SUMMARY
The assembly and function of the mitochondrial respiratory chain (RC) involve the organization of RC enzyme complexes in supercomplexes or respirasomes through an unknown biosynthetic process. This leads to structural interdependences between RC complexes, which are highly relevant from biological and biomedical perspectives, because RC defects lead to severe human disorders. We show that in human cells, respirasome biogenesis involves a complex I assembly intermediate acting as a scaffold for the combined incorporation of complexes III and IV subunits, rather than originating from the association of preassembled individual holoenzymes. The process ends with the incorporation of complex I NADH dehydrogenase catalytic module, which leads to the respirasome activation. While complexes III and IV assemble either as free holoenzymes or by incorporation of free subunits into supercomplexes, the respirasomes constitute the structural units where complex I is assembled and activated, thus explaining the functional significance of the respirasomes for RC function.
INTRODUCTION
The mammalian oxidative phosphorylation (OXPHOS) system is formed by five multiprotein enzyme complexes and two mobile electron carriers (ubiquinone and cytochrome c) embedded in the mitochondrial inner membrane. Complexes I to IV (CI to CIV) form the respiratory chain (RC), which facilitates electron transfer from reducing equivalents to molecular oxygen coupled to proton pumping across the inner membrane. The proton gradient generated is subsequently used by complex V to drive ATP synthesis. With the exception of CII subunits that are all encoded by the nuclear genome, the remaining OXPHOS enzymes contain subunits of dual genetic origin, nuclear and mitochondrial. Nuclear DNA (nDNA)-encoded subunits are synthesized on cytosolic ribosomes and imported into mitochondria, where they assemble together with mitochondrial DNA (mtDNA)-encoded subunits and prosthetic groups to build up OXPHOS complexes with the assistance of specific chaperones or assembly factors (Fernandez-Vizarra et al., 2009).
The structural and functional organization of the respiratory chain has been a matter of debate over more than 50 years. Two models have been historically hypothesized. Following the “fluid state” model, individual OXPHOS complexes diffuse freely in the mitochondrial inner membrane and electron transport occurs when the complexes randomly collide (Hackenbrock et al 1986). Conversely, the “solid state” model proposes that OXPHOS complexes are organized in rigid higher order assemblies (Chance and Williams, 1955) known as supercomplexes or respirasomes (Schagger and Pfeiffer, 2000). It is currently accepted that both organizations probably coexist, giving rise to the “dynamic aggregate” or “plasticity” model. This model suggests that OXPHOS complexes switch from freely moving to fixed structures and viceversa to adapt to changes in cellular metabolism (Acin-Perez et al., 2008). Accordingly, the coexistence of the respirasome unit, composed of at least CI+CIII+CIV, together with free CIII and CIV has been widely described (Althoff et al., 2011; Dudkina et al., 2011; Heinemeyer et al., 2007; Schafer et al., 2006; Schagger and Pfeiffer, 2000). However, the association of complexes II and V with the respirasomes remains controversial (Acin-Perez et al., 2008; Wittig and Schagger, 2009). It has been suggested that association of RC complexes in respirasomes may offer structural or functional advantages, such as the prevention of their destabilization and degradation, the enhancement of electron transport efficiency and substrate channeling, or the decrease of electron or proton leakages (Lenaz and Genova, 2010). As a consequence of their organization in supercomplexes, a structural interdependence exists amongst the individual OXPHOS complexes (Acin-Perez et al., 2004; Diaz et al., 2006; Li et al., 2007; Schagger et al., 2004; Soto et al., 2009). This has major biological as well as biomedical implications, since RC enzyme assembly defects produce severe encephalomyopathies and neurodegenerative disorders in human and have been associated with the aging process. In human mitochondrial diseases, structural alterations primarily affecting one given complex often induce pleiotropic deleterious effects of the other enzymes. For instance, pathogenic mutations in CIII subunits or assembly genes lead to pleiotropic deficiencies of CI and CIV in affected tissues (Fernandez-Vizarra et al., 2007; Lamantea et al., 2002; Moran et al., 2010), mutations in CIV subunits may lead to secondary CI deficiencies (D’Aurelio et al., 2006) and mutations affecting CI-specific genes can produce combined CI and CIII, or CI and CIV, deficiencies in patients (Budde et al., 2000; Saada et al., 2011).
The mechanisms that regulate the biosynthesis of mitochondrial supercomplexes remain unsolved. Pulse-chase experiments that analyzed the time-course incorporation of the thirteen mtDNA-encoded proteins into RC complexes and supercomplexes suggested that the latter originate by the direct association of single pre-assembled complexes (Acin-Perez et al., 2008). However, the observation that in the absence of monomeric holo-CIV newly-imported nDNA-encoded CIV subunits were preferentially integrated into supercomplexes, suggested that the formation of these macromolecular assemblies does not necessarily require the preassembly of individual holocomplexes, but could also be achieved through a coordinated association of partially-assembled complexes and free subunits (Lazarou et al., 2009).
In the present study we have addressed the biosynthetic pathway of mitochondrial RC supercomplexes by partially depleting control cell lines of OXPHOS complexes by treatment with doxycycline, a reversible inhibitor of mitochondrial translation. The synthesis of mtDNA-encoded subunits resumed after drug removal, and we subsequently investigated their time-course incorporation into supercomplex assembly intermediates. Based on the analyses of the composition and function of these intermediates we propose a multistep model for mitochondrial supercomplex assembly. It involves three sequential basic steps: 1-formation of a CI assembly intermediate; 2-incorporation of CIII and CIV subunits and subassemblies; 3- final assembly of CI by incorporation of the NADH dehydrogenase catalytic core and subsequent functional respirasome activation. This model allows us to explain the requirements for CI assembly, the structural interdependences among OXPHOS complexes, and why certain genetic defects affecting a single complex may lead to combined RC enzyme defects in patients.
