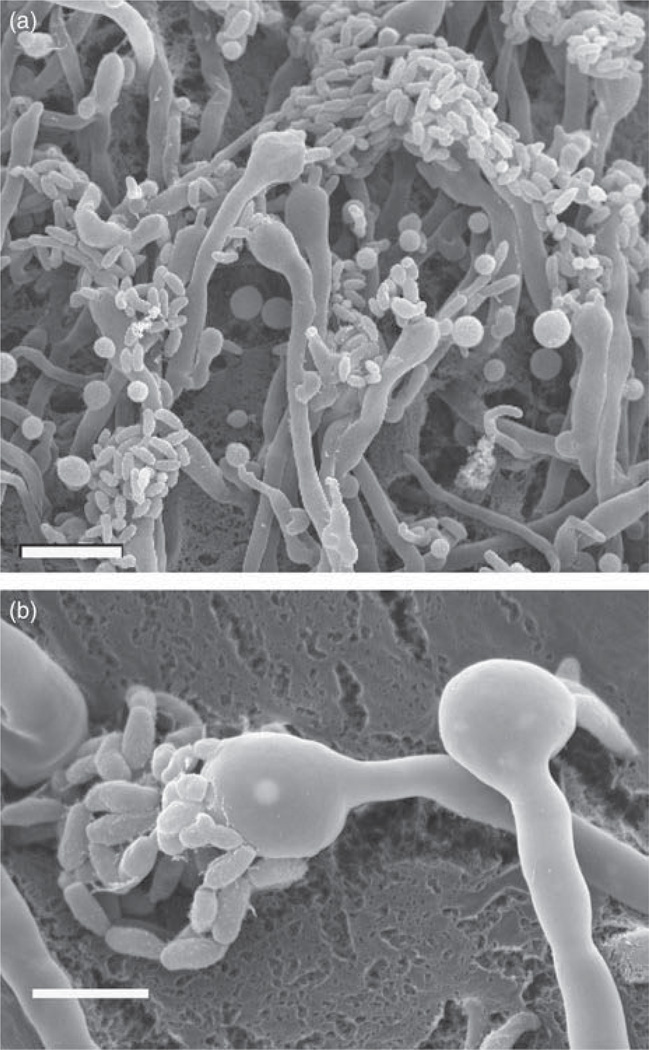
Postmating hyphae (a) of Cryptococcus neoformans serotype A and basidia (b) imaged by scanning electron microscopy. Images were taken by Lukasz Kozubowski (a) and Soo Chan Lee (b) and Valerie Knowlton (a, b).
After fusion has occurred, only one of the parental cells initiates hyphal growth. McClelland et al. (2004) demonstrated that the initial hypha always originates from the a-mating type. This phenomenon is one explanation for the uniparental mitochondrial inheritance observed during mating in C. neoformans (Yan & Xu, 2003). The exclusive inheritance of mitochondria from the a-mating partner is influenced by the migration of the α nucleus to the a cell after cell fusion, an event that could contribute toward limiting or excluding the mitochondria of the α cell from the progeny. Uniparental mitochondrial inheritance requires SXI1α and SXI2a, suggesting that these transcription factors may control nuclear migration within the zygote (Yan et al., 2007).
While it is clear that SXI1α and SXI2a are essential for hyphal development following cell fusion, the mechanism and targets of these transcription factors are relatively unknown (Kruzel & Hull, 2010). One gene target homologous to the CLP1 gene first described in Coprinopsis cinerea is necessary for clamp cell formation (Inada et al., 2001; Ekena et al., 2008). In C. neoformans, CLP1 transcription is upregulated during mating and directly controlled by SXI1α and SXI2a. Following cell fusion, at least one copy of CLP1 is required for hyphal development. CLP1 is also essential for growth after cell fusion during the formation of diploids. This observation supports the hypothesis that Clp1 acts as a cell cycle regulator, enabling growth after cell fusion (Ekena et al., 2008). CLP1, but not SXI1α or SXI2a, is necessary for filamentation during same-sex mating, suggesting that Clp1 responds to an alternative signaling pathway during same-sex mating (Ekena et al., 2008). Because CLP1 deletion prevents hyphal formation similar to SXI1α and SXI2a disruption, it is difficult to establish whether these genes also control developmental events during subsequent hyphal growth. Expression of CLP1 from a regulatable promoter would address this question.
Similar to other basidiomycetes, C. neoformans clamp cells are formed during each hyphal cell division and facilitate proper nuclear distribution and maintenance of the dikaryotic state (Casselton & Olesnicky, 1998) (Fig. 1). The clamp cell develops at the most apical cell of the hypha as a bud, initially bending backwards to produce a hookshaped appendix that eventually fuses with the hyphal cell. Clamp cell development is coordinated with nuclear division of the two genetically distinct haploid nuclei. The nucleus that is closer to the apex of the hypha divides, with the two resulting nuclei moving to the hyphal cell and the clamp cell, whereas the subapical nucleus divides within the hyphal cell, maintaining the dikaryotic state. In basidiomycetes, clamp cell formation and coordinated nuclear divisions are controlled by homeodomain transcription factors, but clamp cell fusion is dependent on pheromone and pheromone receptor genes (Casselton & Olesnicky, 1998). Coprinopsis cinerea clampless 1 protein (Clp1) participates in clamp formation (Inada et al., 2001) and U. maydis Clp1 is required for the formation of clamp-like structures (Scherer et al., 2006); however, further investigations are required to establish whether C. neoformans Clp1 acts during clamp cell formation (Ekena et al., 2008).
Septins, which are filament-forming GTPases, localize to the base of the clamp cell near the point of fusion between the clamp cell and the hypha (Kozubowski & Heitman, 2010). Septin mutants exhibit defects in nuclear distribution and fail to fuse clamp cells in the postmating hyphae, indicating that septins are involved in clamp cell development and nuclear migration. A lack of septins leads to an aberrant nuclear distribution during monokaryotic fruiting, a growth mode characterized by unfused clamp cells. This suggests that septins play a role in nuclear distribution independent of clamp cell fusion. However, it is possible that septins also contribute directly to clamp cell fusion during opposite-sex mating (Kozubowski & Heitman, 2010).
Hsueh et al. (2009) described a constitutively active G protein-coupled receptor in C. neoformans, Cpr2, which protects clamp cells from pheromones generated by other cells and promotes clamp cell fusion. During mating between cpr2Δ mutants, unusual haustorial filaments are produced at sites where clamp cells are normally initiated. Consistent with a hyphae-specific role, Cpr2 expression is highly induced during mating after cell fusion. Although cpr2Δ cells exhibit a modest defect in mating cell fusion assays, this is likely due to the requirement of Cpr2 for cell survival after cell fusion has occurred (Hsueh et al., 2009).
