Summary
Melioidosis is an emerging infectious disease of humans and animals in the tropics caused by the soil bacterium Burkholderia pseudomallei. Despite high fatality rates, the ecology of B. pseudomallei remains unclear. We used a combination of field and laboratory studies to investigate B. pseudomallei colonization of native and exotic grasses in northern Australia. Multivariable and spatial analyses were performed to determine significant predictors for B. pseudomallei occurrence in plants and soil collected longitudinally from field sites. In plant inoculation experiments, the impact of B. pseudomallei upon these grasses was studied and the bacterial load semi-quantified. Fluorescence-in-situ-hybridization and confocal laser-scanning microscopy were performed to localize the bacteria in plants. B. pseudomallei was found to inhabit not only the rhizosphere and roots but also aerial parts of specific grasses. This raises questions about the potential spread of B. pseudomallei by grazing animals whose droppings were found to be positive for these bacteria.
In particular, B. pseudomallei readily colonized exotic grasses introduced to Australia for pasture. The ongoing spread of these introduced grasses creates new habitats suitable for B. pseudomallei survival and may be an important factor in the evolving epidemiology of melioidosis seen both in northern Australia and elsewhere globally.
Introduction
Melioidosis is an infectious disease affecting humans and animals in the tropics. It is caused by the Gram-negative environmental β-Proteobacterium Burkholderia pseudomallei. Melioidosis is a major cause of community-acquired pneumonia and septicemia in endemic areas such as northern Australia and Southeast Asia, with mortality still over 50% in some locations (Cheng et al., 2005; White, 2003). Melioidosis is also seen as an emerging public health threat, with both rising incidence rates in endemic areas (Currie et al., 2010; Limmathurotsakul et al., 2010a) and cases increasingly being described from locations not previously considered to be endemic (Currie et al., 2008). With the high mortality from melioidosis, the potential for airborne transmission of B. pseudomallei and its intrinsic resistance to antibiotics commonly used for sepsis, B. pseudomallei has been classified as a Category B biothreat agent by the US Centers for Disease Control (Rotz et al., 2002).
Since the sequencing of the first genome of B. pseudomallei (Holden et al., 2004), there have been numerous publications analyzing potential pathogenicity determinants of B. pseudomallei (French et al., 2011; Galyov et al., 2010). Nevertheless, the search for why B. pseudomallei can be such a dangerous pathogen has not been matched by studies defining the ecology of B. pseudomallei and many uncertainties remain about its global distribution (Currie et al., 2008) and its habitat in endemic locations. Defining the environmental determinants of B. pseudomallei presence, persistence and propagation is critical to understanding and potentially controlling the local and global spread of B. pseudomallei that is thought to be occurring.
B. pseudomallei is known to prefer moist, slightly acidic, nutrient-rich soil (Inglis et al., 2006; Kaestli et al., 2009; Palasatien et al., 2008; Wuthiekanun et al., 1995) and also occurs in water (Draper et al., 2010; Inglis et al., 2000; Mayo et al., 2011). In a survey on B. pseudomallei occurrence in the Darwin region (12°S latitude) of northern Australia, we found a significant association of these bacteria with grass- and roots-dense areas (Kaestli et al., 2009). This comes as no surprise as bacteria of the genus Burkholderia commonly colonize plants and, in particular, the rhizosphere that is the nutrient-rich zone surrounding the roots (Compant et al., 2008; Fiore et al., 2001). Despite the well-known occurrence of B. pseudomallei in rice fields (Wuthiekanun et al., 1995), to date, only a few publications have reported a direct association of B. pseudomallei with plants: Burkholderia pseudomallei was retrieved from the rhizosphere of a wattle shrub (Acacia colei) in northwest Australia (Inglis et al., 2000) and Levy et al. reported the ability of B. pseudomallei to colonize root-associated mycorrhizal fungi (Levy et al., 2003). In laboratory experiments, B. pseudomallei was found to have phyto-pathogenic traits with infection of tomato plants and intracellular replication (Lee et al., 2010).
The aim of this study was to investigate further our findings of the association of B. pseudomallei with grasses. Grass species found to support the growth of B. pseudomallei might serve as bio-indicators for potential presence of B. pseudomallei and thus, assist in predicting its distribution and spread both locally and globally. Furthermore, the finding of a close interaction of B. pseudomallei with eukaryotic hosts such as plants might have implications for potential future studies on the mechanisms involved in the colonization of other eukaryotic cells by B. pseudomallei and specifically its ability to invade and persist in animal and human tissues.
Results
We investigated the association of B. pseudomallei with various grasses occurring at known hotspots for B. pseudomallei in the Darwin region using a combination of observational longitudinal field studies and laboratory-based grass inoculation experiments. We studied the impact of B. pseudomallei upon these grasses, semi-quantified its load and visualized the colonization of B. pseudomallei in the rhizosphere, roots and aerial parts using fluorescence in situ hybridization (FISH) and confocal laser-scanning microscopy.
