Abstract
Smoking is associated with a wide variety of adverse health outcomes including cancer, chronic obstructive pulmonary disease, diabetes, depression and heart disease. Unfortunately, the molecular mechanisms through which these effects are conveyed are not clearly understood. To examine the potential role of epigenetic factors in these processes, we examined the relationship of smoking to genome wide methylation and gene expression using biomaterial from two independent samples, lymphoblast DNA and RNA (n=119) and lung alveolar macrophage DNA (n=19). We found that in both samples current smoking status was associated with significant changes in DNA methylation, in particular at the aryl hydrocarbon receptor repressor (AHRR), a known tumor suppressor. Both baseline DNA methylation and smoker associated DNA methylation signatures at AHRR were highly correlated (r=0.94 and 0.45, respectively). DNA methylation at the most differentially methylated AHRR CpG residue in both samples, cg0557592, was significantly associated with AHRR gene expression. Pathway analysis of lymphoblast data (genes with most significant methylation changes) demonstrated enrichment in protein kinase C pathways and in TGF beta signaling pathways. For alveolar macrophages, pathway analysis demonstrated alterations in inflammation-related processes. We conclude that smoking is associated with functionally significant genome wide changes in DNA methylation in both lymphoblasts and pulmonary macrophages and that further integrated investigations of these epigenetic effects of smoking on carcinogenesis and other related co-morbidities are indicated.
INTRODUCTION
Despite extensive preventative and treatment interventions, approximately 19% of American adults smoke on a daily basis (Centers for Disease Control 2011). This is a substantial problem because smoking is the leading preventable cause of premature morbidity and mortality. Smoking causes approximately 450,000 premature deaths annually through its effects on the incidence of cancer, heart disease and chronic obstructive pulmonary disease (Center for Disease Control 2005). National data indicate that while both prevalence of smoking and mortality from lung cancer have significantly decreased for men between 1975 and 2007, these rates did not decrease for any racial or ethnic group or for women (Davis and others 2010). In addition, projections suggest that because women who were born around 1960 have higher prevalence of smoking and morbidity than other cohorts, this gender disparity may increase (Kohler and others 2011).
Many of the effects of smoking on the lung are thought to result from the direct effects of cigarette smoke on pulmonary epithelium and alveolar macrophages. However, the exact mechanism(s) through which smoking increases the risk for disease in non-pulmonary tissues such as blood and brain are unclear. Recently, sets of convergent findings have suggested that a portion of that vulnerability may be driven by differential DNA methylation acquired by smoking (Breitling and others 2011; Chang and others 2004; Philibert and others 2010; Suga and others 2008).
Altered DNA methylation that results from genetic lesions present at conception has long been established as a cause of disorders affecting early development of disease in the soma and the CNS. With respect to non-CNS disease, altered imprinting that usually results from maternal monosomy at 15Q causes Prader-Willi syndrome (Gurrieri and Accadia 2009). With respect to the CNS disease, almost all cases of Rett Syndrome result from mutations in MECBP2 which exert their effects by altering DNA methylation (Chahrour and Zoghbi 2007). Guided by clues such as the observations that addition of folate, a methyl donor, to the diets of pregnant women, markedly decreases the frequency of neural tube defects, the field has embraced the concept that alterations in DNA methylation may be associated with acquired early onset developmental disorders as well (Tsankova and others 2007). However, whether environmentally acquired alterations could increase likelihood of disease in adults has been an open question. A number of single gene and genome wide studies provide evidence that altered DNA methylation is associated with smoking and may be a cause of smoking associated illness. In particular, using both genome wide and single gene approaches, we and others have demonstrated that altered DNA methylation is associated with smoking (Breitling and others 2011; Chang and others 2004; Launay and others 2009; Philibert and others 2010; Suga and others 2008). However, these studies have been hindered by low coverage of the total number of genes and CpG residues in the human genome and discrepancies as to the appropriateness of certain forms of biomaterials for studies of epigenetic phenomena.
In this communication, we report our results with respect to smoking status on genome wide methylation and focal gene expression using two independent sets of biomaterials: 1) lymphoblast DNA and RNA derived from 119 female subjects from the Iowa Adoption Studies (IAS) and 2) alveolar macrophage DNA from cells isolated from the lungs of 10 smokers and 9 non-smokers.
METHODS
Human Subjects
The first set of biomaterials was obtained from subjects participating in the Iowa Adoptions Studies (IAS) (Yates and others 1998). In brief, the IAS is a case and control adoption study of the role of genetic, environmental and gene-environment interactions in the etiology of common behavioral illness. The clinical material used in the current study is derived from interviews with the Semi-Structured Interview for the Assessment of the Genetics of Alcoholism, Version II (Bucholz and others 1994), during each of the last two waves of the IAS study (1999–2004 and 2005–2009). The biological material used in this study, lymphoblast cell lines, was derived by Epstein Barr virus (EBV) mediated transformation (Caputo and others 1991) of lymphocytes obtained from blood donated by 165 female subjects during the last wave of the study.
The second set of biomaterials for the current study was alveolar macrophages obtained by bronchoalveolar lavage. Subjects were recruited from the community via advertisements and word-of-mouth. In order to be included, case (smoking) subjects had to be actively smoking with at least 10 pack year history of smoking. To be included as a control, the subject had to deny ever smoking cigarettes. Subjects were excluded if they had any significant co-morbid conditions such as pregnancy, or if a baseline spirometry revealed the Forced Expiratory Volume in the first second (FEV1) was less than 60% of predicted. All of these procedures and protocols were approved by the University of Iowa Institutional Review Board.
Bronchoalveolar Lavage
To obtain human alveolar macrophages a bronchoalveolar lavage was performed. After informed consent was obtained, subjects underwent standard flexible bronchoscopy. After the application of local anesthesia, bronchoalveolar lavage was performed by instilling 20 ml of normal saline into a tertiary bronchus up to five times in three different lung segments. The first collection out of five was discarded for possible contamination from upper airway secretions or by lidocaine, which is used to locally anesthetize the subject during the procedure. The remaining lavage was transported to the laboratory where fluid was filtered through sterile gauze and centrifuged at 200 x g for 5 min to pellet cellular material. The resulting pellet was suspended in phosphate buffered saline and centrifuged at 16,000 x g for one minute. The macrophages were suspended in medium, labeled with Wright stain and microscopically examined to ensure that greater than 95% of the cells were macrophages (Monick and others 2008; Monick and others 2006; Monick and others 2010).
DNA and RNA Isolation
The lymphoblast DNA and RNA used in this study was prepared from growth-entrained cell lines according to our standard procedures (Philibert and others 2008). In brief, on the day before DNA preparation, one-half of the cell media for each culture flask was exchanged. Twenty four hours later, DNA was prepared from the cell lines using cold protein precipitation. Simultaneously, RNA was purified from independent aliquots of the same culture using RNA Midi kits (Invitrogen, USA) according the instructions of the manufacturer. After quantification and purity assessment using a Nanodrop (Thermo Scientific, USA) spectrophotometer, DNA was stored at −20° C and RNA was stored at −80° C until use.
DNA and RNA were isolated from alveolar macrophages using the Qiagen DNAeasy™ kit (Qiagen, Valencia, CA) and MirVana (Applied Biosystems, Austin, TX) reagents according to manufacturer's instructions. Quality assessment was by Nanodrop and Experion (Bio-Rad Experion Automated Electrophoresis Station). After preparation, DNA was stored at −20° C and RNA was stored at −80° C until use.