A septum forms immediately adjacent to the clamp cell to divide individual hyphal compartments. Cryptococcus neoformans possesses a specialized channel in the center of the septum called the dolipore, which facilitates communication between individual compartments of the hyphae and whose precise structure is unknown (Kwon-Chung & Popkin, 1976; Rhodes et al., 1981). Cryptococcus neoformans septin proteins localize to the medial point in the septa, presumably a dolipore, unlike in ascomycete fungi, where septins localize to the entire septum (Westfall & Momany, 2002; Kozubowski & Heitman, 2010). Therefore, the structure and function of the dolipore may depend on septins.
Whereas most hyphae are thick and straight for several days following cell fusion, an outgrowth of a relatively narrow, wandering hypha has been observed in serotype A C. neoformans after the first clamp cell is formed (Kozubowski & Heitman, 2010). These ‘pioneer hyphae’ grow either from hyphal tips or from clamp cells (Kwon-Chung, 1976a). The significance of pioneer hyphae is unknown, but it may allow the hyphal tip to forage for nutrients or contact neighboring hyphae to initiate hyphal connections. Pioneer hyphae are reminiscent of haustorial hyphae described in other fungi and frequently observed in the C. neoformans cpr2Δ mutant (Hsueh et al., 2009). Thus, the deletion of CPR2 may result in an intensification of a physiological phenomenon.
Postmating hyphae of C. neoformans are occasionally connected (Kwon-Chung, 1976a; Kozubowski & Heitman, 2010). Whether such connections are formed based on pheromone-mediated signaling or through self-fusion mechanisms characteristic of other fungi is not known (Read et al., 2009).
Mating filaments in C. neoformans grow significantly slower than in other ‘classic’ filamentous fungi, consistent with the presumed lack of a Spitzenkörper, a vesicle supply center at the growing apex of hyphal cells (Steinberg, 2007). Interestingly, the filamentous sibling species F. depauperata grows at a very slow rate, suggesting that the mode of hyphal growth in this fungus is analogous to the postmating hyphae generated in C. neoformans (Rodriguez-Carres et al., 2010).
Cryptococcus neoformans utilizes polarity-associated proteins to maintain polar extensions. One such protein, Ste20 (a homologue of S. cerevisiae Cla4), maintains filament polarity (Nichols et al., 2004). A bilateral ste20Δ cross results in the formation of aberrant filaments characterized by excessive branching, abnormal tip splitting, and aberrant basidia that do not produce spores (Nichols et al., 2004). Ste20 physically interacts with and is likely responding to the Rho-like GTPases Rac1 and Cdc42 (Vallim et al., 2005). Similar to Ste20, Rac1 is not required for cell fusion during mating and strains lacking Rac1 display an aberrant hyphal morphology in bilateral crosses, including shorter and thicker hyphae and extensive branching (Vallim et al., 2005). However, unlike ste20Δ mutants, rac1Δ mutants have no defect in sporulation, suggesting that the Ste20 PAK kinase plays a more substantial role in mating hyphal morphology than Rac1. Overexpression of either RAC1 or STE20α restores high-temperature growth to the ras1 mutant, suggesting that Ras1 signals through Rac1 and Ste20 (Alspaugh et al., 2000). While Ras1 functions in the initial events of mating, including pheromone production, cell fusion, and the initiation of filamentous growth, Rac1 and its effector kinase Ste20 may act specifically during hyphal growth (Alspaugh et al., 2000; Waugh et al., 2003; Nichols et al., 2004; Vallim et al., 2005).
Staudt et al. demonstrated a conserved role of the microtubule- associated protein Bim1 in hyphal morphology. Crosses between bim1Δ mutants produced unusually short and curved filaments with missegregated nuclei (Staudt et al., 2010). Similarly, in Schizophyllum commune, nuclear migration was associated with microtubule tracks and microtubule-associated motors (Raudaskoski, 1998).
Two types of growth associated with C. neoformans hyphae produce yeast cells. Oval-shaped cells called blastospores can form directly from hyphae by mitotic budding from the edge of the hyphal cell (Lin et al., 2005). Another mode involves yeast cells budding from chlamydospores. Chlamydospores form as a result of the conversion of the hyphal compartment into a large, round structure. Although chlamydospores in Cryptococcus were first reported 40 years ago to be abundant in postmating hyphae, they have only been described recently in C. neoformans (Kurtzman, 1973; Lin & Heitman, 2005). Chlamydospores are enriched in glycogen and may serve as energy stores, but the molecular mechanisms controlling chlamydospore formation are unknown (Lin & Heitman, 2005).
The ultimate stage of sexual development is the production of sexual spores. Basidiomycetes differ from other fungi in that they produce spores outside of the basidium, a specialized, spore-producing cell. Typically, a basidium is a unicellular, bottle-shaped structure originating from the terminal hyphal cell. In the terminal cell, two parental nuclei fuse (karyogamy) and undergo meiosis. The products from a single meiosis and subsequent mitotic divisions give rise to spores that bud from the surface of the basidium in chains (Idnurm, 2010).
Cryptococcus neoformans produces holobasidia from which four spore chains grow (Figs 1 and 3). The mechanism and signals responsible for initiating karyogamy and differentiation of the terminal cell into basidium are not known. Microscopic studies show four nuclei clustered together in some terminal hyphal cells, suggesting that nuclear fusion and meiosis can occur before the terminal cell transforms into the final, globose-like shape characteristic of the basidium (Kozubowski & Heitman, 2010) (Fig. 1). Ploidy-dependent cell growth is a well-established phenomenon (Kondorosi et al., 2000; Larkins et al., 2001); it is possible that nuclear fusion or the subsequent meiosis trigger a signal for the morphological transition. Alternatively, morphological changes may be necessary for nuclear fusion to occur. The deletion of genes encoding the meiosis-specific proteins Dmc1 and Spo11 severely affects sporulation, but basidia form, suggesting that a morphological transition to basidium is not dependent on meiosis (Lin et al., 2005). Whether nuclear fusion with meiosis and the morphological transition from hypha to basidium are dependent on one another will require further studies.
The formation of four spore chains on the basidium is an interesting biological phenomenon. It is unclear what governs the establishment of four areas of polarized growth on the surface of the basidia. This number correlates with four meiotic products, suggesting that the number of nuclei influence the initial protrusions. Even more intriguing is that subsequent mitotic divisions may occur simultaneously within the basidium. In some basidiomycetes, the basidium is divided into four compartments that could potentially provide spatial isolation for the budding of individual protrusions (Boekhout et al., 1991); however, Cryptococcus produces uncompartmentalized holobasidia. Moreover, when nuclei are visualized in the basidium, five or six nuclei are frequently observed in one single cell (Kozubowski & Heitman, 2010). This suggests that postmeiotic mitosis and the budding of subsequent spores are not tightly coordinated.