RESULTS
Distribution of RC subunits in mitochondrial complexes and supercomplexes
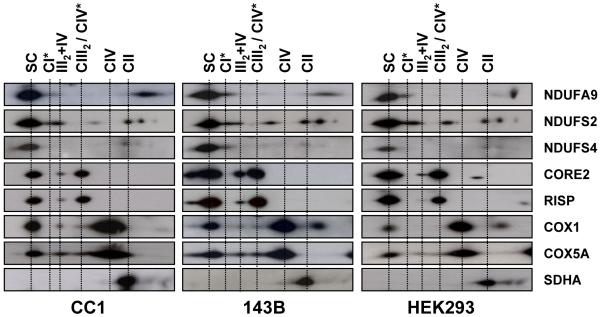
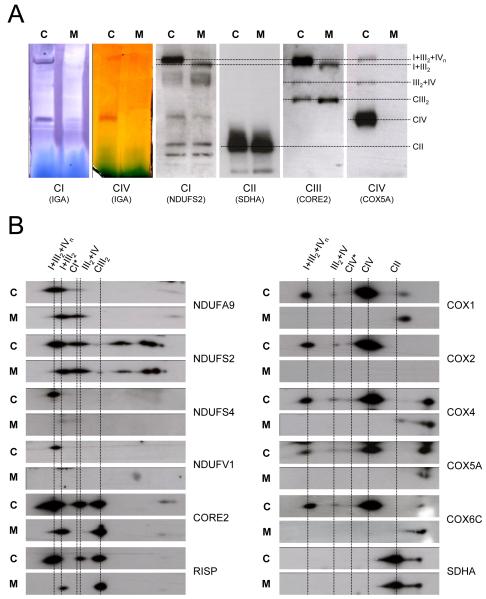
We analyzed the relative distribution of individual RC subunits between large supercomplexes I+III2+IVn (SC), small supercomplexes (III2+IV), and free RC complexes I (CI), III (CIII2), and IV (CIV) in three common human control cell lines (143B206 osteosarcoma cells, HEK293 embryonic kidney cells and control transmitochondrial cybrids). Blue native gel electrophoresis (2D-BN/SDS-PAGE) in combination with western-blot analysis showed comparable protein contents and distribution patterns of all tested RC subunits in the three control cell lines (Figure 1). Between 65-97% of the signals corresponding to CI subunits were localized in supercomplexes larger than ~1500 kDa (SC). The NDUFA9 and NDUFS2 subunits also colocalized in a spot (indicated as CI*) that corresponds to a stable ~830 kDa subcomplex. The NDUFA9, NDUFS2 and NDUFS4 subunits were additionally detected at reduced levels in lower bands that resemble the previously reported pattern of CI subcomplexes (Antonicka et al., 2003; Lazarou et al., 2007; Ugalde et al., 2004a; Vogel et al., 2007a). Concerning CIII subunits, ~50% of CORE2 and Rieske Iron Sulphur Protein (RISP) were detected in large I+III2+IVn supercomplexes, ~40% in free CIII homodimers, and ~5-10% in supercomplex III2+IV. In contrast, ~30% of the signals corresponding to the CIV COX1 and COX5A subunits was detected in I+III2+IVn supercomplexes, ~65-70% in free holo-CIV, and only traces (~1-3%) in supercomplex III2+IV. The signal corresponding to the SDHA subunit appeared mainly at the complex II position (CII), with no clear colocalization with supercomplexes. These results are comparable to those reported in other mammalian cell lines or tissues (Schagger and Pfeiffer, 2001). The signals obtained from three independent western-blots per cell line were quantified and expressed as percentages of the total signal obtained from each antibody (Table S1).The CIII RISP and CIV COX1 subunits were ~50% less abundant within supercomplex III2+IV than their respective partners CORE2 and COX5A. Likewise, the RISP subunit was ~20% less abundant than CORE2 within supercomplex I+III2+IVn, whereas COX1 was ~25% more abundant than COX5A. These results suggest stoichiometric variations among RC subunits of the same complex within the different supercomplex forms.
Figure 1. Steady-state levels of mitochondrial supercomplexes in human cell lines.
Sixty μg of isolated mitochondria were analyzed by 2D-BN/SDS-PAGE. Western-blot analysis was performed using antibodies against the indicated OXPHOS subunits. SC, supercomplexes containing CI, CIII and CIV. CI*, partially-assembled CI. CIII2, complex III dimer. CIV*, putative CIV dimer. III2+IV, supercomplex containing CIII and CIV.
Effects of doxycycline treatment in mitochondrial translation and RC assembly
These variations in the distribution patterns of subunits from the same RC complex could be explained by differences in either the assembly kinetics or the stability of these subunits within the supercomplexes. To discriminate between these two possibilities, we depleted cells of mtDNA-encoded subunits-containing OXPHOS complexes and supercomplexes by treating the cultures with doxycycline, a reversible inhibitor of mitochondrial translation. This strategy was successfully used to follow the assembly kinetics of CI (Ugalde et al., 2004a; Pello et al., 2008). We cultured the 143B cells and control cybrids for 6 days, and the HEK293 cells for 8 days in the presence of 15μg/ml doxycycline. Longer treatments affected cell viability (Ugalde et al., 2004a). Real-time PCR analyses showed that doxycycline induced a partial loss (35-40%) of mtDNA molecules, which recovered back to normal levels 96 h after the treatment. Additionally, doxycycline severely inhibited mitochondrial protein synthesis in 143B cells without affecting overall cytoplasmic translation, although a 10-15% residual synthesis of mitochondrial polypeptides was detectable after quantification (Figure S1). The sporadic mtDNA loss did not further affect mitochondrial translation, since the expression of mitochondrial polypeptides recovered normal levels 72 h after doxycycline treatment.
To follow the accumulation of newly-synthesized RC complexes and their further association into supercomplexes, samples were collected at different time points (0, 6, 15, 24, 48, 72 and 96 hours) after doxycycline removal. Digitonin-solubilized mitochondrial particles were separated by BN-PAGE and analyzed by western-blot. In agreement with the residual synthesis of mtDNA-encoded subunits, we detected an ~80% reduction of CIII and CIV, and a lack of fully-assembled CI (see 0h, Figure S2). These residual amounts were not sufficient to sustain the formation of supercomplexes, in agreement with previous studies suggesting that respirasome assembly proceeds when individual complexes III and IV have accumulated above a critical threshold (D’Aurelio et al., 2006). CII levels were normal after doxycycline treatment, as expected since this complex lacks mtDNA-encoded subunits.
Assembly kinetics of complex I subunits in mitochondrial supercomplexes
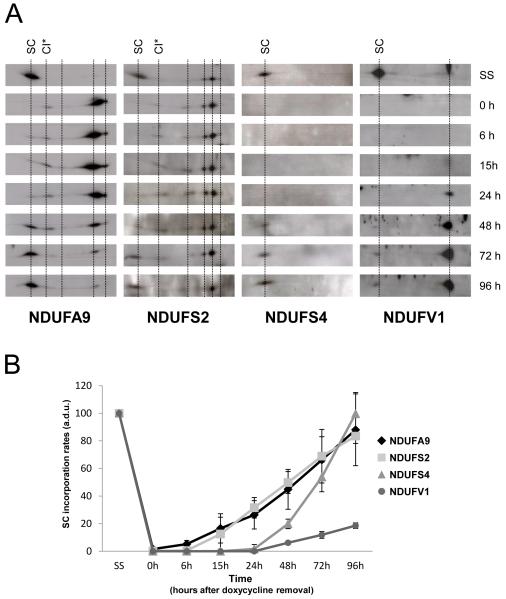
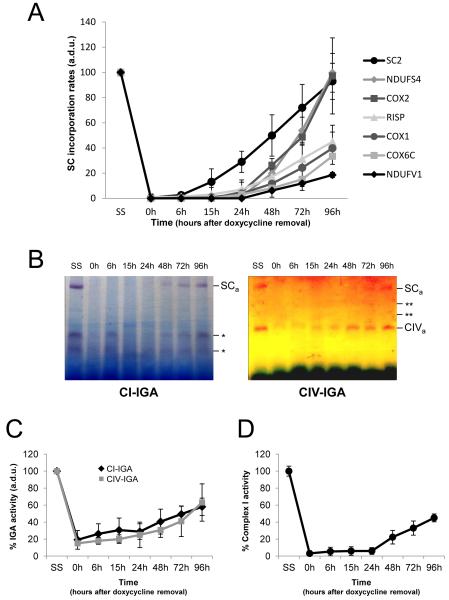
Mammalian CI biogenesis consists in a semi-sequential pathway that involves the preassembly and further association of evolutionarily conserved functional modules (Ugalde et al., 2004a). To follow CI assembly and incorporation into supercomplexes, samples from doxycycline treatment-recovery experiments were separated by 2D-BN/SDS-PAGE and blots were probed with antibodies that recognize subunits representative of different CI assembly stages (McKenzie and Ryan, 2010). NDUFS2 is an early-assembly subunit that forms subcomplexes of the peripheral arm ubiquinone reduction (or Q) module; NDUFA9 is an intermediate-assembly subunit that is thought to be located in the boundary between the peripheral arm and the hydrophobic membrane arm (or P module); and NDUFV1 and NDUFS4 are constituents of the peripheral arm NADH dehydrogenase (or N) catalytic module that are crucial to the final steps of CI assembly. After 6 days of doxycycline treatment (0h in Figure 2A), we did not detect any of the tested CI subunits within the I+III2+IVn supercomplex. At this time, residual amounts of NDUFA9 and NDUFS2 accumulated in spots previously reported as CI subassemblies (Ugalde et al., 2004a; Vogel et al., 2007a). One common spot (indicated as CI*) corresponded to the stable subcomplex of ~830 kDa that lacked the NDUFS4 and NDUFV1 subunits, both of which are part of the CI N module. This subcomplex constitutes a physiological CI assembly intermediate that accumulates in cells from patients with pathogenic mutations in the NDUFS1, NDUFS4 and NDUFV1 genes (Hoefs et al., 2010; Lazarou et al., 2007; Ugalde et al., 2004b; Vogel et al., 2007b). As the translation of mitochondrial proteins restarted, the CI* subcomplex moved towards the position of the large I+III2 or I+III2+IVn supercomplexes and disappeared ~72-96 h after doxycycline removal, indicating that it could be a supercomplex assembly intermediate. Accumulation of the NDUFA9 and NDUFS2 proteins in supercomplexes started to be detected around 15-24 h after doxycycline removal and reached maximum levels at time 96 h. However, the NDUFS4 and NDUFV1 subunits appeared in such structures between 48-72 h after removing the drug, indicating that the initial incorporations of these two subunits within the supercomplexes were delayed in comparison with the NDUFA9 and NDUFS2 subunits. These data suggest that at early supercomplex assembly stages CI is neither fully-assembled nor active when it binds to other RC complexes, since it lacks part of the N module catalytic subunits. The NDUFS4 subunit reached its maximum steady-state levels at time 96 h, indicating that despite of its initial delay this subunit gets rapidly integrated into supercomplexes. However, the NDUFV1 subunit showed much lower levels at 96 h, indicating a marked delay in its incorporation into supercomplexes. A quantitative estimation of the assembly rates for the different CI subunits is shown in Figure 2B.
