Abstract
Low-linear energy transfer (LET) radiation (i.e., γ- and X-rays) induces DNA double-strand breaks (DSBs) that are rapidly repaired (rejoined). In contrast, DNA damage induced by the dense ionizing track of high-atomic number and energy (HZE) particles are slowly repaired or are irreparable. These unrepaired and/or misrepaired DNA lesions may contribute to the observed higher relative biological effectiveness for cell killing, chromosomal aberrations, mutagenesis, and carcinogenesis in HZE particle irradiated cells compared to those treated with low-LET radiation. The types of DNA lesions induced by HZE particles have been characterized in vitro and usually consist of two or more closely spaced strand breaks, abasic sites, or oxidized bases on opposing strands. It is unclear why these lesions are difficult to repair. In this review, we highlight the potential of a new technology allowing direct visualization of different types of DNA lesions in human cells and document the emerging significance of live-cell imaging for elucidation of the spatio-temporal characterization of complex DNA damage. We focus on the recent insights into the molecular pathways that participate in the repair of HZE particle-induced DSBs. We also discuss recent advances in our understanding of how different end-processing nucleases aid in repair of DSBs with complicated ends generated by HZE particles. Understanding the mechanism underlying the repair of DNA damage induced by HZE particles will have important implications for estimating the risks to human health associated with HZE particle exposure.
Keywords: Clustered DNA damage, Ionizing Radiation, HZE particles, low-LET, high-LET, WRN and Artemis
1. Introduction
1.1. Study of biological effects of high-atomic number and energy (HZE) particle radiation is imperative for the safety of manned space missions
Risks to astronauts from exposure to Galactic Cosmic Rays (GCR) have long been of great concern to NASA. These risks are now of even more critical interest with the advent of long-duration space missions on the former Mir Space Station, the present International Space Station, and the contemplation of exploratory missions to the Moon and to Mars. A significant increase over background was observed in numbers of chromosome aberrations, including complex aberrations, in the lymphocytes of eight astronauts who participated in long-duration NASA/Mir missions [1, 2]. Thus the problem is real, and a basic and fundamental understanding of such phenomena is likely to be even more important for the assessment of risks to crews of future missions [3, 4]. The GCR consist primarily of protons, helium nuclei (Z=2), and higher atomic number particles such as iron (Z=26). The relative abundance of protons is the highest (87%), followed by helium (11%) and HZE particles (2%). The energy of these particles can be very high (1000 MeV/n or more), sufficient in many cases to penetrate spacecraft hulls and interior materials. Iron is a good representative particle for research purposes, because it is the heaviest ion present in significant numbers in the GCR, and, due to its high atomic number, its biological effects are potentially severe. During a three-year flight in extramagnetospheric space, 3% of the cells of the human body would be traversed on average by one iron ion [5]. The unique pattern of energy deposition due to HZE particle traversal is of primary interest for evaluating the biological effects of the GCR on astronauts [6].
1.2. Physical characteristics of ionizations induced by a HZE particle
It is well established that HZE particles have a higher (several to many fold greater) relative biological effectiveness (RBE) than X- or γ-rays (sparsely ionizing radiation) [4, 7-11]. It has been predicted that the types of lesions and the complexity of the DNA damage induced by HZE, and ultimately the RBE of the HZE particle, is dependent on its energy [12]. This is because the microscopic pattern of energy deposition will vary as a function of the energy of the HZE particle. Linear energy transfer (LET) varies as a function of velocity (v) and charge (Z), of the HZE particle. The numbers of lesions in a highly localized region of the genome (within twenty base pairs or so) vary with this LET. The microscopic pattern of energy deposition by a very energetic HZE particle presents a complexity in terms of the overall nature of the radiation field. The concept of “core” and “penumbra” has been used to define the physical characteristics of a charged particle track [13]. The “core” region may be defined on the basis of Bohr’s adiabatic principle and can be up to about 0.0015 micron in radius. Within this region, all the excitations of the medium molecules occur. In addition, a large amount of energy is deposited by very low-energy electrons (100 eV – 200 keV) that cannot successfully exit the core. The “penumbra” region is defined by the maximum distance traversed by secondary electrons (termed δ rays) perpendicular to the trajectory. HZE particles deposit their energy inside a cell in two ways: more than 50% of their energy is deposited within the core via direct ionization and excitation of the medium molecule, while secondary electrons (δ rays) emitted from these collisions can extend to significant distances (100s of microns) [13].
1.3. Clustered DNA lesions are associated with HZE ionization tracks
Clustered DNA damage or multiple damage sites (MDS) are DNA lesions generated by a single track of ionizing radiation [14-16] that include two or more individual lesions within one or two helical turns of the DNA [17]. The two basic groups of complex DNA damage are DSBs and non-DSBs. The lesions within the clustered damage sites can be abasic sites (also known as apurinic/apyrimidinic sites or APs), damaged bases (oxidized purines or pyrimidines), single-strand breaks (SSB) and double-strand breaks (DSBs) [18, 19]. The non-DSBs clusters can be a mixture of base modifications and SSBs are found in close proximity to each other [20]. It has been predicted that clustered damage that includes DSBs are probably due to energy depositions of at least two to five ionizations localized within 1-4 nm [14]. The complexity and yield of radiation-induced clustered DNA damage increases with the increasing ionizing density of the radiation [21, 22]. Track structure simulations demonstrate that more than 90% of DSBs induced by densely ionizing radiation are associated with other lesions [23, 24]. However, direct measurement of clustered DNA lesions in cells, with the exception of DSBs, has been very difficult. Only an in vitro method is available to estimate genome-wide clustered DNA lesions from isolated DNA [15, 19]. Using this method, it was shown that among all complex damages induced by γ-ray or HZE particles in mammalian DNA, DSBs comprise ~20% and other lesions the rest [17].
1.4. Clustered DNA lesions inhibit the cell’s capability for DNA repair
It has been clearly demonstrated that clustered DNA damage is more difficult for the cellular machinery to repair than are individual damage sites. Studies with synthetic oligonucleotides containing several types of DNA damage have shown that the efficiency of incision of an AP site, for example, within a region of clustered DNA damage is significantly reduced by the presence of a second AP site or SSB [25]. Similarly, APs or 8-oxoguanine (8-OxoG) sites within clustered DNA damage sites are poorly handled by mammalian cell extracts or purified repair enzymes [25-27]. These unrepaired cluster DNA lesions may generate additional DSBs [28, 29]. Clustered DNA damage sites appear to retard the function of repair enzymes. Theoretical modeling to simulate the dynamics of clustered DNA damage sites containing abasic or 8-OxoG sites suggest that these lesions cause DNA to adopt non-canonical conformations [30]. These conformations may make it difficult for repair enzymes to bind to the region, thereby resulting in a reduction of repair efficiency. Further, model system studies of oligonucleotides and plasmids with defined lesions at specific spacings on opposing strands indicate that clusters may comprise non-repairable, highly repair-resistant, or pre-mutagenic damage [19, 29, 31-33]. Recently, it has been suggested that non-DSBs clusters, if unrepaired, can lead to the formation of mutations and chromosome abnormalities [34].
1.5. HZE particle-induced DNA double-strand breaks are difficult to repair in mammalian cells
Convincing evidence indicates that DNA DSBs are the lesions that result in the cytotoxic, mutagenic, and carcinogenic effects of radiation, whether γ-rays or HZE particles [35, 36]. Thus, the ability or inability of cells to repair DSBs can serve as a very effective measure of the RBE of any form of radiation. Measurements of the rate of repair of DSBs by pulsed-field gel electrophoresis showed that both the half-times of rejoining and the fraction of residual DNA breaks increased with the atomic number (Z) of the particle [37, 38]. This observation has been confirmed recently by monitoring γH2AX foci formation at DSBs sites in situ in various cell types [9, 39-42]. In addition to a slower rate of rejoining, HZE particle-induced DSBs are more frequently misrejoined in mammalian cells than breaks induced by low-LET radiation [8]. The difficulty of the repair of HZE particle-induced DSBs has been attributed to the nature of the complex clustered DNA damage induced by dense ionizations along the HZE particle track [11, 22, 43]; however, there is no in vivo or in situ evidence to support this hypothesis. DNA conformational changes mentioned above due to multiple lesions within two helical turns may result in inefficient repair [27, 30], but it is also possible that non-DSB DNA damage near a DSB retards the function of DNA DSBs repair proteins.