Several C. neoformans mutations have been described that result in defects in spore chain formation. For example, a lack of spore chains was reported in strains lacking CAN2, or CRG2, which encode β-carbonic anhydrase, and a regulator of G protein signaling, respectively (Bahn et al., 2005; Xue et al., 2008). In the presence of 5% CO2, hyphae of the can2Δ mutant do not develop proper basidia and spores, indicating that the conversion of CO2 to by Can2 is necessary for sporulation (Bahn et al., 2005).
In contrast to the can2Δ mutant, postmating hyphae of the crg2Δ mutant form normal basidia, but sporulation efficiency was significantly reduced (Xue et al., 2008). Xue et al. (2008) speculated that hyperactive PKA signaling (via Crg2 or dominant active Gpa1) and inhibition of PKA signaling (via reduced cyclase activation in can2Δ mutants) impair the production of long spore chains. Spore chain production is also affected in mutants lacking Cdc42 homologues (Ballou et al., 2009); these defects may be associated with the inability to assemble septins at sites of polarized growth, given that cdc42Δ mutants exhibit defects in septin organization and septin mutants fail to sporulate properly (Ballou et al., 2009; Kozubowski & Heitman, 2010).
Same-sex mating
When exposed to environmental conditions conducive for mating, a haploid C. neoformans strain can produce hyphae decorated with basidia and spore chains, similar to hyphae resulting from mating between opposite-sex partners (Fig. 1) (Shadomy & Utz, 1966; Erke & Schneidau, 1973). This type of haploid-derived hyphal growth was initially considered asexual haploid fruiting that only occurred in the α-mating type (Wickes et al., 1996), but the exclusive ability of α cells to fruit was subsequently challenged when others demonstrated that mating type a cells can also undergo haploid fruiting (Tscharke et al., 2003; Lin et al., 2006). The discovery of genetic recombination and ploidy changes during haploid fruiting revealed that this form of growth is an alternative mode of sexual reproduction involving only one mating type (Lin et al., 2005; Wang, 2011). Haploid fruiting is also referred to as monokaryotic fruiting, unisexual reproduction, or same-sex mating. The efficiency of same-sex mating is relatively low, and it is not a common phenomenon; in serotype D strains, fruiting of the a-mating type is rare, and neither of the serotype A mating types derived from H99 strain can self-filament under laboratory conditions. Similar to opposite-sex mating, same-sex mating has not been observed directly in the environment. The biological significance of same-sex mating is evident from population genetics studies on environmental and clinical isolates (Lin et al., 2007, 2009; Bui et al., 2008; Saul et al., 2008). These studies demonstrate that same-sex mating plays an important role in generating diversity in C. neoformans populations and contributes to the global spread of cryptococcosis. Given the predominance of the α-mating type in nature, same-sex mating may be one way in which C. neoformans can undergo genetic recombination in the environment (Lin et al., 2005).
It is striking that both same-sex and opposite-sex mating are triggered and regulated by similar environmental conditions and involve the pheromone signaling pathway. However, same-sex mating is a significantly more plastic process compared with opposite-sex mating and relies on alternative signaling pathways in addition to pheromone signaling (Wang, 2011). This is exemplified by several mutants defective in bisexual mating, but capable of unisexual mating (Hsueh & Shen, 2005; Hsueh et al., 2007; Ekena et al., 2008). During opposite-sex mating, a diploid nucleus is formed by nuclear fusion before sporulation, but diploidization in same-sex mating may occur via endoreplication (Lin et al., 2005). Under a low magnification, hyphae produced during same-sex mating resemble those resulting from opposite-sex mating, but appear more sporadically. However, unlike opposite-sex mating hyphae, hyphal cells produced by same-sex mating contain single nuclei (monokaryotic), clamp cells do not fuse, and spores are smaller and rounder (Wickes et al., 1996; Lin et al., 2005).
Sexual development and RNAi
RNAi is a sequence-specific gene silencing mechanism involving small (20–30 nucleotides) RNAs (Hannon, 2002). A novel repeat-silencing mechanism during C. neoformans mating was discovered recently: sex-induced silencing (SIS) (Wang et al., 2010). Wang et al. conducted a comparative transcriptome analysis and identified a group of retrotransposons located in the candidate centromeric regions that were highly induced in the RNAi-deficient strain specifically under mating conditions. Wang et al. demonstrated that an increase of transposition during mating is prevented through SIS, which involves the classic RNAi components Dicer, Argonaute, and an RNA-dependent RNA polymerase. Mating between serotype A strains lacking some RNAi components resulted in significant morphological abnormalities in postmating hyphae, frequently characterized by defects in spore chain formation. Wang et al. hypothesize that SIS plays a dual role as a defense mechanism against introduced repeats of foreign DNA and against sex-induced transposition. Silencing occurred before the onset of meiosis, and regulation was at the level of translation of the RNAi components. This phenomenon likely involves other mating- dependent mechanisms, because the translation of RNAi components was also induced under nutrient-limiting conditions without the mating partner (Wang et al., 2010). This also indicated that the translational regulation of RNAi components is not specific to sexual reproduction and may also occur during stress, but the exact function of RNAi during stress remains to be defined.
Several fungal species in the ascomycete and basidiomycete lineages have lost the RNAi machinery, indicating that this form of gene silencing is not essential. Cryptococcus neoformans serotype A contains all known components of the RNAi machinery, including two Dicer homologues (Dcr1 and Dcr2), one Argonaute homologue (Ago1), and one RNA-dependent RNA polymerase (Rdp1). Whereas the C. gattii VGI strain WM276 genome contains two Argonaute homologues, C. gattii strain VGII R265 lacks an Argonaute homologue, suggesting that this cryptic species may have lost the ability to silence through RNAi. This is striking because the R265 strain is the cause of the recent outbreak of cryptococcosis in the Pacific Northwest. Therefore, increased transposition activity may be one way in which R265 acquired increased virulence (Wang et al., 2010). In contrast, the C. neoformans serotype D strain JEC21 contains two Argonaute homologues. Whether this would result in more robust silencing or more RNAi-driven pathways operating in this strain will require further investigation (Wang et al., 2010).
SIS mechanisms acting to suppress transposable elements have been described in other organisms (Bourc’his & Bestor, 2004; Kelly & Aramayo, 2007). For example, Neurospora crassa has developed a number of genome defense strategies operating at different stages of its life cycle, including sex-specific repeat-induced point mutation and meiotic silencing of unpaired DNA (Selker, 1997; Cogoni & Macino, 1999; Galagan & Selker, 2004; Kelly & Aramayo, 2007). The mating-induced silencing described by Wang et al. represents a novel mechanism that does not involve unpaired DNA and is regulated by the translational induction of RNAi components. It is plausible that similar mechanisms are conserved in other fungal species, especially in those containing all of the RNAi machinery, but missing a meiotic silencing pathway (Kelly & Aramayo, 2007).