Figure 2. Incorporation rates of individual complex I subunits in supercomplexes.
(A) Example of 2D-BN/SDS-PAGE western blot analysis in doxycycline-treated 143B cells using antibodies against the indicated CI subunits. SC, supercomplexes containing CI, CIII and CIV. CI*, partially-assembled CI. (B) Mean incorporation rates of CI subunits in large supercomplexes (SC). The signals corresponding to CI subunits were quantified in duplicate gels per cell line, and normalized by the CII SDHA subunit. (a.d.u.), arbitrary densitometric units. Mean values are expressed as percentages relative to untreated cells (SS). Error bars represent standard deviations (SD).
Assembly kinetics of complex III subunits in mitochondrial supercomplexes
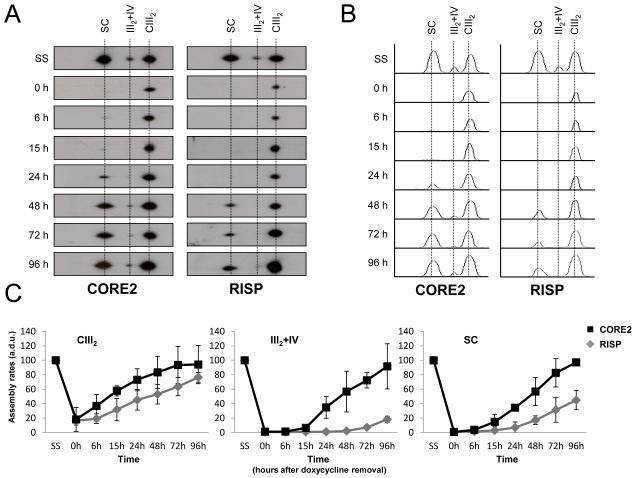
Although CIII assembly has not been well studied in mammals, a model for the biosynthetic pathway of this complex has been proposed in yeast (Zara et al., 2009). Based on this model, we analyzed the integration into CIII of an early-assembly accessory subunit (CORE2), and a late-assembly catalytic subunit (RISP) and their further incorporations into the native supercomplex species III2+IV, I+III2 and I+III2+IVn (the latter two indicated as SC in Figure 3A, B and C). After 6 days of doxycycline treatment (0h), we observed an ~80-90% reduction in the amounts of CORE2 and RISP subunits within the CIII dimer (CIII2) compared to untreated cells (SS in Figure 3B). The accumulation of both subunits within CIII2 increased gradually after the translation of mtDNA-encoded proteins resumed; the CORE2 protein reached maximum steady-state levels after 24-48 h, and the RISP subunit did it after 72-96 h. These data confirmed that in human cells, the RISP subunit also gets assembled at a late stage of CIII assembly. Low levels of the CORE2 protein appeared in the III2+IV and I+III2+IVn supercomplexes around 24-48 h after doxycycline removal, reaching its maximum steady-state levels at 96 h (Figures 3 and S3A). The incorporation of RISP within the I+III2+IVn supercomplexes was delayed compared to CORE2, since it appeared ~48 h after doxycycline removal, keeping low levels (~30-50% of the steady-state) after 96 h. These data indicate that at early supercomplex assembly stages CIII is neither fully-assembled nor active, since it lacks at least the RISP subunit. Surprisingly, RISP started appearing within the III2+IV supercomplex at time 96 h, suggesting that the III2+IV supercomplex gets fully-assembled after the formation of the I+III2+IVn supercomplexes.
Figure 3. Assembly kinetics of individual complex III subunits in supercomplexes.
(A) Example of 2D-BN/SDS-PAGE western blot analysis in doxycycline-treated 143B cells using antibodies against the indicated CIII subunits. (B) Densitometric histogram representing the assembly progress of the CORE2 and RISP subunits in CIII-containing structures. (C) Mean incorporation rates of CORE2 and RISP subunits in CIII-containing structures. The signals were quantified in duplicate gels per cell line, and normalized by CII. CIII2, complex III dimer (left panel); III2+IV, supercomplex containing CIII and CIV (middle panel); SC, supercomplexes containing CI, CIII and CIV (right panel). (a.d.u.), arbitrary densitometric units. Mean values are expressed as percentages relative to untreated cells (SS). Error bars represent standard deviations (SD).