DNA Methylation
Genome wide DNA methylation of the DNA was assessed using the Illumina HumanMethylation450 BeadChip under contract by the University of Minnesota Genome Center using the protocol specified by the manufacturer and the contractor. The resulting microarray data were inspected for complete bisulfite conversion of the DNA, and average beta values (i.e. average methylation) for each CpG residue were determined using the GenomeStudio V2009.2; Methylation module Version 1.5.5., version 3.2 (Illumina, San Diego). The resulting beta values were exported into Microsoft Excel and JMP (SAS Institute, USA) for data analysis. The HumanMethylation450 BeadChip contains 485,577 probes that recognize at least 20216 unique features (i.e. potential transcripts). With respect to this sample, >99.76 % of the 485,577 probes yielded statistically reliable data.
Data Analysis
After logarithmic conversion, data were inspected for outliers or confounding by plate or chip variables, and then the initial data analyses were conducted using genome wide t-tests. Subsequently, beta values for each of the probes were aligned according to their physical location and the data reanalyzed using paired t-tests over a 11-probe sliding window in order to more adroitly capture methylation signatures over larger regions (Dindot and others 2009; Farthing and others 2008). All genome wide comparisons were corrected for multiple comparisons using the method of Benjamini and Hochberg(Benjamini and Hochberg 1995). For select loci, data were analyzed with respect to alcohol use status using ANOVA (Fleiss 1981).
Pathway analysis of differentially methylated genes was conducted using GoMiner™ using default settings (0.05 settings for reports and all gene ontology as the root category setting) using the gene set specified in the text as the “changed” gene set (Zeeberg and others 2003). All values reported include nominal and FDR (false discovery rate) corrected values.
Specific qRT-PCR Analysis of AHRR
The relative expression of the aryl hydrocarbon receptor repressor (AHRR) was determined using primer probe sets from ABI, a Fluidigm BioMark™ System and proprietary BioMark Real-Time Analysis software according to manufacturer's guidelines. Briefly, first, RNA was converted to cDNA using an ABI cDNA archiving kit according to manufacturer’s suggestions. Then after a brief pre-amplification step, each cDNA sample was amplified in quadruplicate with using primer probes for AHRR (Hs01005075) and five housekeeping genes (CALR, RPL7A, PRS19, RPS20 and UBC) obtained from Applied Biosystems (Foster City, USA). The Ct counts exported to the database, normalized using the geometric mean of five housekeeping genes, and then converted to Z scores for statistical analysis.
RESULTS
Iowa Adoption Study Cohort
The demographic and clinical characteristics of the 165 female subjects whose genome wide methylation status was assessed are shown in Table I. Overall, the subjects were largely white and tended to be in their mid-to-late 40s. Consistent with enrichment of the sample for the diathesis of substance use, the majority of the subjects in the study reported daily smoking at some period of their lives (85 of 165). However, many of these individuals (n=46) have quit smoking or were not smoking every day at the time of phlebotomy leaving only 39 subjects reporting daily smoking (i.e. seven days per week every week) at the time of phlebotomy. Because our prior studies have indicated that they methylation signature of those subjects who had recently quit smoking is highly variable, those 46 individuals were excluded from further study (Philibert and others 2010). The number of cigarettes smoked daily by the 39 subjects who smoked daily varied from 4 to 40 with the average number of cigarettes consumed daily being 19 cigarettes or about a pack per day for greater than 20 years. Cigarette smoking tended to be the only form of nicotine use currently being manifested by these 39 subjects with none of the subjects reporting the concomitant use of cigars, chew or other forms of nicotine usage in 2 weeks prior to assessment. There were no significant differences between the three groups (current smokers, never smokers, non-daily smokers/quitters) with respect to alcohol use in the past six months or age.
Table I.
Clinical Characteristics of the 165 Female Iowa Adoptions Studies Probands
| Non-Smoker | Quit or Quitting | Daily Smoker | |
|---|---|---|---|
| N | 80 | 46 | 39 |
| Age | 46 ± 8 | 47 ± 8 | 43 ± 6 |
| Ethnicity | |||
| White | 80 | 44 | 39 |
| Other | 0 | 4 | 0 |
| Alcohol in Past 6 months | |||
| Yes | 58 | 35 | 29 |
| No | 22 | 11 | 7 |
| Daily Cigarette Usage | 19 ± 9 | ||
We contrasted the methylation values for the 39 smokers (average beta value 0.443) with the values for the 80 non-smokers (average beta value 0.446) using single point genome wide t-tests. The results of those analyses are shown in Table II. As the table indicates, only one probe, cg14817490, which maps to intron 3 of the of the aryl hydrocarbon receptor repressor (AHRR) survived genome wide Benjamini-Hochberg correction for multiple comparisons. However, it is interesting to note that 3 other probes from AHRR, cg05575921, cg14454127, and cg03991871, were ranked among the top 13 probes and that none of them were from the rather small promoter associated CpG island. Instead, all 4 of the top AHRR probes target the gene body which contains three (>100 CpG residues) large CpG island according the UCSC genome browser. Finally, we note that cg03636183, a probe that was reported by Breitling and colleagues to be significantly associated with smoking status in lymphocyte DNA (Breitling and others 2011), was also nominally associated (p<0.003; rank 802nd of 485577 probes; smoker average 0.67; non-smoker average 0.74) with smoking status in the current study (Breitling and others 2011).
Table II.
The Top 30 Most Significantly Differentially Methylated Probes in Lymphoblast DNA.