Development and virulence
Spores can infect animals and humans through inhalation, underscoring the critical role of sexual development in the virulence of C. neoformans (Giles et al., 2009; Velagapudi et al., 2009; Botts & Hull, 2010). In addition to spores, small, desiccated yeast cells are also candidates for infectious propagules of C. neoformans. Yeast is the most common morphological form in host tissue, although filamentous Cryptococcus during infection has been reported occasionally (Freed et al., 1971; Anandi et al., 1991;Williamson et al., 1996; Bemis et al., 2000). As described above, the host environment inhibits sexual development and hyphal growth. Experimentally introducing C. neoformans strains growing as hyphae to a model host confirmed the inability of filaments to persist during infection (Shadomy & Utz, 1966; Shadomy & Lurie, 1971; Zimmer et al., 1983). Strains collected from the environment as pseudohyphae were avirulent in a murine model of infection, while yeast cells derived from the same strain were pathogenic (Neilson et al., 1978). Thus, it appears that the formation of hyphae in C. neoformans during infection is rare and not advantageous for pathogenicity.
Several genes involved in the sexual development of Cryptococcus contribute to virulence. Interestingly, the requirement for a given gene for virulence is often dependent on the C. neoformans serotype. For example, the Ste12 transcription factor is needed for full virulence in serotype D, but is dispensable in serotype A (Yue et al., 1999). Conversely, Ste20 is required for virulence in serotype A, but not in D. The latter may be explained by the serotypespecific requirement for Ste20 for growth at 37 °C (Wang et al., 2002). At least in the case of Ste20, this is not due to a difference in the actual protein function between the two serotypes, but is instead associated with differences in related pathways.
Cryptococcus neoformans strains obtained from infected individuals or animals are highly variable with respect to yeast cell size (Cruickshank et al., 1973; Love et al., 1985; Feldmesser et al., 2001). Significant proportions of cells have been described as unusually large, often up to 10 times larger than cells found in the environment or cultured under laboratory conditions. Okagaki and colleagues used a murine inhalation model of cryptococcosis to detect yeast cells ranging from 10 to 100 µm in diameter in the host. These unusually large cells, referred to as giant cells, were mostly present in the lungs and were less abundant in the spleen and the brain (Okagaki et al., 2010). Cells of the a-mating type increased in size when present during coinfection with α-mating-type cells more frequently than either of the mating types alone, and this was dependent on the pheromone receptor Ste3. The authors hypothesize that pheromone sensing by a-mating-type cells during coinfection is necessary for this morphological change. This is particularly intriguing given that a-mating-type cells of serotype D typically swell in response to pheromone, while α-matingtype cells generate conjugation tubes. However, Okagaki and colleagues used a serotype A strain that normally does not swell during mating. Moreover, it is interesting that conditions in the host do not induce conjugation tubes in α-mating-type cells during coinfection. Importantly, giant cells were resistant to phagocytosis and oxidative and nitrosative stress (Okagaki et al., 2010; Zaragoza et al., 2010), which led to the hypothesis that cell gigantism in Cryptococcus may help the fungus avoid the early immune response. Giant cells are polyploid, but did not arrest during the cell cycle and produced daughter cells at a significantly higher rate than regular-sized yeast cells (Okagaki et al., 2010; Zaragoza et al., 2010). Zaragoza and colleagues visualized giant cells with multiple vesicles, a very thick cell wall, and an unusually thick, extensively cross-linked capsule. Zaragoza and colleagues also demonstrated that the cell gigantism is dependent on the cAMP pathway, but not on the Ras1 pathway, although it is conceivable that Ras1 could still be involved, given that the ras1Δ mutant produces larger cells at 37 °C in standard growth media. Nuclear staining and viability assays of ras1Δ giant cells obtained from mice may provide answers. Nonetheless, these data suggest that the pathways involved in mating (the pheromone and cAMP pathways) participate in this morphological transformation and contribute to the survival of C. neoformans during the early stages of infection.
Several features of giant cells are reminiscent of hyphae-derived chlamydospores, including a large size, a thick cell wall, and multiple vacuoles. Both giant cells and chlamydospores may serve a common purpose: to survive harsh environmental conditions. The signaling pathways involved in chlamydospore formation may differ from those involved in giant cell formation, as the formation of chlamydospores does not depend on the cAMP pathway (Lin & Heitman, 2005).
Concluding remarks
The first detailed description of cryptococcosis was made by Otto Busse in 1894 (Busse, 1894). Nearly a century later, with the advent of potent genetic tools and molecular biology techniques, we have started to uncover the biology of Cryptococcus and have learned some of the principles governing its pathogenicity.
The prevalent morphological form of C. neoformans in the environment and the host is a budding yeast. However, sexual differentiation in C. neoformans is well established and spores produced during mating may contribute to pathogenicity. Furthermore, some factors operating during mating contribute to virulence and it is now evident that C. neoformans undergoes a morphological transition to giant cells during infection, which influences disease progression. Despite these important connections, current knowledge of the development of C. neoformans remains limited. So far, the basic principles have been established, and a number of important questions have emerged that should stimulate progress in our understanding of the biology of C. neoformans.
It is apparent that C. neoformans is unique in many respects. While some paradigms from classic fungal model systems apply to its biology, many aspects are strikingly different. For example, mating is orchestrated by pheromone-activated G protein couple receptors and a MAPK pathway, but the detailed architecture of these signaling routes differs from the S. cerevisiae paradigm. Moreover, while we can apply certain principles established from other model yeasts, including S. cerevisiae, C. albicans, and S. pombe, studies on hyphal development must rely on the relatively less complete knowledge from other basidiomycetes.
There are several outstanding questions regarding the sexual development of C. neoformans. The transcriptional programs orchestrating sexual development, including the formation of basidia and sporulation, are largely unknown. Additional downstream targets of Sxi1 and Sxi2 and the mechanisms of how these homeodomain transcription factors govern hyphal growth are also still largely unknown. The mechanistic details of clamp cell formation and septation are lacking, and we do not know how nuclear distribution and dynamics are regulated in hyphae. Although the formation of blastospores and chlamydospores has been described, the exact mechanisms behind their development and the biological significance remain unclear. Despite useful genetic tools, precise evaluation of events leading to spore chain development has been challenging due to difficulties in conducting time-lapse microscopy of hyphae. Unlike yeast, which can be easily grown on a microscope slide, postmating hyphae develop basidia, which are mostly aerial and difficult to image. In addition, light inhibits mating and hyphal development. Overcoming these challenges would allow investigations to uncover the sequence of events that occur during spore chain formation.