2. Quantification and spatio-temporal characterization of complex DNA damage
Detection, identification, and quantification of clustered DNA damages and their processing in cells are essential for evaluating the biological effects of HZE particle radiation on humans. In addition, assessment of the initial damage induced by the HZE particle exposure, determination of the rates and paths of repair; and identification of types and measurements of the levels of damage remaining after cellular processing are essential for understanding the consequences of radiation exposure. Accurate measurement of damage induction and processing requires that damage be assessed immediately after the exposure (before any biological or biochemical processing), and then during and after cellular processing [44]. Undoubtedly, major advances in the DNA repair field have been performed with the help of in vitro systems. The spectrum of these in vitro systems was broadened by the development of a method in which type of damage on each DNA strand (altered bases, abasic sites, SSBs) as well as damage affecting both strands (clustered damage, DSBs) can be quantified in genomic DNA isolated from irradiated human cells by direct measurement using DNA repair enzymes isolated from E. coli as damage probes (Table 1) and constant field or pulsed field gel electrophoresis followed by number-average length analysis [15]. In addition to this approach Georgakilas and co-workers [45, 46], have used polyamines for the detection of very closely spaced abasic sites. Based on the same principle, other groups have also used neutral agarose gel electrophoresis and fraction of activity released assay, hybridization assay or plasmid nicking assay [26, 47]. Further, other groups have developed a modified version of the neutral single cell gel electrophoresis (Comet assay) using again repair enzymes as damage probes [18, 48]. However, in all these in vitro biochemical assays, incomplete cleavage of lesions by the repair enzymes can lead to a detection of only a fraction of the clusters. Further, the sub-cellular localization, dynamics of DNA repair and response proteins and spatial relationship (i.e., distribution of different types of DNA lesions along the dense ionization tracks traversed by HZE particles) cannot be determined despite recent advances in in vitro systems for monitoring repair. Therefore, for an accurate measurement of the biological effects of HZE radiation exposure, an alternative assay sensitive enough to measure complex DNA damage at single-cell level is needed.
Table 1.
DNA repair and response proteins used in the detection of different types of DNA lesions induced by high-LET radiation
| DNA repair and response proteins | Lesion types | Detection Methods | References |
|---|---|---|---|
| Phosphorylated H2AX | DSB | Immunostaining | [9, 40, 49, 63, 66, 129] |
| Phosphorylated DNA-PKcs | DSB | Immunostaining | [9, 50, 129] |
| p53 binding protein 1 | DSB | Immunostaining | [9, 51, 63, 66] |
| Phosphorylated ATM | DSB | Immunostaining | [63] |
| X-ray repair cross complementing 1 | SSB | Immunostaining | This study |
| 8-oxoguanine DNA glycosylase | Base damage | Immunostaining | This study |
| Replication Protein A | ssDNA | Immunostaining | [66] |
| E. coli Fpg protein or human 8-oxoguanine DNA glycosylase | Oxypurines | Biochemical assay | [15, 18, 27, 45-48] |
| E. coli or human Nth1 (Endonuclease III) | Oxypyrimidines | Biochemical assay | [15, 18, 27, 45-48] |
| E. coli Nfo protein or human hAPE1 (endonuclease IV) | Abasic sites | Biochemical assay | [15, 18, 27, 45-48] |
| p53 binding protein 1 | DSB | Live cell imaging | This study and [130] |
| DNA-PKcs | DSB | Live cell imaging | [61] |
| Mediator of DNA damage checkpoint protein 1 | DSB | Live cell imaging | [66] |
| X-ray repair cross complementing 1 | SSB | Live cell imaging | This study and [66] |
| Aprataxin | SSB | Live cell imaging | [66] |
DSB-double-strand breaks
SSB-single-strand breaks
ssDNA-single strand DNA
DNA-PKcs-DNA-dependent protein kinase catalytic subunit
ATM-Ataxia telangiectasia mutated
2.1 Indirect immunostaining approach
Recruitment and retention of DNA repair and response proteins at DNA breaks can be conveniently visualized by fluorescence imaging of repair foci (often called ionizing radiation–induced foci [IRIF]). The H2AX is immediately phosphorylated at the sites of DSBs, and the phosphorylated H2AX (γH2AX) can be visualized in situ by immunostaining with a γH2AX specific antibody [49]. In addition to γH2AX, phosphorylated DNA-dependent protein kinase (DNA-PK) localizes precisely at DSBs and serves as an ideal maker for visualizing DSBs in situ [50]. p53 binding protein 1 (53BP1), a DNA damage response protein, responds to DNA DSBs, forms discrete nuclear foci upon exposure to IR, and co-localizes with γH2AX and DNA-PK; it is also used as a surrogate marker for DSBs [51]. This indirect immunostaining technique has been widely applied to visualize DSBs generated by HZE particles [39-41, 52]. For example, as shown in Fig. 1 (panel A), irradiation of monolayer human skin fibroblasts with iron particle beams perpendicular to the culture surface resulted not only in the formation of γH2AX and DNA-PKcs (pT2609) tracks as early as 10 min after irradiation but also in the co-localization of these markers along the densely ionized track traversed by iron particles. Further, γH2AX, 53BP1, and DNA-PK foci could be observed in iron-particle-irradiated cells even 48 hours post-irradiation [9]. These results together with studies from other laboratories clearly suggest that 53BP1, γH2AX, and DNA-PK are useful predictors of DSBs rejoining kinetics and thus can be used as surrogate markers to study DSBs within the clustered DNA lesions (Table 1).
Figure 1. Clustered DNA lesions induced by iron particles can be visualized by indirect immunostaining with DNA repair and response proteins.

(A) Phosphorylated H2AX and DNA PKcs foci form tracks along the paths traversed by iron particles. Normal human skin fibroblasts were irradiated horizontally with iron particles (1 Gy at 1 GeV/nucleon) and stained with γH2AX and DNA-PKcs (pT2609) antibodies 10 min after irradiation. (B) 53BP1, XRCC1, and OGG1 foci form tracks along the densely ionizing paths traversed by iron particles. HT1080 cells were irradiated horizontally with iron particles (1 Gy at 1 GeV/nucleon) and stained with 53BP1, OGG1, and XRCC1 antibodies 10 min after irradiation.
There is currently no direct approach to detect DNA single-strand breaks (SSB) and base damage in situ. However, essential components of DNA SSB and base damage repair pathways have been identified [53, 54]. Substantial evidence indicates an important role for X-ray repair cross complementing 1 (XRCC1) in SSB repair. XRCC1 acts as a scaffolding protein for other repair factors and has been shown to physically interact with several enzymes known to be involved in the repair of SSBs, including DNA ligase III, DNA polymerase ß, APE1, polynucleotide kinase/phosphatase, and poly(ADP-ribose) polymerases 1 and 2 (PARP-1 and 2) [55]. In addition, it has been shown that XRCC1 is recruited to laser irradiation-induced sites of SSBs [56]. Therefore, SSBs can be visualized in situ by immunostaining with antibodies against XRCC1. For example, as shown in Fig. 1 (panel B), irradiation of monolayer human fibroblasts with iron particle beams perpendicular to the culture surface resulted in the formation of XRCC1 tracks as early as 10 min after irradiation. XRCC1 can be used as a surrogate marker to identify the sites of SSBs within the clustered DNA lesions and serves as a useful predictor of SSBs rejoining kinetics.
Evidence from a number of laboratories indicates that the major DNA glycosylase responsible for the removal of 8-OxoG base lesions in DNA in mammalian cells is 8-oxoguanine DNA glycosylase (OGG1). It has been shown that after UVA treatment of human cells, a fraction of the nuclear OGG1 is specifically recruited to nuclear speckles through a reactive oxygen species–mediated mechanism [57]. Moreover, irradiation of cells with a UVA laser through a microscope lens induces the accumulation of 8-OxoG and subsequently hOGG1 along the laser pathway [56]. These results suggest that damaged base can be visualized in situ by immunostaining with antibodies against OGG1. For example, as shown in Fig. 1 (panel B), irradiation of monolayer human fibroblasts with iron particle beams perpendicular to the culture surface resulted in the formation of OGG1 tracks as early as 10 min after irradiation. OGG1 can be used as a surrogate marker to identify damaged bases within the clustered DNA lesions and serves as a useful predictor of base damage rejoining kinetics. Together, these results clearly demonstrate that different types of DNA lesions can be identified in situ by indirect immunostaining with antibodies against a variety of DNA damage response and repair factors. Further, this approach can be readily applied for verification of whether certain DNA lesions are induced by HZE particles and also to estimate the relative amounts as well as the spatial distribution of different types of DNA lesions within the clustered DNA damage. Although indirect immunostaining approach can provide great insight, similar to in vitro biochemical assay, this approach has some limitations. For example, detection of the exact number of DNA lesions represented by each IRIF may not be accurate. Cellular approaches, including live-cell imaging, in combination with in vitro assay systems, will help to more precisely estimate the amount of clustered DNA lesions induced by HZE particles.