| Probe ID | GENE | Placement | Island Status | N-Smoker Avg | Smoker AVG | T-test | Corrected P-value |
|---|---|---|---|---|---|---|---|
| cg14817490 | AHRR | Body | 0.24 | 0.12 | 2.71E-08 | 0.02 | |
| cg05575921 | AHRR | Body | N Shore | 0.85 | 0.70 | 1.34E-06 | 0.29 |
| cg07313705 | S Shelf | 0.07 | 0.10 | 1.78E-06 | 0.29 | ||
| cg14454127 | AHRR | Body | 0.44 | 0.31 | 2.72E-06 | 0.34 | |
| cg02486161 | NOD2 | 3'UTR | 0.70 | 0.59 | 2.53E-05 | 0.99 | |
| cg14983684 | RAD51L1 | Body | 0.75 | 0.71 | 2.58E-05 | 0.99 | |
| cg23939642 | SLC38A10 | Body | 0.50 | 0.33 | 2.66E-05 | 0.99 | |
| cg25325005 | PLEC1 | Body | N Shelf | 0.63 | 0.41 | 2.96E-05 | 0.99 |
| cg23335946 | C1orf251st | Exon | Island | 0.08 | 0.09 | 3.14E-05 | 0.99 |
| cg20776920 | UNC5D | TSS1500 | N Shore | 0.87 | 0.83 | 3.21E-05 | 0.99 |
| cg26812418 | CPE | TSS200 | Island | 0.05 | 0.07 | 4.09E-05 | 0.99 |
| cg07812589 | 0.26 | 0.20 | 4.59E-05 | 0.99 | |||
| cg03991871 | AHRR | Body | N Shore | 0.78 | 0.67 | 4.97E-05 | 0.99 |
| cg27545205 | Island | 0.02 | 0.02 | 5.26E-05 | 0.99 | ||
| cg10951975 | TRPM4 | Body | Island | 0.35 | 0.22 | 5.49E-05 | 0.99 |
| cg20370184 | SLC44A4 | Body | 0.27 | 0.12 | 5.64E-05 | 0.99 | |
| cg07999887 | CPNE3 | 5'UTR | Island | 0.02 | 0.02 | 5.92E-05 | 0.99 |
| cg08644463 | GNAI3 | Body | 0.87 | 0.83 | 6.83E-05 | 0.99 | |
| cg04366249 | SGCE | 1stExon | Island | 0.05 | 0.07 | 7.34E-05 | 0.99 |
| cg12741529 | C3orf17 | Body | 0.87 | 0.85 | 7.75E-05 | 0.99 | |
| cg08940570 | LOXL3 | 5'UTR | N Shore | 0.80 | 0.66 | 9.09E-05 | 0.99 |
| cg23754924 | RGMA | Body | Island | 0.10 | 0.13 | 9.56E-05 | 0.99 |
| cg24547565 | RUSC1 | TSS1500 | N Shore | 0.51 | 0.62 | 9.85E-05 | 0.99 |
| cg17093877 | MGC16275 | Body | N Shelf | 0.57 | 0.43 | 0.00010 | 0.99 |
| cg21545248 | HMGXB3 | Body | 0.77 | 0.71 | 0.00011 | 0.99 | |
| cg22012583 | LASS2 | TSS1500 | Island | 0.37 | 0.25 | 0.00011 | 0.99 |
| cg17231418 | ESX1 | Body | Island | 0.26 | 0.39 | 0.00011 | 0.99 |
| cg12668122 | TMEM108 | Body | 0.40 | 0.31 | 0.00012 | 0.99 | |
| cg19776793 | SLC38A10 | Body | 0.43 | 0.25 | 0.00013 | 0.99 | |
| cg02724404 | LYSMD4 | TSS1500 | S Shore | 0.88 | 0.84 | 0.00013 | 0.99 |
All average methylation values are non-log transformed beta-values. Island status refers to the position of the probe relative to the island. Classes include: 1) Island, 2) N (north) shore, 3) S (south) shore, 4) N (north shelf), 5) S (south) shelf and 6) blank denoting that the probe does not map to an island.
One possible concern is that some of the differential methylation signature could be secondary to alcohol use. Therefore, even though there were no significant differences between the rate of drinking for smoker and non-smoker groups, we analyzed the data for alcohol-related changes. The relationship of methylation to alcohol intake over the past 6 months to the methylation at loci controlling for alcohol use status was examined. Only two of the top 30 probes, cg07812589 and cg17231418, were even nominally related to amount of alcohol intake in the past 6 months, both at a p-value of 0.04< x<0.05. Hence, there does not appear to be any effect of alcohol intake on the methylation status at the most differentially methylated loci (data available upon request).
Next, as part of our analyses, we conducted a sliding window analysis using an 11-probe window and the same groups of case and control subjects. Table III describes the result of those analyses. The addition of the methylation data immediately flanking each probe increased the overall significance of the findings with 36 comparisons surviving genome wide correction. Not surprisingly, many of the top thirty probes from the analysis tended to lie immediately adjacent to one another. Interestingly, despite the strength of four AHRR probes in the single probe analyses, the gene region containing these probes, which is interrogated by 149 separate markers, was not included in this list of top regions. Inspection of this locus shows that differential methylation was largely confined to the 2 or 3 probe windows surrounding each of these residues with each of these areas being several thousand base pairs apart (Supplemental Table I)..
Table III.
The Top 30 Most Significantly Differentially Methylated Regions in Lymphoblast DNA.
| Average Methylation | |||||||
|---|---|---|---|---|---|---|---|
| Probe ID | GENE | Placement | Island Status | N-Smoker | Smoker | P value* | Corrected P-value |
| cg13581859 | HLA-DPB1 | Body | Island | 0.66 | 0.79 | 2.31E-09 | 0.002 |
| cg25511667 | HLA-DPB1 | Body | Island | 0.69 | 0.85 | 7.34E-09 | 0.002 |
| cg14801692 | HLA-DPB1 | Body | Island | 0.62 | 0.70 | 1.40E-08 | 0.003 |
| cg03636880 | HLA-DPB1 | Body | Island | 0.64 | 0.77 | 1.81E-08 | 0.003 |
| cg01132696 | HLA-DPB1 | Body | Island | 0.64 | 0.81 | 2.30E-08 | 0.003 |
| cg10850215 | HLA-DPB1 | Body | Island | 0.64 | 0.76 | 3.07E-08 | 0.003 |
| cg02692313 | HLA-DPB1 | Body | Island | 0.66 | 0.83 | 4.14E-08 | 0.003 |
| cg03229061 | HLA-DPB1 | Body | Island | 0.62 | 0.71 | 4.53E-08 | 0.003 |
| cg17588455 | HLA-DPB1 | Body | Island | 0.62 | 0.73 | 5.55E-08 | 0.003 |
| cg19990651 | HLA-DPB1 | Body | Island | 0.65 | 0.83 | 6.80E-08 | 0.004 |
| cg14870156 | HLA-DPB1 | Body | Island | 0.66 | 0.79 | 7.47E-08 | 0.004 |
| cg06437840 | HLA-DPB1 | Body | Island | 0.52 | 0.69 | 8.16E-08 | 0.004 |
| cg26645432 | HLA-DPB1 | Body | Island | 0.71 | 0.86 | 1.00E-07 | 0.004 |
| cg20223237 | HLA-DPB1 | Body | Island | 0.73 | 0.88 | 1.24E-07 | 0.005 |
| cg25796439 | ISM1 | TSS1500 | Island | 0.08 | 0.08 | 1.26E-07 | 0.006 |
| cg12893780 | HLA-DPB1 | Body | Island | 0.67 | 0.82 | 1.84E-07 | 0.006 |
| cg19759481 | HOXA5 | TSS200 | Island | 0.63 | 0.54 | 1.99E-07 | 0.007 |
| cg04863892 | HOXA5 | TSS200 | Island | 0.68 | 0.60 | 2.53E-07 | 0.008 |
| cg01992382 | TNXB | Body | Island | 0.42 | 0.47 | 2.74E-07 | 0.008 |
| cg01370449 | HOXA5 | TSS200 | Island | 0.69 | 0.63 | 3.11E-07 | 0.010 |
| cg12746059 | PCDH10 | TSS200 | Island | 0.08 | 0.09 | 3.95E-07 | 0.02 |
| cg13349035 | HLA-DPB1 | Body | N Shore | 0.68 | 0.80 | 4.72E-07 | 0.02 |
| cg09549073 | HOXA5 | 5'UTR | Island | 0.68 | 0.60 | 6.91E-07 | 0.02 |
| cg02916332 | HOXA5 | TSS1500 | Island | 0.64 | 0.58 | 7.89E-07 | 0.02 |
| cg12128839 | HOXA5 | TSS200 | Island | 0.56 | 0.47 | 8.21E-07 | 0.02 |
| cg17569124 | HOXA5 | TSS1500 | Island | 0.57 | 0.48 | 8.90E-07 | 0.02 |
| cg06831576 | CDH8 | TSS200 | Island | 0.11 | 0.15 | 1.00E-06 | 0.02 |
| cg04525757 | FOXG1 | TSS1500 | N Shore | 0.14 | 0.15 | 1.25E-06 | 0.03 |
| cg26242583 | LUZP2 | TSS200 | Island | 0.11 | 0.13 | 1.35E-06 | 0.03 |
| cg19714132 | FOXG1 | TSS1500 | N Shore | 0.19 | 0.21 | 1.58E-06 | 0.03 |
Nominal P-value before Benjamini-Hochberg correction. Corrected value is per Benjamini-Hochberg method.