Some other intriguing aspects of the biology of C. neoformans still remain unexplained. Most notably, the skewed population towards the α-mating type in the environment and clinical samples remains puzzling, despite evident genetic recombination. A better understanding of mechanisms orchestrating unisexual mating will help to resolve this conundrum. Recent findings on RNAi-dependent genome defense mechanisms during mating have opened a new area that needs further exploration. It will be important to establish other possible silencing mechanisms that operate in C. neoformans during mating and in response to cellular stress, including possible responses associated with infection of a human host. Future advances in deciphering the development of C. neoformans will not only help to understand basic biological phenomena, but will also contribute toward better treatments against this emerging pathogen.
Acknowledgements
We are indebted to Cecelia Shertz for careful editing of the manuscript and we also thank Theresa O’Meara for additional editing. We thank all contributors to studies on the development of C. neoformans and we apologize to colleagues whose studies were not cited due to space limitations. Our research was funded by the NIH/NIAID R01 grants AI39115, AI50113, and AI50438.
References
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Gene Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh JA, Perfect JR, Heitman J. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet Biol. 1998;25:1–14. doi: 10.1006/fgbi.1998.1079. [DOI] [PubMed] [Google Scholar]
- Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, Perfect JR, Kronstad JW, Heitman J. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anandi V, Babu PG, John TJ. Infection due to Cryptococcus neoformans of unusual morphology in a patient with AIDS. Mycoses. 1991;34:377–379. doi: 10.1111/j.1439-0507.1991.tb00798.x. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Cox GM, Perfect JR, Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol. 2005;15:2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Ballou ER, Nichols CB, Miglia KJ, Kozubowski L, Alspaugh JA. Two Cdc42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol Microbiol. 2009;75:763–780. doi: 10.1111/j.1365-2958.2009.07019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F. Ustilago maydis, the delightful blight. Trends Genet. 1992;8:174–180. doi: 10.1016/0168-9525(92)90220-x. [DOI] [PubMed] [Google Scholar]
- Bemis DA, Krahwinkel DJ, Bowman LA, Mondon P, Kwon-Chung KJ. Temperature-sensitive strain of Cryptococcus neoformans producing hyphal elements in a feline nasal granuloma. J Clin Microbiol. 2000;38:926–928. doi: 10.1128/jcm.38.2.926-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr Opin Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T, Fonseca A, Batenburg-van der Vegte WH. Bulleromyces genus novum (Tremellales), a teleomorph for Bullera alba, and the occurrence of mating in Bullera variabilis. Antonie van Leeuwenhoek. 1991;59:81–93. doi: 10.1007/BF00445652. [DOI] [PubMed] [Google Scholar]
- Botts MR, Hull CM. Dueling in the lung: how Cryptococcus spores race the host for survival. Curr Opin Microbiol. 2010;13:437–442. doi: 10.1016/j.mib.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, Dromer F, Hoogveld HL, Boekhout T. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- Bovers M, Hagen F, Kuramae EE, Hoogveld HL, Dromer F, St-Germain G, Boekhout T. AIDS patient death caused by novel Cryptococcus neoformans × C gattii hybrid. Emerg Infect Dis. 2008;14:1105–1108. doi: 10.3201/eid1407.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort T, Doehlemann G, Mendoza-Mendoza A, Reissmann S, Djamei A, Kahmann R. Ustilago maydis as a pathogen. Annu Rev Phytopathol. 2009;47:423–445. doi: 10.1146/annurev-phyto-080508-081923. [DOI] [PubMed] [Google Scholar]
- Bui T, Lin X, Malik R, Heitman J, Carter D. Isolates of Cryptococcus neoformans from infected animals reveal genetic exchange in unisexual, alpha mating type populations. Eukaryot Cell. 2008;7:1771–1780. doi: 10.1128/EC.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse O. Uber Parasitare Zelleinschlusse und ihre Zuchtung. Zentralbl Bacteriol. 1894;16:175–180. [Google Scholar]
- Campbell LT, Currie BJ, Krockenberger M, Malik R, Meyer W, Heitman J, Carter D. Clonality and recombination in genetically differentiated subgroups of Cryptococcus gattii. Eukaryot Cell. 2005;4:1403–1409. doi: 10.1128/EC.4.8.1403-1409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton LA, Olesnicky NS. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol Mol Biol R. 1998;62:55–70. doi: 10.1128/mmbr.62.1.55-70.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Miller GF, Kwon-Chung KJ. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect Immun. 2003;71:4953–4960. doi: 10.1128/IAI.71.9.4953-4960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EH, Grote E, Mohler W, Vignery A. Cell–cell fusion. FEBS Lett. 2007;581:2181–2193. doi: 10.1016/j.febslet.2007.03.033. [DOI] [PubMed] [Google Scholar]
- Clarke DL, Woodlee GL, McClelland CM, Seymour TS, Wickes BL. The Cryptococcus neoformans STE11alpha gene is similar to other fungal mitogen-activated protein kinase kinase kinase (MAPKKK) genes but is mating type specific. Mol Microbiol. 2001;40:200–213. doi: 10.1046/j.1365-2958.2001.02375.x. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- Cruickshank JG, Cavill R, Jelbert M. Cryptococcus neoformans of unusual morphology. Appl Microbiol. 1973;25:309–312. doi: 10.1128/am.25.2.309-312.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Moore TD, Odom AR, Heitman J. Characterization of the MFalpha pheromone of the human fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2000;38:1017–1026. doi: 10.1046/j.1365-2958.2000.02213.x. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2003;49:469–485. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- D’Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekena JL, Stanton BC, Schiebe-Owens JA, Hull CM. Sexual development in Cryptococcus neoformans requires CLP1, a target of the homeodomain transcription factors Sxi1alpha and Sxi2a. Eukaryot Cell. 2008;7:49–57. doi: 10.1128/EC.00377-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erke KH. Light microscopy of basidia, basidiospores, and nuclei in spores and hyphae of Filobasidiella neoformans (Cryptococcus neoformans) J Bacteriol. 1976;128:445–455. doi: 10.1128/jb.128.1.445-455.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erke KH, Schneidau JD., Jr Relationship of some Cryptococcus neoformans hypha-forming strains to standard strains and to other species of yeasts as determined by deoxyribonucleic acid base ratios and homologies. Infect Immun. 1973;7:941–948. doi: 10.1128/iai.7.6.941-948.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Casadevall A. Dynamic changes in the morphology of Cryptococcus neoformans during murine pulmonary infection. Microbiology. 2001;147:2355–2365. doi: 10.1099/00221287-147-8-2355. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JA, Diezmann S, Subaran RL, Allen A, Lengeler KB, Dietrich FS, Heitman J. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2004;2:e384. doi: 10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed ER, Duma RJ, Shadomy HJ, Utz JP. Meningoencephalitis due to hyphae-forming Cryptococcus neoformans. Am J Clin Pathol. 1971;55:30–33. doi: 10.1093/ajcp/55.1.30. [DOI] [PubMed] [Google Scholar]