2.2 Direct live cell imaging approach
Cellular approaches, including indirect immunostaining, in combination with in vitro assay systems have significantly improved our understanding of induction and repair of clustered DNA damage; however, some of the challenging questions in the study of HZE particles induced DNA damages cannot yet be answered. For instance, how fast is the recognition of various types of damaged DNA? Which proteins arrive first to the sites of DNA damage? What is the affinity of different repair proteins for clustered DNA lesions? How do distinct proteins recognize clustered DNA lesions? Due to the use of green fluorescent proteins (GFP) and their spectral variants [58] and advancements in microscopy and digital imaging technology, it is now possible to study the spatio-temporal aspects of complex DNA damages in live cells [59]. DNA repair research has been boosted substantially by the development of several methods to locally inflict DNA damage in living cells, enabling the direct visualization of GFP-tagged repair factors. Although quantitative live cell imaging techniques combined with methods to induce local DNA damage in a small region of the nucleus are contributing substantially to unraveling of the molecular mechanisms underlying the cellular response to DNA damage, the use of similar approaches in the study of complex DNA damage has been limited.
The very first insight into the dynamics of protein recruitment to the localized DNA lesions generated by HZE particles came from research conducted at the GSI Helmholtz Centre for Heavy Ion Research [60]. Time-lapse images of the DNA repair protein, aprataxin, and of DNA-PKcs coupled to GFP [61] revealed a very fast recruitment of these proteins to damage sites, demonstrating that the early kinetics of protein recruitment in response to HZE can be determined in live cells (Table 1). Apart from the protein dynamics, specific types DNA lesions generated by HZE particles have also been visualized using fluorescently tagged DNA repair and response factors. For example, YFP-53BP1 (shown in Fig. 2A) and EGFP-XRCC1 (Fig. 2C) are localized along the track within minutes after HZE particle irradiation. Interestingly, similar to immunostaining approach [9], after parallel irradiation of cells with Fe, Si and oxygen ions, we observed distinct fluorescent streaks of 53BP1 signals that were visible as early as 3-10 min after Fe and Si irradiation (Fig. 2A). However, there was no streak-like pattern in cells irradiated with oxygen ions, suggesting a more random distribution of ionizations. Further, in contrast to 53BP1 foci, majority of the XRCC1 foci (Fig. 2C) disappeared within an hour implying that SSBs are repaired earlier than the DSBs in response to HZE particles. An issue that must be considered in interpreting these results is particle fluence. As the Z increases, the LET increases, as does the radial dose fraction. Since all exposures were limited to 1 Gy and were given at a rate of approximately 1 Gy/min, the particle fluence must change as well. For oxygen particles the fluence (4.5 × 107ions/cm2) was approximately 10 times higher than that for iron particles (4.2 × 106ions/cm2). One could expect that with a lower LET combined with higher particle fluence, the DNA damage spectrum would be more random in a cross section of DNA and that it would be less likely to produce multiply damaged sites. Clustered damage generated by electron track ends would remain but would be distributed more evenly within a given nucleus. Conversely, because of the lower fluence and higher LET of iron particles, there would be an increase in energy deposition in more confined regions that would result in larger amounts of multiply damaged sites with a greater density of clustered lesions that complicate DNA damage repair and that would appear as tracks in these assays [9].
Figure 2. DNA double-strand breaks induced by a spectrum of HZE particles can be directly monitored in live cells.

(A) 53BP1 forms foci along the densely ionizing paths traversed by iron (Fe), silicon (Si) and oxygen (O) particles. HT1080 cells stably expressing yellow fluorescent protein (YFP) tagged 53BP1 were imaged prior to irradiation (Pre-IR) and then exposed to Fe, Si and O particles (1 Gy at 1 GeV/nucleon) and were immediately imaged using a Zeiss fluorescent microscope. (B) YFP-53BP1 foci detected in live cells represent the sites of DNA DSBs. HT1080 cells stably expressing yellow fluorescent protein (YFP) tagged 53BP1 were exposed to Fe, Si, and O particles (1 Gy at 1 GeV/nucleon) and were fixed 10 minutes after irradiation. Subsequently, the cells were subjected to indirect immunofluorescence using γH2AX antibodies. (C) XRCC1 forms foci along the densely ionizing paths traversed by iron (Fe), silicon (Si) and oxygen (O) particles. HT1080 cells stably expressing green fluorescent protein (EGFP) tagged XRCC1 were imaged prior to irradiation (Pre-IR) and then exposed to Fe, Si and O particles (1 Gy at 1 GeV/nucleon) and γ-ray and were immediately imaged using a Zeiss fluorescent microscope.
Although live-cell imaging studies are in agreement with immunostaining approaches with different DNA damage surrogate markers, live-cell imaging has several advantages. A major advantage of this approach is that recruitment and formation of DNA damage foci can readily be observed immediately after the induction of DNA damage and post-IR handling of cells is avoided. In addition, by comparing the live-cell images obtained before and after IR, the foci that are generated by IR can be distinguished from background foci. With this simple strategy, one can able to unequivocally and directly identify only the IR-induced DNA lesions and follow their repair kinetics in real time. In addition, as discussed in Section 2.3, it is also possible to obtain very accurate recruitment time constants for different protein pairs or combinations, which can be used to infer interactions among different DNA repair pathways. One can also manipulate the cellular system using inhibitors of specific proteins and observe effects in real time. Therefore, use of live-cell imaging technology to directly follow the induction and repair of various types of DNA damage in the nucleus of live cells provides several notable advantages over analyses of fixed cells.
2.3 Spatio-temporal characterization of clustered DNA lesions
The reasons DNA lesions induced by HZE particles are difficult to repair are still unclear. Though it is believed that HZE particles induce DSBs clustered with other types of DNA lesions along the ion track, but there is no in vivo or in situ evidence to support this theory. Critical questions remain to be answered: What are the actual distributions of DSBs, SSBs and damaged bases within the clustered DNA lesions? Does the spatial distribution of these lesions play any role in the repair process? Do the physical locations of these lesions within nuclear sub-domains affect the cellular ability to repair clustered DNA damages? These important questions must be addressed before the risks of HZE can be accurately determined.
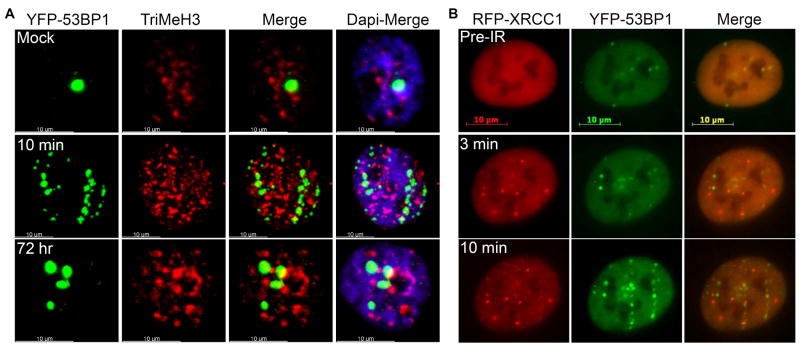
Evidence suggests that chromatin organization regulates the cellular response to DNA damage. For instance, foci induction is much slower in the heterochromatin due to the time it takes to move DSBs to the interface between euchromatin and heterochromatin, and the radiation induced foci (IRIF) resolution is much slower due to the complexity of this damage [62]. Recently, using a novel imaging approach Costes and co-workers have reported that both X-ray and HZE particle radiation induced 53BP1, phospho-H2AX and ATM IRIF are located preferentially at the interface between heterochromatin and euchromatin regions [63]. In addition, it has been shown that the rate of resolution of heterochromatic IRIF is slower than that of the euchromatic IRIF [64]. We recently performed an interesting experiment in which we irradiated HT1080 cells stably expressing YFP-53BP1 with iron particles (1Gy at 1GeV/n) and immunostained the cells with histone H3 (tri methyl K9) antibody that marks the regions of heterochromatin [65]. As shown in Fig. 3 (panel A), of the all initially induced and persistent 53BP1 foci, only a fraction were juxtaposed with heterochromatic regions. The fraction of heterochromatin-associated 53BP1 foci at 10-30 minutes after Fe irradiation was 3.5 per cell. At 72 hours after irradiation with Fe particles only 1.5 53BP1 foci per cell were associated with heterochromatin, demonstrating that most unrepaired lesions were found in euchromatin regions. These results clearly indicate that the difficulties associated with the repair of complex DNA lesions are not due to their physical location within the sub-nuclear domain. Then what makes repair of these complex DNA lesions so difficult? Is it because of the spatial distribution of DSBs, SSBs and base damages within the clustered DNA lesions? Recently, data from a three-dimensional imaging approach revealed that phosphorylated DNA-PKcs (at T2609) was present in only 40–50% of the γH2AX foci [9]. The non-overlapping distribution of γH2AX and DNA-PKcs reflects the partitioning in space of the DNA damage and response proteins. Further, the phospho-DNA-PKcs foci may provide a more realistic picture of DNA DSB than does immunostaining for γH2AX. Presently, we are utilizing a similar approach to examine the distribution of DSBs, SSBs and base damages within the clustered DNA lesions induced by a spectrum of HZE particles. Results obtained from these studies will provide insights into whether the spatial distribution of different kinds of DNA lesions determines the cellular ability to repair HZE particle-induced DNA lesions.