Using GoMiner™, we conducted gene pathway analyses using the information from the 273 probes that were nominally differentially methylated at the p<0.001 level. Table IV shows the top 30 most differentially methylated pathways. Overall, only one pathway, protein kinase C (PKC) activity, survived false discovery rate (FDR) correction at the p<0.05 level. However, a recurrent theme of differential methylation in gene pathways affecting ion transport was found in many of the other less significant top thirty pathways.
Table IV.
The Top 30 Most Differentially Regulated Pathways in Lymphoblast DNA
| Genes | |||||
|---|---|---|---|---|---|
| GO Category | Category Name | Total | Changed | Log10 P-Value | FDR |
| GO:0018107 | peptidyl-threonine phosphorylation | 27 | 5 | −5.03 | 0.01 |
| GO:0018210 | peptidyl-threonine modification | 30 | 5 | −4.80 | 0.01 |
| GO:0060914 | heart formation | 9 | 3 | −4.00 | 0.09 |
| GO:0009653 | anatomical structure morphogenesis | 1490 | 31 | −3.53 | 0.15 |
| GO:0045121 | membrane raft | 160 | 8 | −3.45 | 0.14 |
| GO:0007548 | sex differentiation | 181 | 8 | −3.10 | 0.24 |
| GO:0005024 | TGF beta receptor activity | 18 | 3 | −3.05 | 0.20 |
| GO:0007530 | sex determination | 18 | 3 | −3.05 | 0.20 |
| GO:0003007 | heart morphogenesis | 104 | 6 | −3.03 | 0.18 |
| GO:0004675 | transmembrane receptor protein serine threonine kinase activity | 19 | 3 | −2.97 | 0.19 |
| GO:0003197 | endocardial cushion development | 5 | 2 | −2.94 | 0.22 |
| GO:0005026 | TGF beta receptor activity type II | 5 | 2 | −2.94 | 0.22 |
| GO:0060021 | palate development | 46 | 4 | −2.82 | 0.26 |
| GO:0051015 | actin filament binding | 48 | 4 | −2.75 | 0.33 |
| GO:0030501 | pos. reg. of bone mineralization | 23 | 3 | −2.73 | 0.33 |
| GO:0070169 | pos. reg. of biomineral tissue dev. | 24 | 3 | −2.67 | 0.33 |
| GO:0003128 | heart field specification | 7 | 2 | −2.63 | 0.31 |
| GO:0003129 | heart induction | 7 | 2 | −2.63 | 0.31 |
| GO:0051864 | histone demethylase activity | 7 | 2 | −2.63 | 0.31 |
| GO:0061311 | cell surface receptor linked signaling pathway involved in heart dev. | 7 | 2 | −2.63 | 0.31 |
| GO:0060389 | SMAD protein phosphorylation | 25 | 3 | −2.62 | 0.30 |
| GO:0001649 | osteoblast differentiation | 86 | 5 | −2.62 | 0.29 |
| GO:0005901 | caveola | 53 | 4 | −2.59 | 0.29 |
| GO:0031095 | platelet tubular network membrane | 8 | 2 | −2.51 | 0.32 |
| GO:0035173 | histone kinase activity | 8 | 2 | −2.51 | 0.32 |
| GO:0046541 | saliva secretion | 8 | 2 | −2.51 | 0.32 |
| GO:0045669 | pos. reg. of osteoblast differentiation | 28 | 3 | −2.48 | 0.33 |
| GO:0070838 | divalent metal ion transport | 229 | 8 | −2.45 | 0.32 |
| GO:0045778 | pos. reg. of ossification | 29 | 3 | −2.43 | 0.33 |
| GO:0030154 | cell differentiation | 2041 | 35 | −2.43 | 0.32 |
dev.= development, pos. reg. = positive regulation, FDR=false discovery rate.
Human Alveolar Macrophage Data
Because some may have concerns about the reliability of lymphoblast ability to model the changes found in their cognate lymphocytes and other primary cell types, we repeated these same case and control analyses using DNA from pulmonary alveolar macrophages again using a case and control paradigm. The case macrophages were isolated from the lungs of 10 smokers with at least a 10 year history of ≥ 1ppd smoking (6 male and 3 female) while the control macrophage biomaterial set was isolated from 9 non-smokers (6 male and 4 female). Although these two groups were roughly matched for ethnicity (smokers: 8 White, 2 African Americans; non-smokers: 9 White), the control group was significantly younger than the smoking group (smokers 31 ± 3 yrs, non-smokers 40 ± 4 yrs, p<0.01).
The results of the genome wide single probe contrasts are illustrated in Table V. Overall, the effects of smoking were much more profound with 1381 probes surviving correction for genome wide comparison at a p<0.05 level. Of considerable interest given recent data suggesting a prominent role for AHRR in carcinogenesis, 8 probes from AHRR, including the 3rd ranked probe, cg25648203, were significantly associated after correction for genome wide comparisons. But of the top 4 AHRR probes from the lymphoblast analyses, only cg05575921 was significantly associated after Bonferroni correction.
Table V.
The Top 30 Most Significantly Differentially Methylated Probes in Alveolar Macrophage DNA.
| Probe ID | GENE | Placement | Island Status | N-Smoker Avg | Smoker AVG | T-test | Corrected P-value |
|---|---|---|---|---|---|---|---|
| cg06961313 | MR1 | TSS1500 | 0.80 | 0.57 | 1.06 E-10 | 5.16201E-05 | |
| cg00738897 | S_Shore | 0.71 | 0.55 | 1.90 E-09 | 0.0003 | ||
| cg25648203 | AHRR | Body | 0.38 | 0.72 | 1.97 E-09 | 0.0003 | |
| cg00506299 | RFTN1 Body | 0.23 | 0.46 | 2.67 E-09 | 0.0003 | ||
| cg27229484 | ZC3H12A | Body | 0.26 | 0.53 | 3.34 E-09 | 0.0003 | |
| cg05951221 | Island | 0.28 | 0.42 | 5.85 E-09 | 0.0005 | ||
| cg01432692 | 0.20 | 0.37 | 7.69 E-09 | 0.0005 | |||
| cg09374353 | EHD1 | 3'UTR | N_Shore | 0.12 | 0.39 | 1.05 E-08 | 0.0006 |
| cg14310198 | RAPGEF1 | Body | 0.48 | 0.70 | 1.90 E-08 | 0.0010 | |
| cg21566642 | Island | 0.33 | 0.56 | 2.12 E-08 | 0.0010 | ||
| cg17576603 | DAB2 | 5'UTR | 0.39 | 0.62 | 3.36 E-08 | 0.0013 | |
| cg17574812 | ABHD6 | Body | 0.26 | 0.49 | 3.55 E-08 | 0.0013 | |
| cg06634140 | 0.30 | 0.54 | 3.73 E-08 | 0.0013 | |||
| cg11254522 | FGR | Body | 0.35 | 0.50 | 3.99 E-08 | 0.0013 | |
| cg07457727 | N_Shelf | 0.22 | 0.60 | 4.02 E-08 | 0.0013 | ||
| cg13458803 | CD80 | 5'UTR | 0.36 | 0.16 | 4.86 E-08 | 0.0014 | |
| cg01668352 | SRGAP1 | Body | 0.32 | 0.62 | 4.97 E-08 | 0.0014 | |
| cg04402828 | KIAA1026 | Body | 0.47 | 0.35 | 6.69 E-08 | 0.0018 | |
| cg07650681 | LOC100132354 | Body | 0.66 | 0.40 | 7.19 E-08 | 0.0018 | |
| cg13610455 | LOC388796 | Body | 0.29 | 0.44 | 7.37 E-08 | 0.0018 | |
| cg09127592 | TRIM35 | Body | N_Shelf | 0.33 | 0.73 | 8.72 E-08 | 0.0019 |
| cg14223856 | 0.43 | 0.81 | 9.60 E-08 | 0.0019 | |||
| cg09006487 | RYBP | 3'UTR | 0.35 | 0.50 | 9.69 E-08 | 0.0019 | |
| cg02233197 | TNFAIP8L3 | Body | S_Shelf | 0.29 | 0.72 | 9.85 E-08 | 0.0019 |
| cg05317600 | 0.34 | 0.65 | 9.86 E-08 | 0.0019 | |||
| cg25466245 | SUSD4 | Body | 0.36 | 0.57 | 1.09 E-07 | 0.0020 | |
| cg21418854 | C1orf113 | TSS1500 | N_Shore | 0.42 | 0.58 | 1.13 E-07 | 0.0020 |
| cg02341139 | S_Shelf | 0.34 | 0.60 | 1.17 E-07 | 0.0020 | ||
| cg18030943 | LAMP3 | Body | N_Shelf | 0.23 | 0.38 | 1.20 E-07 | 0.0020 |
| cg05337681 | LIPC | Body | 0.23 | 0.47 | 1.25 -07 | 0.0020 |
All average methylation values are non-log transformed beta-values. Island status refers to the position of the probe relative to the island. Classes include: 1) Island, 2) N (north) shore, 3) S (south) shore, 4) N (north shelf), 5) S (south) shelf and 6) blank denoting that the probe does not map to an island.