- Fu J, Mares C, Lizcano A, Liu Y, Wickes BL. Insertional mutagenesis combined with an inducible filamentation phenotype reveals a conserved STE50 homologue in Cryptococcus neoformans that is required for monokaryotic fruiting and sexual reproduction. Mol Microbiol. 2011;79:990–1007. doi: 10.1111/j.1365-2958.2010.07501.x. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Selker EU. RIP: the evolutionary cost of genome defense. Trends Genet. 2004;20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Gazzoni AF, Severo CB, Barra MB, Severo LC. Atypical micromorphology and uncommon location of cryptococcosis: a histopathologic study using special histochemical techniques (one case report) Mycopathologia. 2009;167:197–202. doi: 10.1007/s11046-008-9169-1. [DOI] [PubMed] [Google Scholar]
- Giles SS, Dagenais TR, Botts MR, Keller NP, Hull CM. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect Immun. 2009;77:3491–3500. doi: 10.1128/IAI.00334-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday CL, Carter DA. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J Clin Microbiol. 2003;41:703–711. doi: 10.1128/JCM.41.2.703-711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Heiman MG, Walter P. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J Cell Biol. 2000;151:719–730. doi: 10.1083/jcb.151.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr Biol. 2006;16:R711–R725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- Heitman J, et al., editors. Cryptococcus from Human Pathogen to Model Yeast. Washington, DC: ASM Press; 2011. [Google Scholar]
- Hiremath SS, Chowdhary A, Kowshik T, Randhawa HS, Sun S, Xu J. Long-distance dispersal and recombination in environmental populations of Cryptococcus neoformans var. grubii from India. Microbiology. 2008;154:1513–1524. doi: 10.1099/mic.0.2007/015594-0. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Heitman J. Orchestration of sexual reproduction and virulence by the fungal mating-type locus. Curr Opin Microbiol. 2008;11:517–524. doi: 10.1016/j.mib.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Shen WC. A homolog of Ste6, the a-factor transporter in Saccharomyces cerevisiae, is required for mating but not for monokaryotic fruiting in Cryptococcus neoformans. Eukaryot Cell. 2005;4:147–155. doi: 10.1128/EC.4.1.147-155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans. Mol Biol Cell. 2007;18:3237–3249. doi: 10.1091/mbc.E07-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Fraser JA, Heitman J. Transitions in sexuality: recapitulation of an ancestral tri- and tetrapolar mating system in Cryptococcus neoformans. Eukaryot Cell. 2008;7:1847–1855. doi: 10.1128/EC.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. A constitutively active GPCR governs morphogenic transitions in Cryptococcus neoformans. EMBO J. 2009;28:1220–1233. doi: 10.1038/emboj.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Davidson RC, Heitman J. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1alpha. Gene Dev. 2002;16:3046–3060. doi: 10.1101/gad.1041402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Boily MJ, Heitman J. Sex-specific homeodomain proteins Sxi1alpha and Sxi2a coordinately regulate sexual development in Cryptococcus neoformans. Eukaryot Cell. 2005;4:526–535. doi: 10.1128/EC.4.3.526-535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics. 2010;185:153–163. doi: 10.1534/genetics.109.113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol. 2005;3:753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- Inada K, Morimoto Y, Arima T, Murata Y, Kamada T. The clp1 gene of the mushroom Coprinus cinereus is essential for A-regulated sexual development. Genetics. 2001;157:133–140. doi: 10.1093/genetics/157.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KW, Kim SY, Okagaki LH, Nielsen K, Bahn YS. Ste50 adaptor protein governs sexual differentiation of Cryptococcus neoformans via the pheromone-response MAPK signaling pathway. Fungal Genet Biol. 2011;48:154–165. doi: 10.1016/j.fgb.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Aramayo R. Meiotic silencing and the epigenetics of sex. Chromosome Res. 2007;15:633–651. doi: 10.1007/s10577-007-1143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent CR, Ortiz-Bermudez P, Giles SS, Hull CM. Formulation of a defined V8 medium for induction of sexual development of Cryptococcus neoformans. Appl Environ Microb. 2008;74:6248–6253. doi: 10.1128/AEM.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BS, Tebbets B. Dimorphism and virulence in fungi. Curr Opin Microbiol. 2007;10:314–319. doi: 10.1016/j.mib.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E, Roudier F, Gendreau E. Plant cell-size control: growing by ploidy? Curr Opin Plant Biol. 2000;3:488–492. doi: 10.1016/s1369-5266(00)00118-7. [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Heitman J. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol Microbiol. 2010;75:658–675. doi: 10.1111/j.1365-2958.2009.06983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11:370–380. doi: 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruzel EK, Hull CM. Establishing an unusual cell type: how to make a dikaryon. Curr Opin Microbiol. 2010;13:706–711. doi: 10.1016/j.mib.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kues U, Casselton LA. Homeodomains and regulation of sexual development in basidiomycetes. Trends Genet. 1992;8:154–155. doi: 10.1016/0168-9525(92)90207-k. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP. Formation of hyphae and chlamydospores by Cryptococcus laurentii. Mycologia. 1973;65:388–395. [PubMed] [Google Scholar]
- Kwon-Chung KJ. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia. 1976a;68:821–833. [PubMed] [Google Scholar]
- Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976b;68:943–946. [PubMed] [Google Scholar]
- Kwon-Chung KJ, Popkin TJ. Ultrastructure of septal complex in Filobasidiella neoformans (Cryptococcus neoformans) J Bacteriol. 1976;126:524–528. doi: 10.1128/jb.126.1.524-528.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Varma A. Do major species concepts support one, two or more species within Cryptococcus neoformans? FEMS Yeast Res. 2006;6:574–587. doi: 10.1111/j.1567-1364.2006.00088.x. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect Immun. 1992;60:602–605. doi: 10.1128/iai.60.2.602-605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkins BA, Dilkes BP, Dante RA, Coelho CM, Woo YM, Liu Y. Investigating the hows and whys of DNA endoreduplication. J Exp Bot. 2001;52:183–192. [PubMed] [Google Scholar]