Figure 3. Spatio-temporal characterization of DNA lesions induced by iron particles.
(A) Not all persistent 53BP1 foci are located in regions of heterochromatin. HT1080 cells stably expressing YFP-tagged 53BP1 were irradiated horizontally with iron particles (1 Gy at 1 GeV/nucleon) and immunostained with histone H3 (tri methyl K9) antibody (TriMeH3) 10 min and 72 hours after irradiation, and the images were acquired using confocal microscopy (Zeiss). 100-120 cells for each time point in three independent experiments were examined. (B) Iron particle-induced SSBs are detected earlier than the DSBs in single cells by the live-cell imaging approach. HT1080 cells stably expressing dual fluorescent proteins (i.e., YFP-tagged 53BP1 and RFP-tagged XRCC1) were imaged prior to irradiation (Pre-IR), exposed to iron particles (1 Gy at 1 GeV/nucleon), and immediately imaged using Zeiss fluorescent microscopy.
Some of the other challenging questions in the study of damage induced by HZE particles are: How do distinct proteins recognize the clustered DNA lesions? How rapid is the recognition of various types of damaged DNA? Which proteins arrive first to the sites of DNA damage? Is the direct physical contact of one protein with damaged DNA is necessary for the subsequent recruitment of other proteins? First insights into the recruitment of repair proteins to the regions of DNA damage came from studies using beamline microscopy developed at GSI [60]. Using this technology combined with detailed time course analysis proved instrumental in addressing one of these questions. Jakob and co-workers have determined the exact kinetics of repair-related proteins after irradiation with different charged particles that induce different lesion densities [66]. The authors showed that DNA-PKcs and XRCC1 were recruited rapidly and 53BP1 and MDC1 were recruited more slowly. We tagged XRCC1 and 53BP1 with red (RFP) and yellow fluorescent proteins (YFP), respectively, and expressed these two proteins in the same cell. Subsequently, we visualized the recruitment of RFP-XRCC1 and YFP-53BP1 to the sites of SSBs and DSBs, respectively, after exposure of cells to iron particles. As shown in Fig. 3 (panel B), RFP-XRCC1 foci were visible as early as 1-3 min following irradiation whereas, YFP-53BP1 foci were observed later. These results clearly demonstrate that SSBs are detected earlier than DSBs. However, these interesting findings raise more questions. What determines the differential recruitment kinetics? Is it because of the sensing characteristics of the repair factors or nature of the lesions? These key questions need to be addressed in order to understand the molecular events that are associated with clustered DNA lesion repair.
3. Pathways of clustered DNA damage repair
3.1. The repair status of the HZE particle-induced DSBs cannot be clearly explained by the current understanding of DSB repair pathways
In mammalian cells, DNA DSBs are repaired mainly by two distinct pathways, non-homologous end joining (NHEJ) and homologous recombination (HR). These two pathways are distinct biochemically, have diverse substrate requirements, operate with different kinetics, and are used differently throughout the cell cycle [67]. HR repair utilizes the sister chromatid as template, resulting in gene conversion [68-70], and, therefore, it is suggested to be an error-free repair pathway preferentially operating in the S-phase of the cell cycle. In contrast to HR, NHEJ utilizes little or no homology to join DNA ends [71]. NHEJ is the primary pathway of DSB repair in G1-/early S-phase cells, whereas both HR and NHEJ contribute to repair of DSBs introduced during late S-/G2 phase of the cell cycle [72]. Evidence clearly indicates that NHEJ is the predominant repair pathway for DSBs induced by low-LET radiation. Mammalian cell mutants deficient in the NHEJ pathway are extremely sensitive to X- and γ-rays and accumulate unrepaired DSBs as function of dose [73]. Mammalian or vertebrate mutants deficient in HR factors exhibit somewhat higher radiosensitivity and their radiosensitivities are much enhanced when mutant cells are synchronized in S phase [74]. Although evidence clearly indicates that NHEJ is the major repair pathway for low-LET radiation induced DSBs, it is not clear which pathways of DSB repair can handle clustered DNA lesions accurately.
3.2. High-LET radiation-induced DSBs are poorly handled by NHEJ
Early studies to determine the relative sensitivities of NHEJ-deficient mammalian cell lines to α-particles indicated that the degree of enhancement of radiosensitivity was less than for X-rays suggesting that NHEJ pathway of DNA repair may only partially repair the complex DSBs induced by high-LET radiation [38, 73, 75]. Similar studies using HZE particles indicated that in NHEJ deficient cells, the RBE did not increase with LET, in contrast to repair-proficient cells, indicating that high-LET radiation induced a higher yield of DNA lesions that cannot be repaired by NHEJ pathway [42]. Moreover, after measuring chromosome fragmentation and γH2AX foci formation, Okayasu and co-workers [42] confirmed that HZE particle-induced DSBs are difficult to repair by NHEJ pathway even at low radiation doses such as 2 Gy. Recently, it was reported that high-LET IR can kill more cells than low-LET IR at the same dose is due to inefficient Ku-dependent NHEJ repair [76]. Recently, these observations were confirmed in NHEJ-deficient mice [77]. More research is required to determine the exact role of newly identified NHEJ pathway factors in the processing of complex DNA lesions.
3.3. The role of homologous recombination in the repair of high-LET radiation-induced DSBs is totally unclear
HR is the predominant pathway for the repair of DSBs in bacteria and yeast [78, 79] and is very efficient in vertebrate pre-B cells [80]. As discussed earlier, both NHEJ and HR are required for the repair of X- and γ-ray induced DSBs throughout the cell cycle. Several laboratories have confirmed that a collaboration between HR and NHEJ is needed for repair of low-LET-induced DNA DSBs [81-86]. However, such studies have not been extended to high-LET radiation and the relative contribution of HR in human cells is not known. Analyses are complicated due to the difficulties in separating these two pathways at a given phase of the cell cycle. Analyzing the nature of hprt mutations induced by low- and high-LET radiation, it was estimated that more than 30% of the IR-induced DSBs might be repaired by HR [87]. However, this estimation may not reflect the actual frequency, as IR-induced large hprt gene deletions on the X-chromosome may not be recoverable [87]. A recent report by Zafar et al. [88] clearly indicates that an intact HR pathway is needed for processing of HZE-induced DNA damage.
HR is initiated by resection of DNA ends at the DSB sites, promoted by the Mre11/Rad50/Nbs1 (MRN) complex, C-terminal binding protein interacting protein (CtIP), and exonuclease 1 (Exo1), to yield 3’-single-stranded DNA (ssDNA) overhangs [82]. This 3’-ssDNA ends are rapidly bound by Replication Protein A (a ssDNA-binding protein), which is required for the subsequent recruitment of the recombinase, Rad51. Once recruited to the DSBs, Rad51 catalyzes strand invasion and exchange into a homologous DNA template, after which the recombination intermediates are resolved. Since the HR, occurs specifically in the late S and G2 phases of the cell cycle, utilizes an identical template for repair, preferably the sister chromatid, and is considered to be a relatively error-free process. Even though the mechanisms of HR are known, future work is still required to validate the role of newly identified HR pathway factors in the repair of complex DNA damages.
4. Novel nucleases might play crucial roles in processing complex DNA ends generated by HZE particles prior to ligation
4.1. Role of nucleases
It is predicted that repair of radiation-induced DNA lesions is dependent upon radiation quality and the structural complexity of DSBs [89]. DSBs induced by high-LET radiation may vary in terms of their overhang configuration and end compatibility; therefore the incompatible DNA ends must be processed in order to make them substrates for a DNA ligase [90]. Various repair proteins may function independently or as complexes to avoid alterations of the base sequence or entanglement of the DNA. In the repair complexes, nucleases are the molecular scissors; these enzymes recognize the damaged moiety and cleave phosphodiester bonds between the sugars and the phosphate moieties of DNA to eliminate the damaged or mismatched nucleotides [91]. In recent years, several nucleases involved in various DNA repair processes have been identified including Mre11 (a multi-functional nuclease), Artemis (a hairpin opening nuclease), and Werner syndrome protein (a RecQ helicase), known to have an end-processing function. The exact involvement of these nucleases in HZE induced DNA repair processing remains unclear.