Assembly kinetics of complex IV subunits in mitochondrial supercomplexes
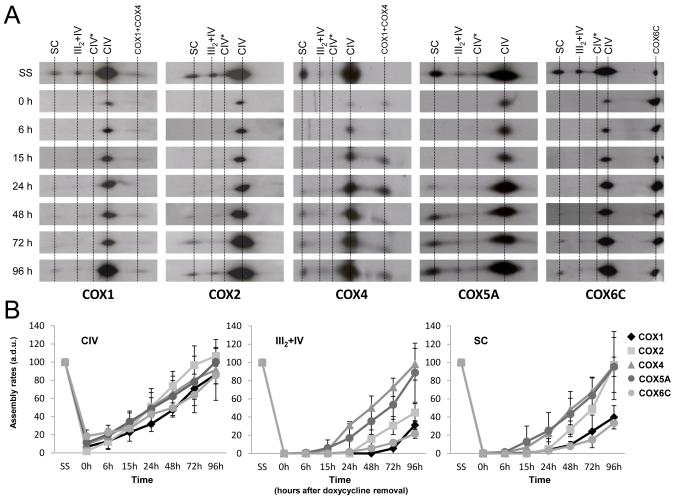
We next analyzed the incorporation of four subunits involved in early steps of human CIV assembly (two mtDNA-encoded catalytic subunits, COX1 and COX2, and two nDNA-encoded structural subunits, COX4 and COX5A) and that of a late-assembly structural subunit (COX6C) into holo-CIV, and into the III2+IV and I+III2+IVn supercomplexes (Fornuskova et al., 2010; Nijtmans et al., 1998)(Figure 4A). The quantification of CIV signals from BN experiments showed that after 6 days of doxycycline treatment (0h), there was a ~90% reduction in the amounts of all COX subunits within holo-CIV compared with the untreated cells (SS) (Figure 4B). The incorporation rates of all subunits within CIV increased steadily after restarting mitochondrial translation and reached maximum steady-state levels at least after 96 h following doxycycline removal. These data confirmed our previous results suggesting that in control human cells, fully-assembled CIV is restored later than CI and CIII (Pello et al., 2008). The COX4 and COX5A subunits started getting incorporated into the I+III2+IVn and III2+IV supercomplexes 24-48 h after doxycycline removal (Figure 4 and S3B). In contrast, the COX1, COX2 and COX6C subunits appeared in the I+III2+IVn supercomplexes after 48-72 h, indicating a delayed incorporation of these three subunits in the large supercomplexes. As shown in Figure 4B, these subunits followed different incorporation rates into I+III2+IVn supercomplexes, since COX2 steady-state levels were fully-restored 96 h after doxycycline removal, while COX1 and COX6C levels remained low (~40-50% of the steady-state). The COX1, COX2 and COX6C subunits were first detected in the III2+IV supercomplex at time 96 h. As for CIII, these data indicate that CIV is neither fully-assembled nor active when its subunits and intermediates bind other RC complexes to form supercomplexes, and confirm that the biosynthesis of the I+III2+IVn supercomplexes precedes the formation of the III2+IV supercomplex.
Figure 4. Assembly kinetics of individual complex IV subunits in supercomplexes.
(A) Example of 2D-BN/SDS-PAGE western blot analysis in doxycycline-treated 143B cells using antibodies against the indicated CIV subunits. (B) Mean incorporation rates of CIV subunits in CIV-containing structures. The signals were quantified in duplicate gels per cell line, and normalized by CII. CIV, complex IV (left panel); III2+IV, supercomplex containing CIII and CIV (middle panel); SC, supercomplexes containing CI, CIII and CIV (right panel). CIV*, putative complex IV dimer. (a.d.u.), arbitrary densitometric units. Mean values correspond to percentages relative to untreated cells (SS). Error bars represent standard deviations (SD).
Assembly kinetics of RC subunits into mitochondrial supercomplexes
To obtain a comprehensive view of the supercomplex assembly pathway, we performed a comparative analysis of the incorporation rates of all tested RC subunits into the large I+III2+IVn supercomplexes (Figure 5 and S3C). Our results showed that CI NDUFA9 and NDUFS2 subunits were initially stabilized in a CI subassembly (CI*, Figure 2) lacking the catalytic N module, which constitutes the first supercomplex assembly intermediate (SC1). In BN gels, this intermediate started moving towards the position of the I+III2+IVn supercomplexes around 24 h after doxycycline removal, in parallel with the incorporation of CIII CORE2 subunit and CIV COX4 and COX5A subunits, reaching maximum levels at time 96 h (Figure S3C). These data imply the formation of a second supercomplex assembly intermediate (SC2) composed of partially-assembled CI with incorporated CIII and CIV subunits or subassemblies (Figure 5A). The CI NDUFS4 and CIV COX2 subunits first appeared in I+III2+IVn supercomplexes around 48 h after removing the drug, but rapidly reached their maximum levels. These data indicate that the incorporation of these two RC subunits within the large supercomplexes occurs in a third assembly stage (SC3). The catalytic CIII RISP and CIV COX1 subunits, and the structural CIV subunit COX6C appeared at low levels in I+III2+IVn supercomplexes after 48-72 h, but these levels remained low at time 96 h, indicating a slower incorporation of these three subunits in a fourth supercomplex assembly stage (SC4). Due to the low relative abundance of COX1 and COX6C within supercomplexes at time 96 h, we cannot differentiate which of these two CIV subunits gets inserted first. This is compatible with a model in which COX1 could get assembled before the final bulk of COX subunits to complete the supercomplex structures. Finally, the catalytic CI subunit NDUFV1 started appearing in I+III2+IVn supercomplexes after 48-72 h, but its assembly kinetics were markedly slower than that of the RISP, COX1 and COX6C subunits, indicating that NDUFV1 incorporates into supercomplexes in a fifth assembly stage (SC5).
Figure 5. Comparative analysis of the incorporation rates of RC subunits into large supercomplexes.
(A) Mean incorporation rates of RC subunits into I+III2+IVn supercomplexes (SC). SC2 represents the mean incorporation rates of NDUFA9, NDUFS2, CORE2, COX4 and COX5A subunits into SC. (B) 60 μg of crude mitochondrial pellets from the doxycycline assays were analyzed by BN-PAGE in combination with in-gel activity (IGA) assays. The left panel shows a CI in-gel activity (IGA) assay, where * indicate unspecific bands with NADH dehydrogenase activity. The right panel shows a CIV IGA assay. ** indicate the putative CIV dimer (CIV*) and supercomplex III2+IV that stained for CIV activity (CIVa). SCa indicates CI and CIV activities in large supercomplexes. (C) Densitometric analysis of the CI and CIV SCa bands performed in three independent experiments. (D) Spectrophotometric CI activities in digitonin-treated 143B cells. Triplicate measurements were normalized by the citrate synthase activity. (a.d.u.), arbitrary densitometric units. Mean values represent percentages relative to untreated cells (SS). Error bars represent standard deviations (SD).
Based on these results, RC supercomplexes should get activated in parallel with the incorporation of the last catalytic subunits (between 48-72 h after doxycycline removal). To confirm this hypothesis, we analyzed samples by BN-PAGE followed by CI and CIV in-gel activity (IGA) assays (Figure 5B). After 6 days of doxycycline treatment (Figure 5B, 0h), we observed a virtual absence of CI and CIV activities within supercomplexes (indicated as SCa) compared to untreated cells (SS, Figure 5B). Densitometric analyses of the IGA assays (Figure 5C) showed that the enzymatic activities of both complexes started increasing gradually in supercomplexes ~48 h after doxycycline removal without reaching their maximum activities after 96 h, in parallel with the incorporation of the NDUFV1 and COX1 subunits. This result was confirmed by spectrophotometric measurements of rotenone-sensitive CI NADH dehydrogenase activity (Figure 5D).