We next repeated the sliding window analyses for the macrophage data using the same method delineated above. Once again, the results (see Table VI) were more robust than those for the lymphoblast data with 40 eleven probe regions being significantly associated after correction for multiple comparisons. Although many highly interesting genes were once again implicated in this analysis, AHRR was once again notable with the 28th ranked 11 probe region being found in the body of the AHRR.
Table VI.
The Top 30 Most Significantly Differentially Methylated Regions in Alveolar Macrophage DNA
| Average Methylation | |||||||
|---|---|---|---|---|---|---|---|
| Probe ID | GENE | Placement | Island Status | N-Smoker | Smoker | P value* | Corrected P-value |
| cg07965566 | 0.60 | 0.30 | 2.40E-28 | 1.16E-22 | |||
| cg14310198 | RAPGEF1 | Body | 0.70 | 0.48 | 4.83E-26 | 1.17E-20 | |
| cg17574812 | ABHD6 | Body | 0.49 | 0.26 | 9.83E-25 | 1.59E-19 | |
| cg01668352 | SRGAP1 | Body | 0.62 | 0.32 | 1.76E-24 | 2.14E-19 | |
| cg17576603 | DAB2 | 5'UTR | 0.62 | 0.39 | 2.52E-23 | 2.45E-18 | |
| cg07457727 | 0.60 | 0.22 | 3.98E-21 | 3.22E-16 | |||
| cg10169462 | 0.13 | 0.06 | 1.30E-15 | 9.05E-11 | |||
| cg24790419 | KIAA1683 | TSS1500 | 0.62 | 0.39 | 1.54E-15 | 9.38E-11 | |
| cg04402828 | KIAA1026; | Body | 0.35 | 0.47 | 3.16E-15 | 1.70E-10 | |
| cg05951221 | 0.42 | 0.28 | 4.80E-12 | 2.33E-07 | |||
| cg16039867 | MKNK1 | Body | 0.58 | 0.78 | 2.50E-11 | 1.10E-06 | |
| cg06634140 | 0.54 | 0.30 | 9.81E-10 | 3.97E-05 | |||
| cg20485084 | FGR | Body | 0.67 | 0.36 | 1.91E-09 | 7.16E-05 | |
| cg02341139 | 0.60 | 0.34 | 6.35E-09 | 0.0002 | |||
| cg22019569 | SMYD3 | Body | 0.60 | 0.75 | 1.38E-08 | 0.0004 | |
| cg14019523 | ASB2 | Body | 0.37 | 0.24 | 2.11E-08 | 0.0006 | |
| cg13675814 | CORO2A | 5'UTR | 0.67 | 0.82 | 6.95E-08 | 0.0018 | |
| cg04307274 | 0.57 | 0.31 | 6.96E-08 | 0.0018 | |||
| cg15149645 | NUPR1 | TSS200 | 0.59 | 0.75 | 9.66E-08 | 0.0024 | |
| cg10192877 | ABCG1 | Body | 0.61 | 0.74 | 1.20E-07 | 0.0028 | |
| cg21566642 | 0.56 | 0.33 | 1.24E-07 | 0.0028 | |||
| cg20568305 | GRAMD4 | Body | 0.68 | 0.52 | 2.95E-07 | 0.0065 | |
| cg01432692 | 0.37 | 0.20 | 3.35E-07 | 0.0070 | |||
| cg24446429 | MBP | Body | 0.60 | 0.39 | 3.88E-07 | 0.0075 | |
| cg14414943 | CHI3L2 | Body | 0.83 | 0.89 | 3.89E-07 | 0.0075 | |
| cg16659773 | 0.56 | 0.43 | 4.16E-07 | 0.0077 | |||
| cg04135110 | AHRR | Body | 0.15 | 0.38 | 5.29E-07 | 0.0095 | |
| cg00738897 | 0.55 | 0.71 | 6.03E-07 | 0.0104 | |||
| cg13458803 | CD80 | 5'UTR | 0.16 | 0.36 | 7.97E-07 | 0.0133 | |
| cg11691844 | SYTL2 | Body | 0.27 | 0.44 | 8.63E-07 | 0.0139 | |
Nominal P-value before Benjamini-Hochberg correction. Corrected value is per Benjamini-Hochberg method.
As a last part of our set of analyses with respect to the macrophage methylation data, we repeated the GoMiner pathway analyses using the list of 1381 probes which were significantly associated in the above analyses as our changed gene set. Table VII shows those results of those analyses. In brief, pathways involved with wound healing, inflammation and G-protein/ras signaling were particularly prominent.
Table VII.