4.2. Werner syndrome protein
Werner Syndrome (WS) is an autosomal recessive disorder that gives rise to multiple progeroid pathologies, including osteoporosis, atherosclerosis, and a greatly increased cancer incidence [92, 93]. WRN, the protein responsible for WS, is unique among all RecQ helicases in having an N-terminal 3’-5’ nuclease activity on dsDNA with 3’ recessed termini [94]. WRN exonuclease functions on a variety of structured DNA substrates, including bubbles, stem-loops, forks, and Holliday junctions, as well as on RNA-DNA duplexes, implying roles for WRN in DNA replication, recombination, and repair [95-97]. WRN 3’-5’ helicase activity shows substrate specificity similar to that of the exonuclease domain, suggesting that the two WRN enzymatic activities have coordinated functions on several classes of DNA structures [98]. The available data indicate an intimate link between WRN and components of the machinery that repairs DSBs by NHEJ, HR, and the base excision repair (BER) pathways.
Essential components of the NHEJ pathway have been found to interact with WRN, namely Ku and DNA-PKcs [99]. Ku stimulates activity of the WRN 3’- to 5’-exonuclease and increases the processivity of the enzyme [100]. DNA-PKcs interact and phosphorylate WRN both in vitro and in vivo and regulates the WRN helicase and exonuclease activities [101]. The substrate specificity of the WRN helicase and its protein interactions suggest that it may function in HR to promote proper intermediate resolution and suppress strand cross-over events. WRN co-localizes and interacts with Rad52 and NBS1 [102, 103] and in vitro shows modest stimulation of Rad52-mediated single-stranded DNA annealing [104, 105]. Recent in vivo data suggest that WRN participates in late stages of HR resolution [106, 107] and might prevent aberrant recombination events at sites of stalled replication forks by dissociating recombination intermediates [108, 109]. Damage to DNA bases resulting from deamination, oxidation, and alkylation are mainly repaired by BER pathways. WRN interacts physically and functionally with many proteins involved in BER [104]. In addition, WRN itself can unwind the BER strand break intermediate. Thus, WRN might function in a number of steps in the BER process, including damage sensing and processing. In addition, recent evidence indicates that WRN participates in the translesion synthesis pathway to prevent genome instability [103].
Given the complexity of the functional domains of the WRN protein, functional interactions with different repair components, and the wide variety of phenotypes of WS, it is likely that WRN participates in repair of complex DNA lesions generated by HZE particles. To determine the role of WRN in the processing of complex DNA lesions, we irradiated WS cells expressing EGFP-WRN with iron particles and found that WRN not only formed foci as early as 30 min after irradiation but also co-localized with γH2AX (Fig. 4A), indicating that WRN is recruited to the sites of DNA lesions. Further, examination of cell survival by clonogenic assay indicated that WRN-deficient human cells [110] are highly sensitive to iron particles as compared with WS cells complemented with wild-type WRN (Fig. 4B). However, the mechanism by which WRN participates in the processing of complex DNA lesions is not clear. We speculate that because WRN is a unique nuclease with three different enzymatic activities with roles in NHEJ, HR, and BER pathways, it might coordinate factors within individual DNA repair pathways or may coordinate activities among the three pathways. This could be achieved either by binding directly to the damaged DNA or by recruitment of other DNA-repair factors.
Figure 4. WRN and Artemis play roles in processing the complex DNA damage induced by iron particles.

(A) WRN is recruited to the sites of DNA damage induced by iron particles. WS cells stably expressing EGFP-WRN were exposed to iron particles (1Gy at 1GeV/nucleon) and immunostained with γH2AX antibody 30 minutes after irradiation. (B) WRN is important for the processing of DNA damage induced by iron particles. WS cells and WS cells expressing wild-type WRN were irradiated with iron particles (1GeV/nucleon) and were subjected to a colony formation assay. (C) Artemis is critical for the processing of DNA damage induced by iron particles. Artemis-deficient and wild-type cells were exposed to different doses of iron particles (1GeV/nucleon) and were subjected to a colony formation assay. The error bars represent STDEV calculated from at least two independent experiments.
4.3. Artemis
The Artemis protein is defective in patients with severe combined immunodeficiency with sensitivity to IR (RS-SCID). It belongs to a sub-group (the β-CASP family) of the metallo-β-lactamase super family [111, 112]. Artemis is a key component of the variable, diversity, joining (V(D)J) recombination machinery and is required for cell survival in response to DSBs [112-115]. When complexed with and/or phosphorylated by DNA-PKcs, Artemis gains endonuclease activity on 5’ and 3- overhangs with base or sugar damage. In addition, it recognizes and cleaves at a variety of DNA structures, including 5’ and 3’ overhangs, hairpins, flaps, gaps, and loops, with range of preferred cutting positions [116, 117]. Artemis interacts with key components of NHEJ pathway [118], but it is not essential for repair of the majority of the γ-ray-induced DSBs. Further, it has been shown that exogenously expressed myc- and GFP-Artemis display uniform nuclear distribution [119, 120]; there is no co-localization of Artemis and γH2AX [120]. Importantly, despite lack of visible concentration at damaged sites, Artemis is rapidly activated as evidenced by increase in its total phosphorylation after IR treatment [121]. It has been proposed recently that a subset (approximately 10%) of the DSB ends generated by IR require Artemis nuclease activity as these ends have single-stranded tails containing damaged bases or sugars [122, 123]. The magnitude of Artemis-dependent DSB rejoining events correlates with the complexity of the DSB. To determine whether Artemis indeed participates in the processing of complex DNA damage, we exposed Artemis-deficient and wild-type cells to iron particles [121] and determined their survival efficiency. As shown in Fig. 4 (panel C), examination of cell survival by clonogenic assay indicated that Artemis-deficient human cells [121] are highly sensitive to iron particles as compared with wild-type cells. However, the mechanism by which Artemis participates in the process of complex DNA lesions is not clear. We can speculate that given the diversity of substrate configurations that arise after HZE radiation, Artemis might be needed to process damaged ends at DSBs prior to rejoining. This might require DNA-PK- and ATM-dependent phosphorylation of Artemis followed by its activity modification from exonuclease to endonuclease [124].
5. Concluding remarks and future perspectives
DNA repair capability is one of the key indicators reflecting the biological consequences of cellular exposure to genotoxic insults. Determining the biochemical nature of the DNA damage induced by HZE particles and identifying the factors that are required for the repair of these DNA lesions will provide tools to assess individual radiation susceptibility and also to validate risk assessment for human exposure to HZE particles. Cellular approaches, including live-cell imaging, in combination with in vitro assay systems, have significantly improved our molecular understanding of clustered DNA damage. We now know about several basal factors that are involved in the processing of each specific DNA lesion type along the track of dense ionization produced by HZE particles. Although these techniques have provided great insight, future work is required to identify the multiprotein complexes that are involved in processing of complex DNA lesions. Developments in imaging, especially those that will increase resolution in time and space, will provide new opportunities to explore the mechanisms of complex DNA lesions repair. In particular, multicolor tracking, in which several fluorescently labeled proteins are expressed in the cell, would allow the synchronous tracking of several factors that associate with complex DNA lesions in living cells. Further, multicolor immunofluorescence will greatly benefit from the use of quantum dots that have superior optical properties (including brightness, fine emission profiles, and resistance to photobleaching). Indeed, the application of different techniques to different repair factors will answer key questions regarding HZE particle-induced genome instability.
There are several lines of evidence indicate a degree of competition between NHEJ and HR, new evidence strongly suggests that the two pathways collaborate to enhance overall DNA repair and safeguard genomic integrity [82-84]. DSB repair pathway choice is regulated by several factors, including the nature of the lesion and the cell cycle phase. Although NHEJ factors are recruited to DSBs more rapidly than HR factors, and factors in each pathway appear to be recruited independently of factors in the other pathway, there is a significant period of time when both sets of factors can be detected at damage sites. While proteins that catalyze HR and NHEJ are largely distinct, a few proteins have been implicated in both pathways. For example, defects in the yeast Mre11/Rad50/Xrs2 complex affect both HR and NHEJ [125], reflecting the importance of Mre11 nuclease and Rad50 end-tethering functions for DSB repair. DNA-PK has a well-characterized role in NHEJ, but it has also been implicated in regulating the choice between NHEJ and HR [85, 86]. However, the mechanisms underlying repair pathway choice and the precise role of proteins responsible for this process in response to clustered DNA lesions remain unclear.
The repair of lesions and gaps in DNA is performed by different pathways mediated by specific proteins and complexes. Post-translational modifications in many of these proteins govern their activities and interactions, ultimately determining whether a particular pathway is followed [126, 127]. Prominent among these modifications are the additions of phosphate or ubiquitin (and ubiquitin-like) moieties; these modifications are sources of specificity, strength, and timing and also confer new binding surfaces and conformational states on the modified proteins. Apart from a few notable exceptions, the significance of these multiple protein modifications in cellular responses to HZE particles remains elusive.