B. pseudomallei occurs significantly more often in areas with exotic grasses
In the Darwin region, four sites next to streams were chosen to study the occurrence of B. pseudomallei in areas dense in grasses commonly occurring in the Darwin region. These sites had previously been positive for B. pseudomallei (Kaestli et al., 2009). Native grass species (including Sorghum spp. and Whiteochloa spp.) were present at each study site. Exotic grasses were found at three of these four sites, such as Brachiaria humidicola cv Tully (Tully Grass), Pennisetum pedicellatum and polystachion (Mission Grass) and Paspalum plicatulum (Paspalum).
Using a grid-like sampling approach over two consecutive dry and wet seasons, soil and grasses of these sites were repeatedly screened for presence of B. pseudomallei by an enriched soil/plant DNA extraction method and TTS1 real-time PCR (Kaestli et al., 2007). We found 37% (142/380) of soil samples, 18% (38/217) of collected grass roots and 23% (31/135) of foliage samples positive for B. pseudomallei at these sites (see Table 1). A significant difference was evident in B. pseudomallei load in grass foliage between the dry and wet season, with a higher proportion of leaves being B. pseudomallei positive in the dry (42% of screened leaves (13/31)) as opposed to the wet season (17% (18/104), Fisher’s Exact, P=0.007).
Table 1.
Overview of B. pseudomallei prevalence in soil and grasses at four field sites
| Sample No. (Soil; Grasses) | Soil around roots % (B.ps positive/total samples) | Grass (roots+aerial) % (B.ps positive/total samples) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Dry Season | Wet Season | Dry Season | Wet Season | ||||||
| Nativ | Exot | Nativ | Exot | Nativ | Exot | Nativ | Exot | ||
| Total | (380; 217) | 20 % ** | 71 % | 31 % ** | 63 % | 10 % ** | 55 % | 12 % ** | 39 % |
| Site B Pasp+MG | (106; 53) | 44 % ** | 96 % | 59 % ** | 86 % | 13% ** | 75% | 14 % ** | 67 % |
| Site E Tully | (103; 61) | 19 % ** | 79 % | 31 % ** | 85 % | 8 % ** | 54 % | 8 % * | 55 % |
| Site P Tully | (96; 73) | 14 % | 40 % | 9 % | 32 % | 5 % | 33 % | 11 % | 15 % |
| Site S No Exotics | (75; 30) | 6 % | na | 23 % | na | 19 % | na | 7 % | na |
P<0.01 and
P<0.05 for difference of prevalence between native and exotic grasses.
B.ps = B. pseudomallei; MG=Mission Grass; Pasp=Paspalum; Nativ=native grasses; Exot=exotic grasses
There was strong evidence that more root and foliage samples of exotic grasses were B. pseudomallei positive (47% of screened exotic grasses; 43/91) than native grasses (10%; 13/126)(Fisher’s Exact, P<0.001). Soil associated with native grasses (mostly annuals) showed a higher B. pseudomallei detection rate in the wet (32% (34/107)) as opposed to the dry season (20% (31/157), Fisher’s Exact, P=0.030). No such trend was evident for the soil associated with exotic grasses (mostly perennials) with year-round high B. pseudomallei prevalence, 71% (40/56) in the dry and 62% (37/60) in the wet season.
A multivariable logistic regression analysis clustered for sites was performed to identify environmental factors significantly contributing to the occurrence of B. pseudomallei in soil at these sites (see Table 2). For the dry season, exotic grasses were the most dominant predictor for presence of B. pseudomallei followed by moist to wet soil and a less acidic soil pH (pH at B. pseudomallei positive sites in the dry season: 95 % confidence interval 5.1–5.6; at negative sites 5.0–5.3). In the wet season, these sites were partially inundated and significant predictors for presence of B. pseudomallei in the soil were road drainage, exotic grasses and the soil colours reddish grey and reddish yellow. Reddish grey and yellow soil colours have previously been associated with the presence of B. pseudomallei (Kaestli et al., 2007; Thomas et al., 1979). These soil colours are commonly found in the Top End of Australia, often indicating imperfect drainage and oxidized iron. Indeed, this was also confirmed in our study by the redoxic hydrosol soil profiles of these sites with a ferricrete layer at site E (see Supplement Table 1). Sandy clay loam topsoil of site E was deposited from upland red massive earths which have formed under alternating tropical wet and dry conditions with weathering and leaching of mainly siliceous parent materials and segregation of iron and manganese oxides (Stace et al., 2011). Topsoil (n=6) of site E also had a high reactive iron content with a mean above 3000 mg/kg (see Supplement Table 2).
Table 2.