- Lengeler KB, Fox DS, Fraser JA, Allen A, Forrester K, Dietrich FS, Heitman J. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot Cell. 2002;1:704–718. doi: 10.1128/EC.1.5.704-718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shen G, Zhang ZG, Wang YL, Thompson JK, Wang P. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol Biol Cell. 2007;18:4201–4209. doi: 10.1091/mbc.E07-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol. 2009;9:401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Lin X, Heitman J. Chlamydospore formation during hyphal growth in Cryptococcus neoformans. Eukaryot Cell. 2005;4:1746–1754. doi: 10.1128/EC.4.10.1746-1754.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- Lin X, Hull CM, Heitman J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature. 2005;434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- Lin X, Huang JC, Mitchell TG, Heitman J. Virulence attributes and hyphal growth of C. neoformans are quantitative traits and the MATalpha allele enhances filamentation. PLoS Genet. 2006;2:e187. doi: 10.1371/journal.pgen.0020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Litvintseva AP, Nielsen K, Patel S, Floyd A, Mitchell TG, Heitman J. alpha AD alpha hybrids of Cryptococcus neoformans: evidence of same-sex mating in nature and hybrid fitness. PLoS Genet. 2007;3:1975–1990. doi: 10.1371/journal.pgen.0030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Patel S, Litvintseva AP, Floyd A, Mitchell TG, Heitman J. Diploids in the Cryptococcus neoformans serotype a population homozygous for the alpha mating type originate via unisexual mating. PLoS Pathog. 2009;5:e1000283. doi: 10.1371/journal.ppat.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Marra RE, Nielsen K, Heitman J, Vilgalys R, Mitchell TG. Evidence of sexual recombination among Cryptococcus neoformans serotype A isolates in sub-Saharan Africa. Eukaryot Cell. 2003;2:1162–1168. doi: 10.1128/EC.2.6.1162-1168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Kestenbaum L, Vilgalys R, Mitchell TG. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J Clin Microbiol. 2005a;43:556–564. doi: 10.1128/JCM.43.2.556-564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in sub-Saharan Africa. J Infect Dis. 2005b;192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- Love GL, Boyd GD, Greer DL. Large Cryptococcus neoformans isolated from brain abscess. J Clin Microbiol. 1985;22:1068–1070. doi: 10.1128/jcm.22.6.1068-1070.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YK, Sun KH, Shen WC. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol Microbiol. 2005;56:480–491. doi: 10.1111/j.1365-2958.2005.04549.x. [DOI] [PubMed] [Google Scholar]
- Lurie HI, Shadomy HJ. Morphological variations of a hypha-forming strain of Cryptococcus neoformans (Coward strain) in tissues of mice. Sabouraud. 1971;9:10–14. doi: 10.1080/00362177185190041. [DOI] [PubMed] [Google Scholar]
- McClelland CM, Chang YC, Varma A, Kwon-Chung KJ. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 2004;12:208–212. doi: 10.1016/j.tim.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Morrow CA, Fraser JA. Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res. 2009;9:161–177. doi: 10.1111/j.1567-1364.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- Nadal M, Garcia-Pedrajas MD, Gold SE. Dimorphism in fungal plant pathogens. FEMS Microbiol Lett. 2008;284:127–134. doi: 10.1111/j.1574-6968.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- Neilson JB, Ivey MH, Bulmer GS. Cryptococcus neoformans: pseudohyphal forms surviving culture with Acanthamoeba polyphaga. Infect Immun. 1978;20:262–266. doi: 10.1128/iai.20.1.262-266.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. P Natl Acad Sci USA. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CB, Fraser JA, Heitman J. PAK kinases Ste20 and Pak1 govern cell polarity at different stages of mating in Cryptococcus neoformans. Mol Biol Cell. 2004;15:4476–4489. doi: 10.1091/mbc.E04-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols CB, Perfect ZH, Alspaugh JA. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2007;63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Heitman J. Sex and virulence of human pathogenic fungi. Adv Genet. 2007;57:143–173. doi: 10.1016/S0065-2660(06)57004-X. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, De Obaldia AL, Heitman J. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot Cell. 2007;6:949–959. doi: 10.1128/EC.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- Raudaskoski M. The relationship between B-mating-type genes and nuclear migration in Schizophyllum commune. Fungal Genet Biol. 1998;24:207–227. doi: 10.1006/fgbi.1998.1069. [DOI] [PubMed] [Google Scholar]
- Raudaskoski M, Kothe E. Basidiomycete mating type genes and pheromone signaling. Eukaryot Cell. 2010;9:847–859. doi: 10.1128/EC.00319-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read ND, Lichius A, Shoji JY, Goryachev AB. Self-signalling and self-fusion in filamentous fungi. Curr Opin Microbiol. 2009;12:608–615. doi: 10.1016/j.mib.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Rhodes JC, Kwon-Chung KJ, Popkin TJ. Ultrastructure of the septal complex in hyphae of Cryptococcus laurentii. J Bacteriol. 1981;145:1410–1412. doi: 10.1128/jb.145.3.1410-1412.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Carres M, Findley K, Sun S, Dietrich FS, Heitman J. Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic Cryptococcus species complex. PLoS One. 2010;5:e9620. doi: 10.1371/journal.pone.0009620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua D, Tobe BT, Kron SJ. Cell cycle control of yeast filamentous growth. Curr Opin Microbiol. 2001;4:720–727. doi: 10.1016/s1369-5274(01)00274-0. [DOI] [PubMed] [Google Scholar]
- Rutherford JC, Lin X, Nielsen K, Heitman J. Amt2 permease is required to induce ammonium-responsive invasive growth and mating in Cryptococcus neoformans. Eukaryot Cell. 2008;7:237–246. doi: 10.1128/EC.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul N, Krockenberger M, Carter D. Evidence of recombination in mixed-mating-type and alpha-only populations of Cryptococcus gattii sourced from single eucalyptus tree hollows. Eukaryot Cell. 2008;7:727–734. doi: 10.1128/EC.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer M, Heimel K, Starke V, Kamper J. The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell. 2006;18:2388–2401. doi: 10.1105/tpc.106.043521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. Epigenetic phenomena in filamentous fungi: useful paradigms or repeat-induced confusion? Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- Shadomy HJ, Lurie HI. Histopathological observations in experimental cryptococcosis caused by a hypha-producing strain of Cryptococcus neoformans (Coward strain) in mice. Sabouraud. 1971;9:6–9. doi: 10.1080/00362177185190031. [DOI] [PubMed] [Google Scholar]
- Shadomy HJ, Utz JP. Preliminary studies on a hypha-forming mutant of Cryptococcus neoformans. Mycologia. 1966;58:383–390. [PubMed] [Google Scholar]