Until now, many aspects of the cellular response to complex DNA damage have been investigated in monolayer cell culture. Although these systems have provided useful and powerful information, monolayer cell cultures do not recapitulate the three-dimensional structural organization or functional differentiation of the cells in vivo. Recently, radiation-induced cellular responses in three-dimensional (3D) cell culture models have been reported [128, 129]. Though cells grown in 3D culture recapitulate some of the in vivo tissue architecture, they do not truly reflect the in vivo tissue physiology. Therefore, investigation of DNA repair in various tissues will be needed to unravel tissue-specific effects and to explain why certain organ regions are differentially affected by HZE particle radiation.
Acknowledgments
This research was partially supported by the Office of Science (BER), U.S. Department of Energy (DE-AI02-05ER64048), NASA (NNJ05HD36 and NNZ07AU42G) and National Institutes of Health (CA134991) to David J. Chen. We thank Adam Rusek, Peter Guida and Angela Kim of NSRL, Brookhaven National Laboratory, for their help in particle irradiation.
Abbreviations
- IR
ionizing radiation
- DSB
double strand break
- LET
linear energy transfer
- EGFP
enhanced green fluorescent protein
- NHEJ
non-homologous end-joining
- HR
homologous recombination
- WRN
Werner syndrome protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Letaw JR, Silberberg R, Tsao CH. Radiation hazards on space missions. Nature. 1987;330:709–710. doi: 10.1038/330709a0. [DOI] [PubMed] [Google Scholar]
- 2.George K, Durante M, Wu H, Willingham V, Badhwar G, Cucinotta FA. Chromosome aberrations in the blood lymphocytes of astronauts after space flight. Radiat Res. 2001;156:731–738. doi: 10.1667/0033-7587(2001)156[0731:caitbl]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 3.Goodhead DT. New radiobiological, radiation risk and radiation protection paradigms. Mutat Res. 2010;687:13–16. doi: 10.1016/j.mrfmmm.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Durante M, Cucinotta FA. Heavy ion carcinogenesis and human space exploration. Nat Rev Cancer. 2008;8:465–472. doi: 10.1038/nrc2391. [DOI] [PubMed] [Google Scholar]
- 5.Curtis SB, Letaw JR. Galactic cosmic rays and cell-hit frequencies outside the magnetosphere. Adv Space Res. 1989;9:293–298. doi: 10.1016/0273-1177(89)90452-3. [DOI] [PubMed] [Google Scholar]
- 6.Fry RJ, Nachtwey DS. Radiation protection guidelines for space missions. Health Phys. 1988;55:159–164. doi: 10.1097/00004032-198808000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Kramer M, Weyrather WK, Scholz M. The increased biological effectiveness of heavy charged particles: from radiobiology to treatment planning. Technol Cancer Res Treat. 2003;2:427–436. doi: 10.1177/153303460300200507. [DOI] [PubMed] [Google Scholar]
- 8.Rydberg B, Cooper B, Cooper PK, Holley WR, Chatterjee A. Dose-dependent misrejoining of radiation-induced DNA double-strand breaks in human fibroblasts: experimental and theoretical study for high- and low-LET radiation. Radiat Res. 2005;163:526–534. doi: 10.1667/rr3346. [DOI] [PubMed] [Google Scholar]
- 9.Asaithamby A, Uematsu N, Chatterjee A, Story MD, Burma S, Chen DJ. Repair of HZE-particle-induced DNA double-strand breaks in normal human fibroblasts. Radiat Res. 2008;169:437–446. doi: 10.1667/RR1165.1. [DOI] [PubMed] [Google Scholar]
- 10.George K, Durante M, Willingham V, Wu H, Yang TC, Cucinotta FA. Biological effectiveness of accelerated particles for the induction of chromosome damage measured in metaphase and interphase human lymphocytes. Radiat Res. 2003;160:425–435. doi: 10.1667/rr3064. [DOI] [PubMed] [Google Scholar]
- 11.Goodhead DT. Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int J Radiat Biol. 1994;65:7–17. doi: 10.1080/09553009414550021. [DOI] [PubMed] [Google Scholar]
- 12.Chatterjee A, Holley WR. Biochemical mechanisms and clusters of damage for high-LET radiation. Adv Space Res. 1992;12:33–43. doi: 10.1016/0273-1177(92)90087-e. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee A, Schaefer HJ. Microdosimetric structure of heavy ion tracks in tissue. Radiat Environ Biophys. 1976;13:215–227. doi: 10.1007/BF01330766. [DOI] [PubMed] [Google Scholar]
- 14.Brenner DJ, Ward JF. Constraints on energy deposition and target size of multiply damaged sites associated with DNA double-strand breaks. Int J Radiat Biol. 1992;61:737–748. doi: 10.1080/09553009214551591. [DOI] [PubMed] [Google Scholar]
- 15.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc Natl Acad Sci U S A. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hada M, Georgakilas AG. Formation of clustered DNA damage after high-LET irradiation: a review. J Radiat Res (Tokyo) 2008;49:203–210. doi: 10.1269/jrr.07123. [DOI] [PubMed] [Google Scholar]
- 17.Sutherland BM, Bennett PV, Schenk H, Sidorkina O, Laval J, Trunk J, Monteleone D, Sutherland J. Clustered DNA damages induced by high and low LET radiation, including heavy ions. Phys Med. 2001;17(Suppl 1):202–204. [PubMed] [Google Scholar]
- 18.Blaisdell JO, Harrison L, Wallace SS. Base excision repair processing of radiation-induced clustered DNA lesions. Radiat Prot Dosimetry. 2001;97:25–31. doi: 10.1093/oxfordjournals.rpd.a006634. [DOI] [PubMed] [Google Scholar]
- 19.Harrison L, Hatahet Z, Purmal AA, Wallace SS. Multiply damaged sites in DNA: interactions with Escherichia coli endonucleases III and VIII. Nucleic Acids Res. 1998;26:932–941. doi: 10.1093/nar/26.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gollapalle E, Wang R, Adetolu R, Tsao D, Francisco D, Sigounas G, Georgakilas AG. Detection of oxidative clustered DNA lesions in X-irradiated mouse skin tissues and human MCF-7 breast cancer cells. Radiat Res. 2007;167:207–216. doi: 10.1667/rr0659.1. [DOI] [PubMed] [Google Scholar]
- 21.Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 22.Goodhead DT. Energy deposition stochastics and track structure: what about the target? Radiat Prot Dosimetry. 2006;122:3–15. doi: 10.1093/rpd/ncl498. [DOI] [PubMed] [Google Scholar]
- 23.Nikjoo H, O’Neill P, Terrissol M, Goodhead DT. Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat Environ Biophys. 1999;38:31–38. doi: 10.1007/s004110050135. [DOI] [PubMed] [Google Scholar]
- 24.Nikjoo H, O’Neill P, Wilson WE, Goodhead DT. Computational approach for determining the spectrum of DNA damage induced by ionizing radiation. Radiat Res. 2001;156:577–583. doi: 10.1667/0033-7587(2001)156[0577:cafdts]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 25.David-Cordonnier MH, Cunniffe SM, Hickson ID, O’Neill P. Efficiency of incision of an AP site within clustered DNA damage by the major human AP endonuclease. Biochemistry. 2002;41:634–642. doi: 10.1021/bi011682l. [DOI] [PubMed] [Google Scholar]
- 26.Gulston M, Fulford J, Jenner T, de Lara C, O’Neill P. Clustered DNA damage induced by gamma radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomax ME, Salje H, Cunniffe S, O’Neill P. 8-OxoA inhibits the incision of an AP site by the DNA glycosylases Fpg, Nth and the AP endonuclease HAP1. Radiat Res. 2005;163:79–84. doi: 10.1667/rr3284. [DOI] [PubMed] [Google Scholar]
- 28.Gulston M, de Lara C, Jenner T, Davis E, O’Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eccles LJ, Lomax ME, O’Neill P. Hierarchy of lesion processing governs the repair, double-strand break formation and mutability of three-lesion clustered DNA damage. Nucleic Acids Res. 2010;38:1123–1134. doi: 10.1093/nar/gkp1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujimoto H, Pinak M, Nemoto T, O’Neill P, Kume E, Saito K, Maekawa H. Molecular dynamics simulation of clustered DNA damage sites containing 8-oxoguanine and abasic site. J Comput Chem. 2005;26:788–798. doi: 10.1002/jcc.20184. [DOI] [PubMed] [Google Scholar]
- 31.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J Mol Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 32.Malyarchuk S, Castore R, Harrison L. DNA repair of clustered lesions in mammalian cells: involvement of non-homologous end-joining. Nucleic Acids Res. 2008;36:4872–4882. doi: 10.1093/nar/gkn450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellon S, Shikazono N, Cunniffe S, Lomax M, O’Neill P. Processing of thymine glycol in a clustered DNA damage site: mutagenic or cytotoxic. Nucleic Acids Res. 2009;37:4430–4440. doi: 10.1093/nar/gkp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, Bonner WM. Role of oxidatively induced DNA lesions in human pathogenesis. Mutat Res. 2010;704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodhead DT, Thacker J, Cox R. Weiss Lecture. Effects of radiations of different qualities on cells: molecular mechanisms of damage and repair. Int J Radiat Biol. 1993;63:543–556. doi: 10.1080/09553009314450721. [DOI] [PubMed] [Google Scholar]