The Top 30 Most Differentially Regulated Gene Ontology Pathways in Macrophage DNA
| Genes | ||||||
|---|---|---|---|---|---|---|
| GO Category | Category Name | Total | Changed | Log10 | P-Value | FDR |
| GO:0005737 | cytoplasm | 7845 | 429 | −8.40 | 0 | |
| GO:0007165 | signal transduction | 2324 | 159 | −7.89 | 0 | |
| GO:0005515 | protein binding | 6815 | 378 | −7.62 | 0 | |
| GO:0023052 | signaling | 3788 | 233 | −7.56 | 0 | |
| GO:0023033 | signaling pathway | 2813 | 183 | −7.48 | 0 | |
| GO:0007264 | small GTPase mediated signal trans. | 566 | 54 | −6.90 | 0 | |
| GO:0023060 | signal transmission | 2728 | 173 | −6.29 | 0 | |
| GO:0023046 | signaling process | 2730 | 173 | −6.27 | 0 | |
| GO:0009611 | response to wounding | 828 | 68 | −6.06 | 0 | |
| GO:0030234 | enzyme regulator activity | 880 | 71 | −6.02 | 0 | |
| GO:0030695 | GTPase regulator activity | 437 | 43 | −5.96 | 0.001 | |
| GO:0035466 | reg. of sig. pathway | 1158 | 87 | −5.96 | 0.001 | |
| GO:0051056 | reg. of GTPase mediated sig. trans. | 339 | 36 | −5.85 | 0.001 | |
| GO:0023034 | intracellular sig. pathway | 1708 | 117 | −5.81 | 0.001 | |
| GO:0060589 | nucleoside-triphosphatase reg. activity | 446 | 43 | −5.73 | 0.001 | |
| GO:0006928 | cellular component movement | 687 | 58 | −5.61 | 0.001 | |
| GO:0044444 | cytoplasmic part | 5629 | 311 | −5.60 | 0.001 | |
| GO:0007265 | Ras protein signal transduction | 335 | 35 | −5.54 | 0.001 | |
| GO:0016192 | vesicle-mediated transport | 743 | 61 | −5.48 | 0.001 | |
| GO:0010876 | lipid localization | 188 | 24 | −5.44 | 0.001 | |
| GO:0005085 | guanyl-nucleotide exchange factor act. | 152 | 21 | −5.38 | 0.001 | |
| GO:0035556 | intracellular signal transduction | 1454 | 101 | −5.31 | 0.001 | |
| GO:0006869 | lipid transport | 167 | 22 | −5.26 | 0.001 | |
| GO:0010885 | reg. of cholesterol storage | 12 | 6 | −5.24 | 0.001 | |
| GO:0016477 | cell migration | 564 | 49 | −5.16 | 0.001 | |
| GO:0042060 | wound healing | 470 | 43 | −5.15 | 0.001 | |
| GO:0007166 | cell surface receptor linked sig. path | 1745 | 116 | −5.14 | 0.001 | |
| GO:0005089 | Rho guanyl-nucleotide exchange act. | 68 | 13 | −5.08 | 0.001 | |
| GO:0051179 | localization | 3540 | 207 | −5.02 | 0.002 | |
| GO:0001816 | cytokine production | 240 | 27 | −4.99 | 0.002 | |
pos. reg. = positive reg., sig. =sig., trans. = transduction, FDR=false discovery rate.
Comparison of Lymphoblast and Macrophage Data
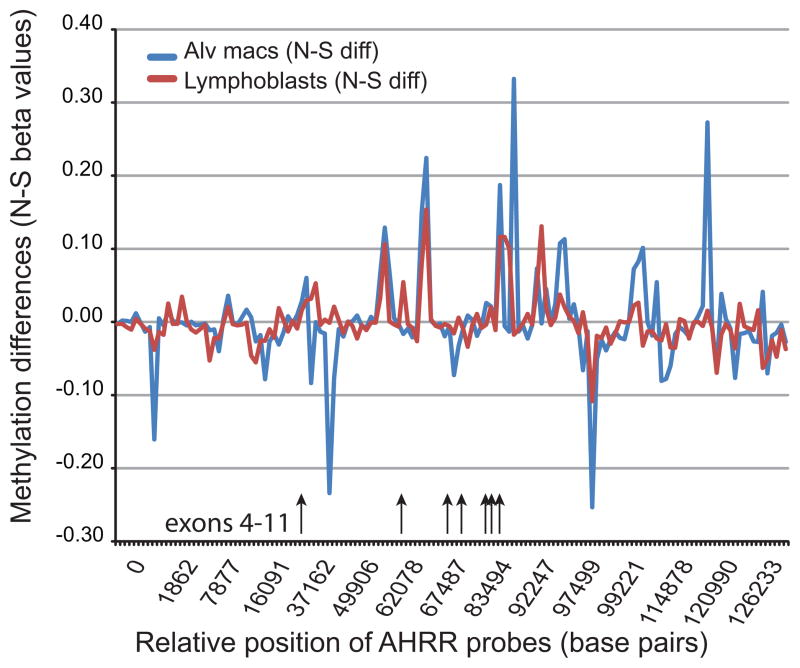
In both the macrophage and lymphoblast analyses, probes from AHRR were repeatedly associated with smoking status. Therefore, we compared the methylation signatures from these two biomaterials with respect to smoking status. Supplemental Table I details the average methylation and single point analyses for each of the 146 probes for the gene for each biomaterial. In brief, 14 probes in the lymphoblast analyses and 40 of the probes in the macrophage analyses were associated with smoking status at a p<0.05 with 8 of the 14 probes in the lymphoblast analyses also being nominally significantly associated with smoking status in the macrophages with the direction of methylation being consistent at each probe (greater methylation in smokers). The overall methylation signature between the control lymphoblasts and macrophages at AHRR was highly correlated (r=0.95). Figure 1 illustrates the relationship between the differential methylation at each of the 146 residues listed in Supplementary Table I for the lymphoblast and macrophage DNA samples. As the figure shows, the differential methylation signature was also highly correlated across the gene with over 20% of the differential methylation signature that was associated with smoking status being shared between the two DNA sources (r=0.45; p<0.001).
Figure 1.
An advantage of lymphoblasts is the ability to easily create high-quality RNA for gene expression studies. Therefore, to determine whether this differential methylation had functional consequences on lymphoblast gene expression, we then analyzed the relationship between AHRR gene expression and methylation status at cg05575921, the AHRR probe with the most consistent associations in the two analyses, using RNA prepared from the case and control samples. Interestingly, increasing methylation at this probe was associated with decreasing lymphoblast AHRR gene expression (p<0.03, n=108) which suggests that the CpG residues in this region may have a functional in vivo role in regulating gene expression at this locus.
DISCUSSION
In summary, we report that cigarette smoking is associated with significant changes in genome wide methylation, and in particular, AHRR methylation, in DNA derived from pulmonary alveolar macrophages and lymphoblasts. Strengths of this manuscript include confirmation of the findings from lymphoblast DNA, which are immortalized lymphocytes, with data from primary tissue from the lungs of smokers and the presentation of evidence that these changes at AHRR may be functional. Possible limitations include the relative poor matching of the subjects who contributed lymphoblast and pulmonary macrophage DNA, occasional mis-annotations in the probe descriptor files, possible unaccounted effects of polymorphisms in the regions containing him the probes, and the fact that we did not verify the results with bisulfite sequencing.