Altered assembly of mitochondrial supercomplexes in COX2 mutant cybrids
To verify our supercomplex assembly model, we investigated the effect of pathogenic mutations that affect the assembly of single RC complexes in the formation of supercomplexes. We analyzed a 143B mutant cybrid carrying the homoplasmic m.7896 G>A mutation in the COX2 gene, described in a CIV-deficient patient (Campos et al., 2001). BN-IGA assays showed severe reductions of CI and CIV enzyme activities in the mutant cybrids compared to an isogenic control (Figure 6A). Western-blot analyses of duplicate BN gels showed the mutants lack fully-assembled CIV and COX-containing supercomplexes, although they accumulate the I+III2 supercomplex and CIII dimer (CIII2) (Figure 6A). Analyses by the sensitive 2D-BN/SDS-PAGE method confirmed that mutant cybrids (M in Figure 6B) completely lack holo-CIV due to the absence of COX2, as a consequence of which, most of the remainder CIV subunits were presumably degraded and those detected accumulated as monomers or small CIV assembly intermediates. This lack of CIV led to the absence of I+III2+IVn and III2+IV supercomplexes, and an accumulation of CIII2, the I+III2 supercomplex, and the ~830 kDa supercomplex assembly intermediate (CI* or SC1). We observed a severe reduction of the NDUFS4 and NDUFV1 subunits within the I+III2 supercomplex, in contrast with the relative abundance of other CI subunits such as NDUFA9 or NDUFS2 (Figure 6B). These results suggest that in the absence of CIV most I+III2 supercomplex is probably partially-assembled and inactive due to a hampered assembly or stability of the NDUFS4 and NDUFV1 subunits. At longer exposures (Figure S4A) the COX1 and COX4 subunits were detected at low levels in the I+III2 supercomplex, although the COX5A and COX6C subunits were not so clearly distinguished. These data suggest that certain free COX subunits or subassemblies may bind directly to supercomplex intermediates in the absence of holo-CIV, as previously proposed (Lazarou et al., 2009).
Figure 6. Analysis of mitochondrial supercomplexes in COX2 mutant cybrids.
(A) BN-PAGE was performed in COX2 mutant cybrids carrying the homoplasmic m.7896G>A mutation (M) and its corresponding isogenic control (C), followed by CI and CIV IGA assays, or alternatively, blotted on nitrocellulose and incubated with the indicated antibodies. (B) 2D-BN/SDS-PAGE western-blot analysis in control (C) and the COX2 mutant (M) cybrids using antibodies against the indicated OXPHOS subunits. CI*, partially-assembled CI. CIII2, complex III dimer. CIV*, putative complex IV dimer.
The steady-state levels of CII were similar in COX2 mutant and control cells, and did not colocalize with supercomplexes (Figure 6). After long exposures, CII traces were detected in a high supramolecular structure that could represent a supercomplex (Figure S4B). However, the abundance and electrophoretic mobility of this particular spot (indicated as SCII) was unaffected by the lack of CIV in the mutant cell line. This result argues against CII being a functional constituent of human I+III2+IVn supercomplexes.
DISCUSSION
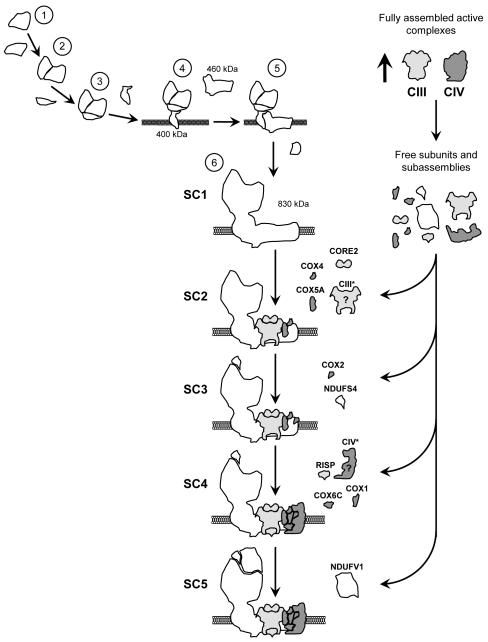
We a i med to elucidate the biosynthetic pathway of mitochondrial supercomplexes, to better understand the molecular mechanisms underlying the interdependence between RC complexes that would aid explaining the severe RC assembly defects found in patients with mitochondrial disorders. To address the dynamics of assembly, we depleted control cell lines of OXPHOS complexes by treatment with doxycycline to reversibly inhibit mitochondrial translation, and upon drug removal we followed complex and supercomplex assembly by BN-PAGE and immunodetection. Based on the analyses of the assembly kinetics of OXPHOS subunits, we propose a multi-step model of mitochondrial supercomplex assembly (Figure 7). The conceptual novelty of this model is that the biogenesis of the respirasome involves the coordinated and sequential association of specific combinations of partially-assembled RC complexes and free subunits. In a first stage, CIII and CIV assemble as individual entities until reaching a threshold supposed to trigger the accumulation of free subunits and assembly intermediates from these two complexes. At this early stage, the assembly of CI probably also starts taking place, in agreement with reports demonstrating that the assembly of CI and CIII launch in parallel (Pello et al., 2008), until building up a CI intermediate of ~830 kDa that lacks the NADH dehydrogenase module and perhaps additional subunits. Up to this stage, the pattern of CI subcomplexes is in agreement with previous reports (Antonicka et al., 2003; Lazarou et al., 2007; Ugalde et al., 2004a; Vogel et al., 2007a). We propose that the ~830 kDa CI subassembly actually constitutes the first supercomplex assembly intermediate (SC1), since it remains in a stable competent state for the subsequent combined incorporation of either individual subunits or subassemblies from CI, CIII and CIV in at least five sequential assembly steps (SC1 to SC5 in Figure 7). The latest supercomplex assembly step involves the association of catalytic subunits from the CI NADH dehydrogenase module prior to the respirasome activation cascade, although we cannot exclude additional intermediate steps between these two events. Our results not only imply that SC1 is the core structure necessary for the formation of the respirasomes, but also that the respirasomes constitute the structural units where CI gets fully-assembled and activated, thus explaining the essentiality of the respirasomes for CI function (Schagger et al., 2004).
Figure 7. Model for the assembly of mitochondrial supercomplexes.
In a first stage, the syntheses of fully-assembled and active CIII and CIV take place until reaching a threshold that probably triggers the accumulation of free subunits and assembly intermediates from these two complexes. At this early stage the assembly of CI also initiates (steps 1 to 6) to building up a CI subassembly of ~830 kDa that constitutes the first supercomplex assembly intermediate (SC1). This subcomplex remains in a stable assembly-competent state for the subsequent binding of CIII subunit CORE2 and CIV subunits COX4 and COX5A, to form a second supercomplex assembly intermediate (SC2). The incorporations of CI NDUFS4 and CIV COX2 subunits, and maybe other free RC subunits or subassemblies, take place in a third stage (SC3). The catalytic CIII RISP and CIV COX1 subunits and the structural CIV subunit COX6C incorporate to the supercomplexes in a fourth stage (SC4). The latest supercomplex assembly step involves the association of catalytic subunits from the CI NADH dehydrogenase module prior to the respirasome activation (SC5).
The mechanism we propose contrasts with the previously suggested pathway based on the association of individual fully-assembled RC complexes (Acin-Perez et al., 2008). To demonstrate their hypothesis, the authors analyzed the time course incorporation of the13 mtDNA-encoded OXPHOS proteins into RC complexes and supercomplexes by BN-PAGE, convincingly showing that there is a temporal gap between the incorporation of mtDNA-encoded subunits into the individual complexes and that into supercomplexes. Our results agree with this observation, but the additional analyses of nuclear-encoded OXPHOS subunits provided here do not support the interpretation that supercomplexes are formed by the ordered association of preassembled complexes. The faster kinetics of assembly of supercomplexes under their conditions is probably due to the fact that a large pool of translated RC subunits already exists in preassembled subcomplexes and supercomplex intermediates (Fernandez-Vizarra et al., 2009), which remain in a competent state for the efficient channeling of newly translated mtDNA-encoded subunits into assembly intermediates and subsequently in RC complexes or supercomplexes. In our supercomplex assembly model, the biogenesis of CIII and CIV take place prior to the formation of CI, which can only occur in the context of the respirasome. In this regard, our model does not contradict the plasticity model, since it does not exclude the assembly of holo-CIII and CIV as independent pathways, but it excludes the formation of fully-assembled CI as an independent unit. Rather, we propose that there are at least two regulatory steps in the biogenesis of the OXPHOS system: first, the independent regulation of the assembly of individual holo-CIII and CIV, and second, the association of free CIII and CIV subunits or subassemblies with a CI assembly intermediate to form the respirasome. There must be additional regulatory steps, since the functional meaning of intermediate supercomplex forms remains unknown and deserve future attention. For example, supercomplexes I+III2 or III2+IV could constitute either intermediate structures that would aid the assembly of newly-synthesized supercomplexes to adapt to changes in cellular metabolism or maybe just supercomplex degradation products.