- 36.Dianov GL, O’Neill P, Goodhead DT. Securing genome stability by orchestrating DNA repair: removal of radiation-induced clustered lesions in DNA. Bioessays. 2001;23:745–749. doi: 10.1002/bies.1104. [DOI] [PubMed] [Google Scholar]
- 37.Heilmann J, Rink H, Taucher-Scholz G, Kraft G. DNA strand break induction and rejoining and cellular recovery in mammalian cells after heavy-ion irradiation. Radiat Res. 1993;135:46–55. [PubMed] [Google Scholar]
- 38.Taucher-Scholz G, Heilmann J, Kraft G. Induction and rejoining of DNA double-strand breaks in CHO cells after heavy ion irradiation. Adv Space Res. 1996;18:83–92. doi: 10.1016/0273-1177(95)00794-f. [DOI] [PubMed] [Google Scholar]
- 39.Jakob B, Scholz M, Taucher-Scholz G. Biological imaging of heavy charged-particle tracks. Radiat Res. 2003;159:676–684. doi: 10.1667/0033-7587(2003)159[0676:biohct]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Desai N, Davis E, O’Neill P, Durante M, Cucinotta FA, Wu H. Immunofluorescence detection of clustered gamma-H2AX foci induced by HZE-particle radiation. Radiat Res. 2005;164:518–522. doi: 10.1667/rr3431.1. [DOI] [PubMed] [Google Scholar]
- 41.Karlsson KH, Stenerlow B. Focus formation of DNA repair proteins in normal and repair-deficient cells irradiated with high-LET ions. Radiat Res. 2004;161:517–527. doi: 10.1667/rr3171. [DOI] [PubMed] [Google Scholar]
- 42.Okayasu R, Okada M, Okabe A, Noguchi M, Takakura K, Takahashi S. Repair of DNA damage induced by accelerated heavy ions in mammalian cells proficient and deficient in the non-homologous end-joining pathway. Radiat Res. 2006;165:59–67. doi: 10.1667/rr3489.1. [DOI] [PubMed] [Google Scholar]
- 43.Sutherland BM, Bennett PV, Saparbaev M, Sutherland JC, Laval J. Clustered DNA damages as dosemeters for ionising radiation exposure and biological responses. Radiat Prot Dosimetry. 2001;97:33–38. doi: 10.1093/oxfordjournals.rpd.a006635. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland BM, Georgakilas AG, Bennett PV, Laval J, Sutherland JC. Quantifying clustered DNA damage induction and repair by gel electrophoresis, electronic imaging and number average length analysis. Mutat Res. 2003;531:93–107. doi: 10.1016/j.mrfmmm.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Georgakilas AG, Bennett PV, Sutherland BM. High efficiency detection of bi-stranded abasic clusters in gamma-irradiated DNA by putrescine. Nucleic Acids Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Georgakilas AG, Bennett PV, Wilson DM, 3rd, Sutherland BM. Processing of bistranded abasic DNA clusters in gamma-irradiated human hematopoietic cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leloup C, Garty G, Assaf G, Cristovao A, Breskin A, Chechik R, Shchemelinin S, Paz-Elizur T, Livneh Z, Schulte RW, Bashkirov V, Milligan JR, Grosswendt B. Evaluation of lesion clustering in irradiated plasmid DNA. Int J Radiat Biol. 2005;81:41–54. doi: 10.1080/09553000400017895. [DOI] [PubMed] [Google Scholar]
- 48.Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen BP, Chan DW, Kobayashi J, Burma S, Asaithamby A, Morotomi-Yano K, Botvinick E, Qin J, Chen DJ. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem. 2005;280:14709–14715. doi: 10.1074/jbc.M408827200. [DOI] [PubMed] [Google Scholar]
- 51.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Asaithamby A, Chen DJ. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res. 2009;37:3912–3923. doi: 10.1093/nar/gkp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okano S, Kanno S, Nakajima S, Yasui A. Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J Biol Chem. 2000;275:32635–32641. doi: 10.1074/jbc.M004085200. [DOI] [PubMed] [Google Scholar]
- 54.Bennett PV, Cintron NS, Gros L, Laval J, Sutherland BM. Are endogenous clustered DNA damages induced in human cells? Free Radic Biol Med. 2004;37:488–499. doi: 10.1016/j.freeradbiomed.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 55.Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res. 2008;18:48–63. doi: 10.1038/cr.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:13738–13743. doi: 10.1073/pnas.0406048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Campalans A, Amouroux R, Bravard A, Epe B, Radicella JP. UVA irradiation induces relocalisation of the DNA repair protein hOGG1 to nuclear speckles. J Cell Sci. 2007;120:23–32. doi: 10.1242/jcs.03312. [DOI] [PubMed] [Google Scholar]
- 58.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 59.Dinant C, Luijsterburg MS, Hofer T, von Bornstaedt G, Vermeulen W, Houtsmuller AB, van Driel R. Assembly of multiprotein complexes that control genome function. J Cell Biol. 2009;185:21–26. doi: 10.1083/jcb.200811080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jakob B, Rudolph JH, Gueven N, Lavin MF, Taucher-Scholz G. Live cell imaging of heavy-ion-induced radiation responses by beamline microscopy. Radiat Res. 2005;163:681–690. doi: 10.1667/rr3374. [DOI] [PubMed] [Google Scholar]
- 61.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costes SV, Chiolo I, Pluth JM, Barcellos-Hoff MH, Jakob B. Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutat Res. 2010;704:78–87. doi: 10.1016/j.mrrev.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Costes SV, Ponomarev A, Chen JL, Nguyen D, Cucinotta FA, Barcellos-Hoff MH. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Comput Biol. 2007;3:e155. doi: 10.1371/journal.pcbi.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 65.Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tobias F, Durante M, Taucher-Scholz G, Jakob B. Spatiotemporal analysis of DNA repair using charged particle radiation. Mutat Res. 2010;704:54–60. doi: 10.1016/j.mrrev.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 67.Tamulevicius P, Wang M, Iliakis G. Homology-directed repair is required for the development of radioresistance during S phase: interplay between double-strand break repair and checkpoint response. Radiat Res. 2007;167:1–11. doi: 10.1667/RR0751.1. [DOI] [PubMed] [Google Scholar]
- 68.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 70.Wilson DM, 3rd, Thompson LH. Molecular mechanisms of sister-chromatid exchange. Mutat Res. 2007;616:11–23. doi: 10.1016/j.mrfmmm.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 71.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagasawa H, Little JB, Inkret WC, Carpenter S, Raju MR, Chen DJ, Strniste GF. Response of X-ray-sensitive CHO mutant cells (xrs-6c) to radiation. II. Relationship between cell survival and the induction of chromosomal damage with low doses of alpha particles. Radiat Res. 1991;126:280–288. [PubMed] [Google Scholar]
- 74.Hinz JM, Yamada NA, Salazar EP, Tebbs RS, Thompson LH. Influence of double-strand-break repair pathways on radiosensitivity throughout the cell cycle in CHO cells. DNA Repair (Amst) 2005;4:782–792. doi: 10.1016/j.dnarep.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 75.Tucker JD, Marples B, Ramsey MJ, Lutze-Mann LH. Persistence of chromosome aberrations in mice acutely exposed to 56Fe+26 ions. Radiat Res. 2004;161:648–655. doi: 10.1667/rr3177. [DOI] [PubMed] [Google Scholar]
- 76.Wang H, Wang X, Zhang P, Wang Y. The Ku-dependent non-homologous end-joining but not other repair pathway is inhibited by high linear energy transfer ionizing radiation. DNA Repair (Amst) 2008;7:725–733. doi: 10.1016/j.dnarep.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 77.Wang H, Zhang X, Wang P, Yu X, Essers J, Chen D, Kanaar R, Takeda S, Wang Y. Characteristics of DNA-binding proteins determine the biological sensitivity to high-linear energy transfer radiation. Nucleic Acids Res. 38:3245–3251. doi: 10.1093/nar/gkq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 80.Adachi N, So S, Iiizumi S, Nomura Y, Murai K, Yamakawa C, Miyagawa K, Koyama H. The human pre-B cell line Nalm-6 is highly proficient in gene targeting by homologous recombination. DNA Cell Biol. 2006;25:19–24. doi: 10.1089/dna.2006.25.19. [DOI] [PubMed] [Google Scholar]