The most significant and consistent finding in the current study is with respect to AHRR locus. AHHR is a feedback inhibition modulator of the aryl hydrocarbon receptor (AHR) that exerts its effects by competing with AHR for binding with its cognate nuclear receptor dimer partner (AHR nuclear translocator) or at xenobiotic response elements in AHR regulated genes (Haarmann-Stemmann and others 2007). This feedback modulation plays a pivotal role in AHR regulation and may be critical in moderating AHR role in oncogenesis and altered immune function (Opitz and others 2011). Our finding of smoking associated methylation at AHRR is highly plausible for several reasons. First and foremost, smoking is the leading preventable cause of cancer. Hence, this association may explain part of the connection. Second, the direction of the differential methylation was consistent among the 8 AHRR probes with nominal significance in both lymphoblast and macrophage comparisons with a high degree of shared smoking associated differential methylation (see supplemental table 1). Third, AHRR was the only gene locus that had significant localizations in both studies after correction for multiple comparisons. Fourth, previous studies have shown that smoking induces production of the AHR (Martey and others 2005; Meek and Finch 1999), a process which is thought to be critical for certain forms of smoking related forms of carcinogenesis (Andersson and others 2002; Gumus and others 2008; Shimizu and others 2000). Assuming that the decreased methylation at AHRR seen in smokers in the current study may result from a feedback mechanism associated with smoking induction of AHR transcription, the current findings are very consistent with previous findings and suggest potential avenues for addressing AHR mediated neoplastic transformation. Unfortunately, even given the promising gene expression findings, rigorous testing of this hypothesis may be difficult because review of the Ensembl and University of Santa Clara (UCSC) genome browser databases demonstrates the presence of three large CpG islands that are interspersed throughout the gene and at least 11 AHRR transcripts, each of which codes for a differently sized protein that may have unique competitive properties with respect to AHR. Hence, while the current findings are encouraging, a more definitive understanding of relationship between AHHR methylation and both AHRR gene expression and AHR function may require more complex and detailed examination of this region.
The pathway analyses of the macrophage data were illuminating and consistent with our understanding of the effects of smoking. The macrophage data was characterized by changes in inflammation, wound healing and Ras/G-protein signaling pathways. The repeated finding of altered methylation in Ras/G-protein signaling pathways seems logical since activation of these proteins are thought to be part of the oncogenic process for many types of cancers (Lewinski and Wojciechowska 2007; Tchevkina and others 2004). Similarly, the recurrent identification of wound healing and inflammatory pathways seems logical since smoking is the leading cause of Chronic Obstructive Pulmonary Disease (COPD), a syndrome in which the vast morbidity of the pathology is secondary to inflammatory moderated remodeling of the lung epithelium (Shapiro and Ingenito 2005). In contrast, the results of the lymphoblast analyses were less robust with only two pathways, related to peptidyl-threonine modification, surviving FDR correction. However, it is important to note that while both pathways are closely related with the basis of their significance in our analyses relying on the same five probes with the omission of one probe from either of these comparisons would result in nonsignificant findings.
The comparative weakness of the methylation findings in lymphoblasts as compared to macrophages highlight the importance of incorporating studies of primary tissues directly exposed to the substance in question. Overall, the smoking associated differential methylation was markedly more pronounced in the alveolar macrophage DNA than in the lymphoblast DNA. This is probably because circulating lymphocytes are less exposed to the direct effects of smoke than the macrophages resident in the lung. During cell replication, DNA methyltransferase 1 (DNMT1) stably copies cellular DNA methylation patterns (Suzuki and Bird 2008). However, it is possible that our conversion of these same lymphocytes into the transformed lymphoblast cell lines may further weaken the smoking induced signal. The latter possibility needs to be considered because although lymphoblast cell lines are excellent models of the lymphocytes from which they are derived, lymphoblast lines are vulnerable to clonal selection artifacts and there are well documented differences between lymphocyte and lymphoblast gene expression that occur as a function of EBV mediated transformation (Grafodatskaya and others 2009; Rollins and others 2010). Therefore, even though Vawter and colleagues have demonstrated that once transformed, gene expression profiles of lymphoblasts are relatively stable (Rollins and others 2010), the fact that the lymphoblasts by definition proliferate in non-smoking conditions, probably impact the data. To certain a certain extent this makes sense, if exposure to smoke induces an epigenetic change, the continued in vitro replication in the absence of smoking associated chemicals may mute the findings. This supports the importance of examining primary cells along with lymphoblasts.
It should also be recognized that most investigators, including Breitling and colleagues, use Ficoll separated mononuclear cell pellets rather than purified lymphocytes(Breitling and others 2011). Since these “lymphocyte pellets” contain a variety of cell types including B-lymphocytes, T-lymphocytes, monocytes and Natural Killer T-cell, it may well be that use of this heterogeneous cell mix may have obscured other potential findings which may explain why Breitling and colleagues only identified one differentially methylated probe in their study despite using a similar number of subjects.
Beyond the relative merits of lymphocyte and lymphoblast preparations, the current findings suggest that the lymphoblast lines paired with primary pulmonary macrophages will be useful in other investigations of the epigenetics of smoking because: 1) smoking has a broad effect on tissues throughout the body including the blood, and 2) integration of histone modification and gene expression status with DNA methylation status will require large numbers of cells. Some types of histone modification examinations necessitate relatively larger amounts of fresh cellular material. This suggests the utility of lymphoblasts in histone modification studies. A clear picture of lymphoblast gene expression and DNA methylation data relative to a primary smoking-relevant cell (alveolar macrophages) data will be needed for these potential future studies. In this respect, our convergent finding in lymphocytes and macrophages with respect to AHRR are reassuring.
One potential direction for future work is the determination of the specific AHRR transcripts that are differentially affected by differential methylation. The Taqman™ gene expression probe for AHRR used in this study (Hs01005075) recognizes the exon 3-4 exon boundary that is included in most splice variants. However, given the numerous splice variants produced by this gene, the epigenetic complexity of the gene (e.g. three large CpG islands not associated with the promoter), and its putative role in oncogenesis, future studies that examine specific splice variants altered by smoking is warranted.
The relationship of gene methylation to histone code modification should also be explored. In particular, the relationship of H3K4 and H3K27 methylation and H3K27 acetylation to AHRR gene expression should be examined because of the strong relationship of these modifications to gene expression (Heintzman and others 2009; Kharchenko and others). Though DNA methylation is thought to have a weaker relationship to gene expression (Pai and others 2011; Wu and others 2010), if we can establish a stronger understanding of the histone-DNA modification relationship on a genome wide level, it well may be that we can use DNA methylation at loci such as AHRR as a proxy for histone status, and thereby gene expression status. Studies of DNA methylation are much cheaper and easier to conduct than histone modification studies. A better understanding of the relationship of peripheral blood methylation to methylation in other tissues, such as brain, may allow more informative studies of the role of DNA methylation and other forms of epigenetic changes in normal and disease related human development.
In summary, we report that cigarette smoking is associated with genome wide changes in lymphoblast and pulmonary macrophage DNA methylation, in particular at AHRR. We suggest replication and extension of the current findings and further investigations of the role of epigenetic changes in smoking altered gene expression.