The current available literature regarding OXPHOS-deficient patients and animal models support the key steps of our respirasome assembly model. It was described in human primary fibroblasts that the 830 kDa CI intermediate (SC1) contains most CI subunits, but neither NDUFS4 nor the subunits from the N catalytic module (Lazarou et al., 2007). In agreement, the 830 kDa intermediate is usually accumulated in patients’ fibroblasts with mutations in the CI NDUFS4 subunit (or mice lacking this subunit) or in subunits from the N catalytic module, such as NDUFS1 or NDUFS6 (Calvaruso et al., 2011; Hoefs et al., 2010; Kirby et al., 2004; Ugalde et al., 2004b). More importantly, this 830 kDa subassembly has been found associated with CIII subunits CORE1 and CORE2 in partially-assembled supercomplexes from patients’ cells and mutant mice tissues (Calvaruso et al., 2011; Lazarou et al., 2007). Tissues from patients harboring mutations in the BCS1L gene, involved in the assembly of the RISP subunit into CIII, revealed the accumulation of the CORE proteins in high-molecular-weight respirasome intermediates that lacked the RISP protein in mutant but not in wild-type cells, suggesting that these subunits could get assembled into supercomplexes prior to the incorporation of RISP (Fernandez-Vizarra et al., 2009). Mitochondria from patients in whom CIV biogenesis was compromised displayed a preferential accumulation of certain CIV subunits in supercomplexes (Lazarou et al., 2009), as observed in our COX2 mutant cybrids. These similarities between human and mice primary cell cultures or tissues and the established cell lines shown in our work support the generality in mammals of the respirasome assembly mechanisms that we propose.
Our results concerning the composition of mitochondrial supercomplexes in human control cell lines agree with previous observations in other mammalian systems. Approximately 90% of CI is associated with a CIII dimer (CIII2) and several CIV units to form two major supercomplexes: I+III2 and I+III2+IV1-4 (Schagger and Pfeiffer, 2001). In contrast, ~50-60% of CIII and 70-80% of CIV remain individual, although both dynamically interact in an intermediate III2+IV supercomplex. Incidentally, our data do not support CII colocalization with mitochondrial supercomplexes. Tiny amounts of CII that migrated as a smear up to 400 kDa, below the CIII2 position, could correspond to CII trimers as shown in bacteria (Sousa et al., 2011). Another tiny spot detected in a supramolecular structure above supercomplex I+III2+IVn could represent CII as a component of the mitoKATP channel (Ardehali et al., 2004). None of these spots clearly colocalized with any RC complexes or supercomplexes. This is in disagreement with a previous study based on co-IP and BN analyses and the same experimental conditions used here, which claimed binding of some CII to supercomplexes (Acin-Perez et al., 2008). These discrepancies could be due to technical differences in the tissue-specific mitochondrial protein extraction methods or the electrophoretic conditions used, which could affect the mobility of the samples. However, most studies using techniques as diverse as BN-PAGE, single particle electron microscopy, cryoelectron tomography, flux control analyses, or studies based on time-dependent localization of fluorescent RC subunits failed to show structural or functional evidences supporting the presence of CII in mammalian RC supercomplexes in normal physiological conditions (Dudkina et al., 2011; Lenaz and Genova, 2010; Muster et al., 2010; Quarato et al., 2011; Schafer et al., 2006; Schagger and Pfeiffer, 2000, 2001).
Importantly, our respirasome assembly model may explain why certain genetic defects affecting a single complex lead to combined RC enzyme deficiencies in patients. For instance, it explains why CIV deficiences due to mutations in COX1 and COX2 lead to pleiotropic CI defects (this work and D’Aurelio et al., 2006). The insertions of these two CIV subunits into supercomplexes occur prior or in parallel with the incorporation of subunits from the CI N catalytic module (NDUFV1 and NDUFS4). Consequently, the lack of COX1/COX2 or the total absence of holo-CIV affects the assembly or stability of NDUFS4 and NDUFV1 within supercomplexes. This in turn leads to the accumulation of the inactive ~830 kDa supercomplex assembly intermediate and a I+III2 intermediate that partially lacks the CI N catalytic module, thus explaining the severe reduction in CI activity detected in the COX mutant cells. A similar assembly phenotype has been recently described in NDUFS4 knock-out mice (Calvaruso et al., 2011). The same argument serves to explain why failures in the insertion of the CIII subunit RISP may lead to secondary CI and CIV deficiencies in human tissues (Fernandez-Vizarra et al., 2007; Moran et al., 2010).
The fact that CIII and CIV can get assembled either as individual holoenzymes or by direct binding of free subunits to supercomplex assembly intermediates supports the existence of several independently-regulated assembly pathways for the biosynthesis of these two complexes. This would explain why decreased CI levels do not usually lead to CIII and CIV assembly and functional defects in mammals (Acin-Perez et al., 2004; Schagger et al., 2004). Exceptions have been described in CI-deficient patients with mutations in the NDUFS4 subunit (Budde et al., 2000; Ugalde et al., 2004b) and C20ORF7 (Saada et al., 2011), a mitochondrial methyltransferase involved in the assembly or stability of an early CI assembly intermediate (Sugiana et al., 2008). While the reason remains unclear, the possibility exists that the CIII and CIV deficiencies observed in these patients could be secondary phenomena. Alternatively, NDUFS4 could function in the assembly or stability of individual CIII, besides its role in supercomplex biogenesis. Similarly, C20ORF7 could participate in the assembly or stability of monomeric CIV, as well as in the post transcriptional modification of proteins important for respirasome structural stability. In this regard, C20ORF7 has been suggested to play a role in the methylation of CI subunit NDUFB3, and it could potentially modify additional OXPHOS components (Sugiana et al., 2008).
Our model indirectly clarifies the possible roles of known chaperones in the assembly of RC supercomplexes. The most evident case is the NDUFAF2 (B17.2L) assembly factor, which appears to function in a late stage of CI assembly. This protein binds to the ~830 kDa subcomplex and participates in the insertion of the N module into CI (Ogilvie et al., 2005; Vogel et al., 2007b). Because the ~830 kDa subcomplex is a supercomplex assembly intermediate, and the insertion of the N module is one of the last steps in the respirasome assembly pathway, it seems appropriate to consider NDUFAF2 as the first known supercomplex assembly factor. The roles of other chaperones involved in late steps of CI assembly, or assembly factors that regulate the biosynthesis of RC CIII and CIV could be also redefined within the frame of the supercomplex assembly pathway described here.
Additional work is required to explain in detail the assembly of all OXPHOS subunits into respirasomes, identify the mechanisms that regulate the diverse biosynthetic pathways for the formation of RC complexes and supercomplexes, and their modulation in response to environmental and physiological conditions. Nonetheless, our model for supercomplexes assembly represents a starting point to further elucidate this intricate process, and provides a framework to better understand RC assembly defects in patients with mitochondrial disorders.