- 81.Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, Alt FW, Jasin M. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 84.You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010;20:402–409. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allen C, Kurimasa A, Brenneman MA, Chen DJ, Nickoloff JA. DNA-dependent protein kinase suppresses double-strand break-induced and spontaneous homologous recombination. Proc Natl Acad Sci U S A. 2002;99:3758–3763. doi: 10.1073/pnas.052545899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Olsson G, Czene S, Jenssen D, Harms-Ringdahl M. Induction of homologous recombination in the hprt gene of V79 Chinese hamster cells in response to low- and high-LET irradiation. Cytogenet Genome Res. 2004;104:227–231. doi: 10.1159/000077494. [DOI] [PubMed] [Google Scholar]
- 88.Zafar F, Seidler SB, Kronenberg A, Schild D, Wiese C. Homologous recombination contributes to the repair of DNA double-strand breaks induced by high-energy iron ions. Radiat Res. 173:27–39. doi: 10.1667/RR1910.1. [DOI] [PubMed] [Google Scholar]
- 89.Pastwa E, Neumann RD, Mezhevaya K, Winters TA. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiat Res. 2003;159:251–261. doi: 10.1667/0033-7587(2003)159[0251:roridd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 90.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 91.Nishino T, Morikawa K. Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene. 2002;21:9022–9032. doi: 10.1038/sj.onc.1206135. [DOI] [PubMed] [Google Scholar]
- 92.Goto M. Hierarchical deterioration of body systems in Werner’s syndrome: implications for normal ageing. Mech Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 93.Rossi ML, Ghosh AK, Bohr VA. Roles of Werner syndrome protein in protection of genome integrity. DNA Repair (Amst) 2010;9:331–344. doi: 10.1016/j.dnarep.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3’-->5’ exonuclease. Nat Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 96.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perry JJ, Asaithamby A, Barnebey A, Kiamanesch F, Chen DJ, Han S, Tainer JA, Yannone SM. Identification of a coiled coil in werner syndrome protein that facilitates multimerization and promotes exonuclease processivity. J Biol Chem. 2010;285:25699–25707. doi: 10.1074/jbc.M110.124941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Opresko PL, Sowd G, Wang H. The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation. PLoS One. 2009;4:e4825. doi: 10.1371/journal.pone.0004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cooper MP, Machwe A, Orren DK, Brosh RM, Ramsden D, Bohr VA. Ku complex interacts with and stimulates the Werner protein. Genes Dev. 2000;14:907–912. [PMC free article] [PubMed] [Google Scholar]
- 100.Karmakar P, Piotrowski J, Brosh RM, Jr, Sommers JA, Miller SP, Cheng WH, Snowden CM, Ramsden DA, Bohr VA. Werner protein is a target of DNA-dependent protein kinase in vivo and in vitro, and its catalytic activities are regulated by phosphorylation. J Biol Chem. 2002;277:18291–18302. doi: 10.1074/jbc.M111523200. [DOI] [PubMed] [Google Scholar]
- 101.Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ. Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase. J Biol Chem. 2001;276:38242–38248. doi: 10.1074/jbc.M101913200. [DOI] [PubMed] [Google Scholar]
- 102.Sakamoto S, Nishikawa K, Heo SJ, Goto M, Furuichi Y, Shimamoto A. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001;6:421–430. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- 103.Kobayashi J, Okui M, Asaithamby A, Burma S, Chen BP, Tanimoto K, Matsuura S, Komatsu K, Chen DJ. WRN participates in translesion synthesis pathway through interaction with NBS1. Mech Ageing Dev. 2010;131:436–444. doi: 10.1016/j.mad.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Baynton K, Otterlei M, Bjoras M, von Kobbe C, Bohr VA, Seeberg E. WRN interacts physically and functionally with the recombination mediator protein RAD52. J Biol Chem. 2003;278:36476–36486. doi: 10.1074/jbc.M303885200. [DOI] [PubMed] [Google Scholar]
- 105.Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA. Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci. 2006;119:5137–5146. doi: 10.1242/jcs.03291. [DOI] [PubMed] [Google Scholar]
- 106.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol Cell Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Swanson C, Saintigny Y, Emond MJ, Monnat RJ., Jr The Werner syndrome protein has separable recombination and survival functions. DNA Repair (Amst) 2004;3:475–482. doi: 10.1016/j.dnarep.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 108.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner’s syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. Embo J. 2010 doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Perry JJ, Yannone SM, Holden LG, Hitomi C, Asaithamby A, Han S, Cooper PK, Chen DJ, Tainer JA. WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat Struct Mol Biol. 2006;13:414–422. doi: 10.1038/nsmb1088. [DOI] [PubMed] [Google Scholar]
- 111.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 112.Moshous D, Callebaut I, de Chasseval R, Poinsignon C, Villey I, Fischer A, de Villartay JP. The V(D)J recombination/DNA repair factor artemis belongs to the metallo-beta-lactamase family and constitutes a critical developmental checkpoint of the lymphoid system. Ann N Y Acad Sci. 2003;987:150–157. doi: 10.1111/j.1749-6632.2003.tb06043.x. [DOI] [PubMed] [Google Scholar]
- 113.de Villartay JP. V(D)J recombination deficiencies. Adv Exp Med Biol. 2009;650:46–58. doi: 10.1007/978-1-4419-0296-2_4. [DOI] [PubMed] [Google Scholar]
- 114.Kurosawa A, Adachi N. Functions and Regulation of Artemis: A Goddess in the Maintenance of Genome Integrity. J Radiat Res (Tokyo) 2010 doi: 10.1269/jrr.10017. [DOI] [PubMed] [Google Scholar]
- 115.Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 116.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 117.Gu J, Li S, Zhang X, Wang LC, Niewolik D, Schwarz K, Legerski RJ, Zandi E, Lieber MR. DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair (Amst) 2010;9:429–437. doi: 10.1016/j.dnarep.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pannicke U, Ma Y, Hopfner KP, Niewolik D, Lieber MR, Schwarz K. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. Embo J. 2004;23:1987–1997. doi: 10.1038/sj.emboj.7600206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Soubeyrand S, Pope L, De Chasseval R, Gosselin D, Dong F, de Villartay JP, Hache RJ. Artemis phosphorylated by DNA-dependent protein kinase associates preferentially with discrete regions of chromatin. J Mol Biol. 2006;358:1200–1211. doi: 10.1016/j.jmb.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 121.Wang J, Pluth JM, Cooper PK, Cowan MJ, Chen DJ, Yannone SM. Artemis deficiency confers a DNA double-strand break repair defect and Artemis phosphorylation status is altered by DNA damage and cell cycle progression. DNA Repair (Amst) 2005;4:556–570. doi: 10.1016/j.dnarep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 122.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 123.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 124.Dahm K. Functions and regulation of human artemis in double strand break repair. J Cell Biochem. 2007;100:1346–1351. doi: 10.1002/jcb.21226. [DOI] [PubMed] [Google Scholar]
- 125.Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G, Lee SE. Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. Embo J. 2010 doi: 10.1038/emboj.2010.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Morris JR. More modifiers move on DNA damage. Cancer Res. 2010;70:3861–3863. doi: 10.1158/0008-5472.CAN-10-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Thomson TM, Guerra-Rebollo M. Ubiquitin and SUMO signalling in DNA repair. Biochem Soc Trans. 2010;38:116–131. doi: 10.1042/BST0380116. [DOI] [PubMed] [Google Scholar]
- 128.Lin YF, Nagasawa H, Peng Y, Chuang EY, Bedford JS. Comparison of several radiation effects in human MCF10A mammary epithelial cells cultured as 2D monolayers or 3D acinar stuctures in matrigel. Radiat Res. 2009;171:708–715. doi: 10.1667/RR1554.1. [DOI] [PubMed] [Google Scholar]
- 129.Roig AI, Hight SK, Shay JW. Two- and three-dimensional models for risk assessment of radiation-enhanced colorectal tumorigenesis. Radiat Res. 2009;171:33–40. doi: 10.1667/RR1415.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jakob B, Splinter J, Durante M, Taucher-Scholz G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc Natl Acad Sci U S A. 2009;106:3172–3177. doi: 10.1073/pnas.0810987106. [DOI] [PMC free article] [PubMed] [Google Scholar]