Supplementary Material
Acknowledgments
The work in this study was supported by DA015789 and MH080898 to Dr. Philibert and DA021898 and DA018871. Additional support for these studies was derived from the Center for Contextual Genetics and Prevention Science (Grant Number P30 DA027827, GB) funded by the National Institute on Drug Abuse. The University of Iowa has filed intellectual property right claims on some of the material related to this manuscript on behalf of Dr. Philibert. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Andersson P, McGuire J, Rubio C, Gradin K, Whitelaw ML, Pettersson S, Hanberg A, Poellinger L. A constitutively active dioxin/aryl hydrocarbon receptor induces stomach tumors. 2002. pp. 9990–9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B, Methodological. 1995;57:289–300. [Google Scholar]
- Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-Smoking-Related Differential DNA Methylation: 27K Discovery and Replication. American journal of human genetics. 2011;88(4):450–7. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–58. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Caputo J, Thompson A, McClintock P, Reid Y, Hay R. An Effective Method for Establishing Human B Lymphoblastic Cell Lines Using Epstein Barr Virus. J Tiss Cult Meth. 1991;13:39–44. [Google Scholar]
- Center for Disease Control. Annual Smoking-Attributable Mortality, Years of Potential Life Lost, and Productivity Losses --- United States, 1997--2001. Morbidity and Mortality Weekly. 2005;54(25):625– 628. [PubMed] [Google Scholar]
- Centers for Disease Control. Vital signs: current cigarette smoking among adults aged >/=18 years--United States, 2005–2010. MMWR. 2011;60:1207–12. [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56(3):422–37. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chang HW, Ling GS, Wei WI, Yuen AP. Smoking and drinking can induce p15 methylation in the upper aerodigestive tract of healthy individuals and patients with head and neck squamous cell carcinoma. Cancer. 2004;101(1):125–32. doi: 10.1002/cncr.20323. [DOI] [PubMed] [Google Scholar]
- Davis W, Hartman A, Gibson J. Trends in Smoking Prevalence by Race based on the Tobacco Use Supplement to the Current Population Survey. 2010. [Google Scholar]
- Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res. 2009;19(8):1374–83. doi: 10.1101/gr.089185.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan C-F, Andrews S, Dean W, Hemberger M, Reik W. Global Mapping of DNA Methylation in Mouse Promoters Reveals Epigenetic Reprogramming of Pluripotency Genes. PLoS Genet. 2008;4(6):e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss JL. Statistical Methods for Rates and Proportions. New York, NY: John Wiley & Sons Inc; 1981. [Google Scholar]
- Grafodatskaya D, Choufani S, Ferreira JC, Butcher DT, Lou Y, Zhao C, Scherer SW, Weksberg R. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 2009;95(2):73–83. doi: 10.1016/j.ygeno.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Gumus ZH, Du B, Kacker A, Boyle JO, Bocker JM, Mukherjee P, Subbaramaiah K, Dannenberg AJ, Weinstein H. Effects of tobacco smoke on gene expression and cellular pathways in a cellular model of oral leukoplakia. Cancer Prev Res (Phila) 2008;1(2):100–11. doi: 10.1158/1940-6207.CAPR-08-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurrieri F, Accadia M. Genetic imprinting: the paradigm of Prader-Willi and Angelman syndromes. Endocr Dev. 2009;14:20–8. doi: 10.1159/000207473. [DOI] [PubMed] [Google Scholar]
- Haarmann-Stemmann T, Bothe H, Kohli A, Sydlik U, Abel J, Fritsche E. Analysis of the Transcriptional Regulation and Molecular Function of the Aryl Hydrocarbon Receptor Repressor in Human Cell Lines. 2007. pp. 2262–2269. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 471(7339):480–5. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler BA, Ward E, McCarthy BJ, Schymura MJ, Ries LA, Eheman C, Jemal A, Anderson RN, Ajani UA, Edwards BK. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. Journal of the National Cancer Institute. 2011;103(9):714–36. doi: 10.1093/jnci/djr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launay J-M, Del Pino M, Chironi G, Callebert J, Peoc'h K, Mégnien J-L, Mallet J, Simon A, Rendu F. Smoking Induces Long-Lasting Effects through a Monoamine-Oxidase Epigenetic Regulation. PLoS ONE. 2009;4(11):e7959. doi: 10.1371/journal.pone.0007959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinski A, Wojciechowska K. Genetic background of carcinogenesis in the thyroid gland. Neuro endocrinology letters. 2007;28(2):77–105. [PubMed] [Google Scholar]
- Martey CA, Baglole CJ, Gasiewicz TA, Sime PJ, Phipps RP. The aryl hydrocarbon receptor is a regulator of cigarette smoke induction of the cyclooxygenase and prostaglandin pathways in human lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2005;289(3):L391–9. doi: 10.1152/ajplung.00062.2005. [DOI] [PubMed] [Google Scholar]
- Meek MD, Finch GL. Diluted mainstream cigarette smoke condensates activate estrogen receptor and aryl hydrocarbon receptor-mediated gene transcription. Environ Res. 1999;80(1):9–17. doi: 10.1006/enrs.1998.3872. [DOI] [PubMed] [Google Scholar]
- Monick MM, Powers LS, Barrett CW, Hinde S, Ashare A, Groskreutz DJ, Nyunoya T, Coleman M, Spitz DR, Hunninghake GW. Constitutive ERK MAPK activity regulates macrophage ATP production and mitochondrial integrity. J Immunol. 2008;180(11):7485–96. doi: 10.4049/jimmunol.180.11.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monick MM, Powers LS, Gross TJ, Flaherty DM, Barrett CW, Hunninghake GW. Active ERK Contributes to Protein Translation by Preventing JNK-Dependent Inhibition of Protein Phosphatase 1. J Immunol. 2006;177(3):1636–45. doi: 10.4049/jimmunol.177.3.1636. [DOI] [PubMed] [Google Scholar]
- Monick MM, Powers LS, Walters K, Lovan N, Zhang M, Gerke A, Hansdottir S, Hunninghake GW. Identification of an autophagy defect in smokers' alveolar macrophages. J Immunol. 2010;185(9):5425–35. doi: 10.4049/jimmunol.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011 doi: 10.1038/nature10491. advance online publication. [DOI] [PubMed] [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues. PLoS Genet. 2011;7(2):e1001316. doi: 10.1371/journal.pgen.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Beach SR, Gunter TD, Brody GH, Madan A, Gerrard M. The effect of smoking on MAOA promoter methylation in DNA prepared from lymphoblasts and whole blood. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):619–28. doi: 10.1002/ajmg.b.31031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Gunter TD, Beach SR, Brody GH, Madan A. MAOA methylation is associated with nicotine and alcohol dependence in women. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(5):565–70. doi: 10.1002/ajmg.b.30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins B, Martin MV, Morgan L, Vawter MP. Analysis of whole genome biomarker expression in blood and brain. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2010;153B(4):919–936. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro SD, Ingenito EP. The Pathogenesis of Chronic Obstructive Pulmonary Disease: Advances in the Past 100 Years. Am J Respir Cell Mol Biol. 2005;32(5):367–372. doi: 10.1165/rcmb.F296. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Nakatsuru Y, Ichinose M, Takahashi Y, Kume H, Mimura J, Fujii-Kuriyama Y, Ishikawa T. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. 2000. pp. 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga Y, Miyajima K, Oikawa T, Maeda J, Usuda J, Kajiwara N, Ohira T, Uchida O, Tsuboi M, Hirano T, et al. Quantitative p16 and ESR1 methylation in the peripheral blood of patients with non-small cell lung cancer. Oncol Rep. 2008;20(5):1137–42. [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature reviews Genetics. 2008;9(6):465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Tchevkina E, Agapova L, Dyakova N, Martinjuk A, Komelkov A, Tatosyan A. The small G-protein RalA stimulates metastasis of transformed cells. Oncogene. 2004;24(3):329–335. doi: 10.1038/sj.onc.1208094. [DOI] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8(5):355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-Dependent Nonpromoter DNA Methylation Facilitates Transcription of Neurogenic Genes. Science. 2010;329(5990):444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates W, Cadoret R, Troughton E. The Iowa Adoption Studies Methods and Results. In: LaBuda M, Grigorenko E, editors. On the way to individuality: Methodological Issues in Behavioral Genetics. Hauppauge NY: Nova Science Publishers; 1998. pp. 95–125. [Google Scholar]
- Zeeberg B, Feng W, Wang G, Wang M, Fojo A, Sunshine M, Narasimhan S, Kane D, Reinhold W, Lababidi S, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biology. 2003;4(4):1–8. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.