EXPERIMENTAL PROCEDURES
Cell lines and culture conditions
Cybrids were constructed using the osteosarcoma 143B TK-206 rho zero cell line and enucleated control fibroblasts as described (King and Attardi, 1989). Cells were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM, Life Technologies) supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, 1mM sodium pyruvate and antibiotics. To block mitochondrial translation, 15 μg/ml doxycycline was added to the culture medium. Cells were grown in exponential conditions and harvested at the indicated time points.
Quantification of mtDNA copy number
The relative quantification of mtDNA versus nuclear DNA (nDNA) was performed by real-time PCR in a HT 7500 Real Time PCR System (Applied Biosystems) as previously described (Pello et al., 2008).
Mitochondrial protein preparation
Mitochondrial pellets were isolated from cell cultures as described (Pello et al., 2008). To prepare native mitochondrial proteins, pellets were solubilized in 200 μl buffer containing 1.5 M aminocaproic acid, 50 mM Bis-Tris, pH 7.0. After optimizing solubilization conditions, we decided to use digitonin at a concentration of 4 g/g protein. Solubilized samples were incubated on ice for 15 min, centrifuged for 30 min at 13000 rpm at 4°C, and the supernatant was combined with 20 μl of sample buffer (750mM aminocaproic acid, 50mM Bis-Tris, 0.5mM EDTA, 5% Serva Blue G-250) prior to loading.
Blue Native electrophoresis and in-gel activity assays
Blue Native 3-13% gradient gels were loaded with 60 μg of mitochondrial protein and processed as described (Nijtmans et al., 2002). After electrophoresis, proteins were transferred to a PROTAN® nitrocellulose membrane (Schleicher & Schuell) at 35 V, overnight, and probed with specific antibodies. Duplicate gels were further used for in-gel enzyme activity (IGA) assays and for second dimension 10% SDS-PAGE gels.
Antibodies
Western-blot was performed using primary antibodies raised against the following human OXPHOS subunits: NDUFV1 (Sigma), NDUFA9, NDUFS2, NDUFS4, CORE2, RISP, COX1, COX2, COX4, COX5A, COX6C and SDHA (Mitosciences). Peroxidase-conjugated anti-mouse IgG was used as a secondary antibody (Molecular Probes). The signal was detected with ECL® plus (Amersham Biosciences).
Spectrophotometric assays
Rotenone-sensitive CI (NADH: ubiquinone oxidoreductase) activity was measured with a DU-650 spectrophotometer (Beckman) at 37°C by incubating 150 μg of mitochondrial protein in 1 ml of assay medium (20 mM KP pH 8.0, 0.1 % BSA-EDTA, 100 μM CoQ1, 1 mM NaN3, 0.2 mM NADH without and with 5 μM rotenone). The absorbance decrease at 340 nm due to the NADH oxidation was measured, and expressed as the percentage relative to the citrate synthase activity.
Statistical analysis
Densitometry was performed with the ImagePro-Plus 4.1 image analysis software (Media Cybernetics). Graphs represent mean values of at least three independent measurements. Error bars represent standard deviations (SD).
Supplementary Material
HIGHLIGHTS.
Mitochondrial respirasome biogenesis is a multistep biosynthetic process
It does not require the associations of independent pre-assembled RC complexes
A CI intermediate acts as a scaffold for the incorporation of free CIII+CIV subunits
Respirasomes are the structural units where CI is fully-assembled and activated
ACKNOWLEDGEMENTS
We thank Dr. JA Enríquez for the kind gift of the COX2 mutant cybrids. Our study was supported by FIB Hospital Universitario 12 de Octubre and Instituto de Salud Carlos III to CU (grants # PI08-0021 and # PI11-00182), Muscular Dystrophy Association to FF and AB (grant # 158547) and NIH Grant GM071775A to AB.
REFERENCES
- Acin-Perez R, Bayona-Bafaluy MP, Fernandez-Silva P, Moreno-Loshuertos R, Perez-Martos A, Bruno C, Moraes CT, Enriquez JA. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell. 2004;13:805–815. doi: 10.1016/s1097-2765(04)00124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I(1)III(2)IV(1) EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka H, Ogilvie I, Taivassalo T, Anitori RP, Haller RG, Vissing J, Kennaway NG, Shoubridge EA. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 2003;278:43081–43088. doi: 10.1074/jbc.M304998200. [DOI] [PubMed] [Google Scholar]
- Ardehali H, Chen Z, Ko Y, Mejia-Alvarez R, Marban E. Multiprotein complex containing succinate dehydrogenase confers mitochondrial ATP-sensitive K+ channel activity. Proc. Natl. Acad. Sci. USA. 2004;101:11880–11885. doi: 10.1073/pnas.0401703101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde SM, van den Heuvel LP, Janssen AJ, Smeets RJ, Buskens CA, DeMeirleir L, Van Coster R, Baethmann M, Voit T, Trijbels JM, et al. Combined enzymatic complex I and III deficiency associated with mutations in the nuclear encoded NDUFS4 gene. Biochem. Biophys. Res. Commun. 2000;275:63–68. doi: 10.1006/bbrc.2000.3257. [DOI] [PubMed] [Google Scholar]
- Calvaruso MA, Willems P, van den Brand M, Valsecchi F, Kruse S, Palmiter R, Smeitink J, Nijtmans L. Mitochondrial complex III stabilizes complex I in the absence of NDUFS4 to provide partial activity. Hum. Mol. Genet. 2011;21:115–120. doi: 10.1093/hmg/ddr446. [DOI] [PubMed] [Google Scholar]
- Campos Y, Garcia-Redondo A, Fernandez-Moreno MA, Martinez-Pardo M, Goda G, Rubio JC, Martin MA, del Hoyo P, Cabello A, Bornstein B, et al. Early-onset multisystem mitochondrial disorder caused by a nonsense mutation in the mitochondrial DNA cytochrome C oxidase II gene. Ann. Neurol. 2001;50:409–413. doi: 10.1002/ana.1141. [DOI] [PubMed] [Google Scholar]
- Chance B, Williams GR. A simple and rapid assay of oxidative phosphorylation. Nature. 1955;175:1120–1121. doi: 10.1038/1751120a0. [DOI] [PubMed] [Google Scholar]
- D’Aurelio M, Gajewski CD, Lenaz G, Manfredi G. Respiratory chain supercomplexes set the threshold for respiration defects in human mtDNA mutant cybrids. Hum. Mol. Genet. 2006;15:2157–2169. doi: 10.1093/hmg/ddl141. [DOI] [PubMed] [Google Scholar]
- Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc. Natl. Acad. Sci. USA. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Vizarra E, Bugiani M, Goffrini P, Carrara F, Farina L, Procopio E, Donati A, Uziel G, Ferrero I, Zeviani M. Impaired complex III assembly associated with BCS1L gene mutations in isolated mitochondrial encephalopathy. Hum. Mol. Genet. 2007;16:1241–1252. doi: 10.1093/hmg/ddm072. [DOI] [PubMed] [Google Scholar]
- Fernandez-Vizarra E, Tiranti V, Zeviani M. Assembly of the oxidative phosphorylation system in humans: what we have learned by studying its defects. Biochim. Biophys. Acta. 2009;1793:200–211. doi: 10.1016/j.bbamcr.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Fornuskova D, Stiburek L, Wenchich L, Vinsova K, Hansikova H, Zeman J. Novel insights into the assembly and function of human nuclear-encoded cytochrome c oxidase subunits 4, 5a, 6a, 7a and 7b. Biochem. J. 2010;428:363–374. doi: 10.1042/BJ20091714. [DOI] [PubMed] [Google Scholar]