- Shen G, Wang YL, Whittington A, Li L, Wang P. The RGS protein Crg2 regulates pheromone and cyclic AMP signaling in Cryptococcus neoformans. Eukaryot Cell. 2008;7:1540–1548. doi: 10.1128/EC.00154-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WC, Davidson RC, Cox GM, Heitman J. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot Cell. 2002;1:366–377. doi: 10.1128/EC.1.3.366-377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia RA, Lengeler KB, Heitman J. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol. 2000;29:153–163. doi: 10.1006/fgbi.2000.1192. [DOI] [PubMed] [Google Scholar]
- Staib F. The perfect state of Cryptococcus neoformans, Filobasidiella neoformans, on pigeon manure filtrate agar. Zentralbl Bakteriol A. 1981;248:575–578. [PubMed] [Google Scholar]
- Staib F, Blisse A. Bird manure filtrate agar for the formation of the perfect state of Cryptococcus neoformans, Filobasidiella neoformans. A comparative study of the agars prepared from pigeon and canary manure. Zentralbl Bakteriol Mikrobiol Hyg A. 1982;251:554–562. [PubMed] [Google Scholar]
- Stanton BC, Giles SS, Kruzel EK, Warren CL, Ansari AZ, Hull CM. Cognate site identifier analysis reveals novel binding properties of the sex inducer homeodomain proteins of Cryptococcus neoformans. Mol Microbiol. 2009;72:1334–1347. doi: 10.1111/j.1365-2958.2009.06719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BC, Giles SS, Staudt MW, Kruzel EK, Hull CM. Allelic exchange of pheromones and their receptors reprograms sexual identity in Cryptococcus neoformans. PLoS Genet. 2010;6:e1000860. doi: 10.1371/journal.pgen.1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt MW, Kruzel EK, Shimizu K, Hull CM. Characterizing the role of the microtubule binding protein Bim1 in Cryptococcus neoformans. Fungal Genet Biol. 2010;47:310–317. doi: 10.1016/j.fgb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G. Hyphal growth: a tale of motors, lipids, and the Spitzenkörper. Eukaryot Cell. 2007;6:351–360. doi: 10.1128/EC.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G, Perez-Martin J. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 2008;18:61–67. doi: 10.1016/j.tcb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Torres-Guererro H, Edman JC. Melanin-deficient mutants of Cryptococcus neoformans. J Med Vet Mycol. 1994;32:303–313. [PubMed] [Google Scholar]
- Tscharke RL, Lazera M, Chang YC, Wickes BL, Kwon-Chung KJ. Haploid fruiting in Cryptococcus neoformans is not mating type alpha-specific. Fungal Genet Biol. 2003;39:230–237. doi: 10.1016/s1087-1845(03)00046-x. [DOI] [PubMed] [Google Scholar]
- Vallim MA, Nichols CB, Fernandes L, Cramer KL, Alspaugh JA. A Rac homolog functions downstream of Ras1 to control hyphal differentiation and high-temperature growth in the pathogenic fungus Cryptococcus neoformans. Eukaryot Cell. 2005;4:1066–1078. doi: 10.1128/EC.4.6.1066-1078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi R, Hsueh Y, Geunes-Boyer S, Wright JR, Heitman J. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma-Gaur J, Deshpande S, Sadhale PP. RAM pathway contributes to Rpb4 dependent pseudohyphal differentiation in Saccharomyces cerevisiae. Fungal Genet Biol. 2008;45:1373–1379. doi: 10.1016/j.fgb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- Walton FJ, Heitman J, Idnurm A. Conserved elements of the RAM signaling pathway establish cell polarity in the basidiomycete Cryptococcus neoformans in a divergent fashion from other fungi. Mol Biol Cell. 2006;17:3768–3780. doi: 10.1091/mbc.E06-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lin X. Mechanisms of unisexual mating in Cryptococcus neoformans. Fungal Genet Biol. 2011;48:651–660. doi: 10.1016/j.fgb.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Wang P, Heitman J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 1999;2:358–362. doi: 10.1016/S1369-5274(99)80063-0. [DOI] [PubMed] [Google Scholar]
- Wang P, Perfect JR, Heitman J. The G-protein beta subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol Cell Biol. 2000;20:352–362. doi: 10.1128/mcb.20.1.352-362.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Nichols CB, Lengeler KB, Cardenas ME, Cox GM, Perfect JR, Heitman J. Mating-type-specific and nonspecific PAK kinases play shared and divergent roles in Cryptococcus neoformans. Eukaryot Cell. 2002;1:257–272. doi: 10.1128/EC.1.2.257-272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hsueh YP, Li W, Floyd A, Skalsky R, Heitman J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Gene Dev. 2010;24:2566–2582. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh MS, Vallim MA, Heitman J, Alspaugh JA. Ras1 controls pheromone expression and response during mating in Cryptococcus neoformans. Fungal Genet Biol. 2003;38:110–121. doi: 10.1016/s1087-1845(02)00518-2. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Momany M. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol Biol Cell. 2002;13:110–118. doi: 10.1091/mbc.01-06-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM, Rose MD. Yeast mating: getting close to membrane merger. Curr Biol. 2001;11:R16–R20. doi: 10.1016/s0960-9822(00)00036-1. [DOI] [PubMed] [Google Scholar]
- Wickes BL, Mayorga ME, Edman U, Edman JC. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the alpha-mating type. P Natl Acad Sci USA. 1996;93:7327–7331. doi: 10.1073/pnas.93.14.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Edman U, Edman JC. The Cryptococcus neoformans STE12alpha gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- Williamson JD, Silverman JF, Mallak CT, Christie JD. Atypical cytomorphologic appearance of Cryptococcus neoformans: a report of five cases. Acta Cytol. 1996;40:363–370. doi: 10.1159/000333769. [DOI] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, Heitman J. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host Microbe. 2007;1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Xue C, Hsueh YP, Chen L, Heitman J. The RGS protein Crg2 regulates both pheromone and cAMP signalling in Cryptococcus neoformans. Mol Microbiol. 2008;70:379–395. doi: 10.1111/j.1365-2958.2008.06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Xu J. Mitochondria are inherited from the MATa parent in crosses of the basidiomycete fungus Cryptococcus neoformans. Genetics. 2003;163:1315–1325. doi: 10.1093/genetics/163.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hull CM, Sun S, Heitman J, Xu J. The mating type-specific homeodomain genes SXI1 alpha and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans. Curr Genet. 2007;51:187–195. doi: 10.1007/s00294-006-0115-9. [DOI] [PubMed] [Google Scholar]
- Yeh YL, Lin YS, Su BJ, Shen WC. A screening for suppressor mutants reveals components involved in the blue light-inhibited sexual filamentation in Cryptococcus neoformans. Fungal Genet Biol. 2009;46:42–54. doi: 10.1016/j.fgb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, Heitman J. The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010;6:e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer BL, Hempel HO, Goodman NL. Pathogenicity of the hyphae of Filobasidiella neoformans. Mycopathologia. 1983;81:107–110. doi: 10.1007/BF00436987. [DOI] [PubMed] [Google Scholar]