- Hackenbrock CR, Chazotte B, Gupte SS. The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J. Bioenerg. Biomembr. 1986;18:331–368. doi: 10.1007/BF00743010. [DOI] [PubMed] [Google Scholar]
- Heinemeyer J, Braun HP, Boekema EJ, Kouril R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J. Biol. Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- Hoefs SJ, Skjeldal OH, Rodenburg RJ, Nedregaard B, van Kaauwen EP, Spiekerkotter U, von Kleist-Retzow JC, Smeitink JA, Nijtmans LG, van den Heuvel LP. Novel mutations in the NDUFS1 gene cause low residual activities in human complex I deficiencies. MOL. GENET. METAB. 2010;100:251–256. doi: 10.1016/j.ymgme.2010.03.015. [DOI] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Kirby DM, Salemi R, Sugiana C, Ohtake A, Parry L, Bell KM, Kirk EP, Boneh A, Taylor RW, Dahl HH, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J. Clin. Invest. 2004;114:837–845. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamantea E, Carrara F, Mariotti C, Morandi L, Tiranti V, Zeviani M. A novel nonsense mutation (Q352X) in the mitochondrial cytochrome b gene associated with a combined deficiency of complexes I and III. Neuromuscul. Disord. 2002;12:49–52. doi: 10.1016/s0960-8966(01)00244-9. [DOI] [PubMed] [Google Scholar]
- Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Smith SM, Thorburn DR, Ryan MT, McKenzie M. Assembly of nuclear DNA-encoded subunits into mitochondrial complex IV, and their preferential integration into supercomplex forms in patient mitochondria. FEBS J. 2009;276:6701–6713. doi: 10.1111/j.1742-4658.2009.07384.x. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML. Structure and organization of mitochondrial respiratory complexes: a new understanding of an old subject. Antioxid. Redox Signal. 2010;12:961–1008. doi: 10.1089/ars.2009.2704. [DOI] [PubMed] [Google Scholar]
- Li Y, D’Aurelio M, Deng JH, Park JS, Manfredi G, Hu P, Lu J, Bai Y. An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem. 2007;282:17557–17562. doi: 10.1074/jbc.M701056200. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Ryan MT. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life. 2010;62:497–502. doi: 10.1002/iub.335. [DOI] [PubMed] [Google Scholar]
- Moran M, Marin-Buera L, Gil-Borlado MC, Rivera H, Blazquez A, Seneca S, Vazquez-Lopez M, Arenas J, Martin MA, Ugalde C. Cellular pathophysiological consequences of BCS1L mutations in mitochondrial complex III enzyme deficiency. Hum. Mutat. 2010;31:930–941. doi: 10.1002/humu.21294. [DOI] [PubMed] [Google Scholar]
- Muster B, Kohl W, Wittig I, Strecker V, Joos F, Haase W, Bereiter-Hahn J, Busch K. Respiratory chain complexes in dynamic mitochondria display a patchy distribution in life cells. PloS One. 2010;5:e11910. doi: 10.1371/journal.pone.0011910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans LG, Henderson NS, Holt IJ. Blue Native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Taanman JW, Muijsers AO, Speijer D, Van den Bogert C. Assembly of cytochrome-c oxidase in cultured human cells. Eur. J. Biochem. 1998;254:389–394. doi: 10.1046/j.1432-1327.1998.2540389.x. [DOI] [PubMed] [Google Scholar]
- Ogilvie I, Kennaway NG, Shoubridge EA. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Invest. 2005;115:2784–2792. doi: 10.1172/JCI26020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pello R, Martin MA, Carelli V, Nijtmans LG, Achilli A, Pala M, Torroni A, Gomez-Duran A, Ruiz-Pesini E, Martinuzzi A, et al. Mitochondrial DNA background modulates the assembly kinetics of OXPHOS complexes in a cellular model of mitochondrial disease. Hum. Mol. Genet. 2008;17:4001–4011. doi: 10.1093/hmg/ddn303. [DOI] [PubMed] [Google Scholar]
- Quarato G, Piccoli C, Scrima R, Capitanio N. Variation of flux control coefficient of cytochrome c oxidase and of the other respiratory chain complexes at different values of protonmotive force occurs by a threshold mechanism. Biochim. Biophys. Acta. 2011;1807:1114–1124. doi: 10.1016/j.bbabio.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Saada A, Edvardson S, Shaag A, Chung WK, Segel R, Miller C, Jalas C, Elpeleg O. Combined OXPHOS complex I and IV defect, due to mutated complex I assembly factor C20ORF7. J. Inherit. Metab. Dis. 2011;35:125–131. doi: 10.1007/s10545-011-9348-y. [DOI] [PubMed] [Google Scholar]
- Schafer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J. Architecture of active mammalian respiratory chain supercomplexes. J. Biol. Chem. 2006;281:15370–15375. doi: 10.1074/jbc.M513525200. [DOI] [PubMed] [Google Scholar]
- Schagger H, de Coo R, Bauer MF, Hofmann S, Godinot C, Brandt U. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 2004;279:36349–36353. doi: 10.1074/jbc.M404033200. [DOI] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 2001;276:37861–37867. doi: 10.1074/jbc.M106474200. [DOI] [PubMed] [Google Scholar]
- Soto IC, Fontanesi F, Valledor M, Horn D, Singh R, Barrientos A. Synthesis of cytochrome c oxidase subunit 1 is translationally downregulated in the absence of functional F1F0-ATP synthase. Biochim. Biophys. Acta. 2009;1793:1776–1786. doi: 10.1016/j.bbamcr.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa PM, Silva ST, Hood BL, Charro N, Carita JN, Vaz F, Penque D, Conrads TP, Melo AM. Supramolecular organizations in the aerobic respiratory chain of Escherichia coli. Biochimie. 2011;93:418–425. doi: 10.1016/j.biochi.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Sugiana C, Pagliarini DJ, McKenzie M, Kirby DM, Salemi R, Abu-Amero KK, Dahl HH, Hutchison WM, Vascotto KA, Smith SM, et al. Mutation of C20orf7 disrupts complex I assembly and causes lethal neonatal mitochondrial disease. Am. J. Hum. Genet. 2008;83:468–478. doi: 10.1016/j.ajhg.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugalde C, Vogel R, Huijbens R, Van Den Heuvel B, Smeitink J, Nijtmans L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 2004a;13:2461–2472. doi: 10.1093/hmg/ddh262. [DOI] [PubMed] [Google Scholar]
- Ugalde C, Janssen RJ, van den Heuvel LP, Smeitink JA, Nijtmans LG. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 2004b;13:659–667. doi: 10.1093/hmg/ddh071. [DOI] [PubMed] [Google Scholar]
- Vogel RO, Dieteren CE, van den Heuvel LP, Willems PH, Smeitink JA, Koopman WJ, Nijtmans LG. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 2007a;282:7582–7590. doi: 10.1074/jbc.M609410200. [DOI] [PubMed] [Google Scholar]
- Vogel RO, van den Brand MA, Rodenburg RJ, van den Heuvel LP, Tsuneoka M, Smeitink JA, Nijtmans LG. Investigation of the complex I assembly chaperones B17.2L and NDUFAF1 in a cohort of CI deficient patients. Mol. Genet. Metab. 2007b;91:176–182. doi: 10.1016/j.ymgme.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim. Biophys. Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Zara V, Conte L, Trumpower BL. Biogenesis of the yeast cytochrome bc1 complex. Biochim. Biophys. Acta. 2009;1793:89–96. doi: 10.1016/j.bbamcr.2008.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.