(A) Unadjusted and (B) Multivariable logistic regression analysis (odds ratio OR (95% CI) P value) of environmental factors contributing at 5 % significance level to the presence of B. pseudomallei in soil at the four field sites in the dry and wet season. Italics in unadjusted models indicate non-significant odds ratios. The following environmental factors were included in the initial model: presence of introduced grass, presence of native spear grass such as Sorghum spp., soil pH, soil water status, soil colour, sampling round. Soil texture was consistent both within and between sites and was not included in the analysis (see text and Supplement Table 1). Road drainage was included as a confounder covariate. Controlling for road drainage, the Mantel-Haenszel estimate of the odds ratio of presence of B. pseudomallei associated with exotic grasses in the dry season was 4.7 (1.8–12.3) P<0.001. The regression analysis was clustered for sites and the final multivariable models with significant predictor variables were specified correctly as tested by a linktest and the fit of models was declared good (Pearson goodness-of-fit, P>0.36).
| Dry Season(n=213) | Wet Season(n=166) | |||
|---|---|---|---|---|
|
|
|
|||
| Unadjusted Model | Multivariable Model | Unadjusted Model | Multivariable Model | |
|
|
|
|||
| Exotic Grass |
10.2 (2.8–37.5) P=0.001 |
11.7 (3.0–44.7) P<0.001 |
3.5 (1.5–8.0) P=0.004 |
1.8 (1.1–2.8) P=0.013 |
|
|
|
|||
| Moist to Wet Soil |
5.8 (1.2–28.1) P=0.028 |
7.0 (2.2–22.6) P=0.001 |
1.2 (0.3–4.3) P=0.779 |
|
|
|
|
|||
| Road Drainage |
8.3 (3.8–18.1) P<0.001 |
4.7 (1.6–13.8) P=0.005 |
3.3 (1.2–8.8) P=0.019 |
|
|
|
|
|||
| Soil pH |
1.3 (0.9–2.0) P=0.128 |
1.5 (1.2–1.9) P=0.002 |
1.0 (0.7–1.4) P=0.838 |
|
|
|
|
|||
| Soil Color Reddish Grey |
0.5 (0.1–3.6) P=0.531 |
1.8 (0.7–4.6) P=0.242 |
2.6 (1.1–5.8) P=0.025 |
|
|
|
|
|||
| Soil Color Reddish Yellow |
2.3 (0.6–8.7) P=0.237 |
2.6 (1.3–5.0) P=0.005 |
1.3 (1.1–1.4) P<0.001 |
|
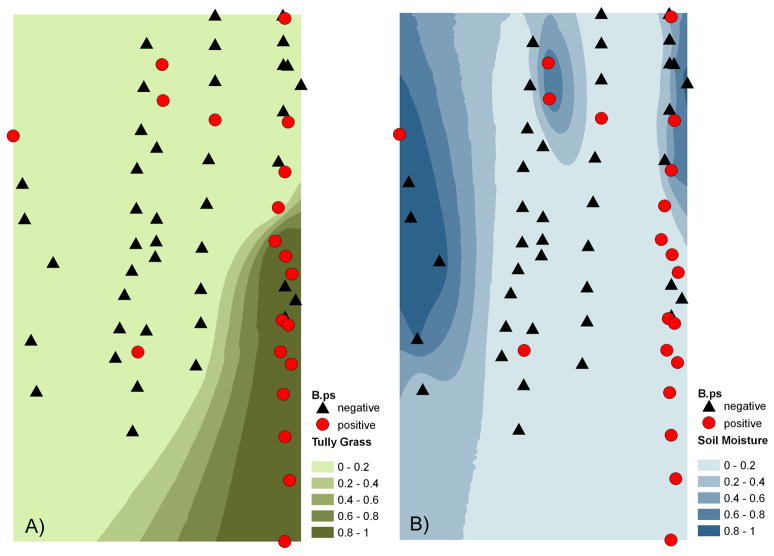
We found the occurrence of B. pseudomallei in soil to be clustered at sites with exotic grasses; at site E there was a significant positive spatial autocorrelation in the 0–0.0005 degree (0–50 meters) band (Geary’s c 0.67–0.88, P<0.001) which matched the patchy distribution of Tully Grass which also showed a significant positive spatial correlation (Geary’s c 0.17–0.65, P<0.001). This clustering was also visualized using indicator kriging to map Tully Grass as well as soil moisture against B. pseudomallei positive and negative holes (see Figure 1).
Figure 1.
At field site E which was rich in exotic Tully Grass, soil samples positive for B. pseudomallei are shown together with clusters of Tully Grass (A) or moist soil (B). The circles indicate the results of soil screening for presence of B. pseudomallei over two consecutive dry seasons. A) The cluster of Tully Grass along the southeast edge is shown by a map based on Indicator Kriging which predicts the probability that the threshold value 0 for no presence of Tully Grass is exceeded. B) Probability map for moist to wet soil (approx. >200 mV or >4% vol soil water content).
B. pseudomallei culture isolates from these field sites were subject to molecular typing, namely multilocus sequence typing (MLST) (Godoy et al., 2003). Sequence types (STs) commonly retrieved had previously been isolated from clinical cases and environmental sources in rural Darwin (e.g. ST 109, 144, 266).
FISH/CLSM and real-time PCR studies show colonization of B. pseudomallei of roots and aerial parts of grasses
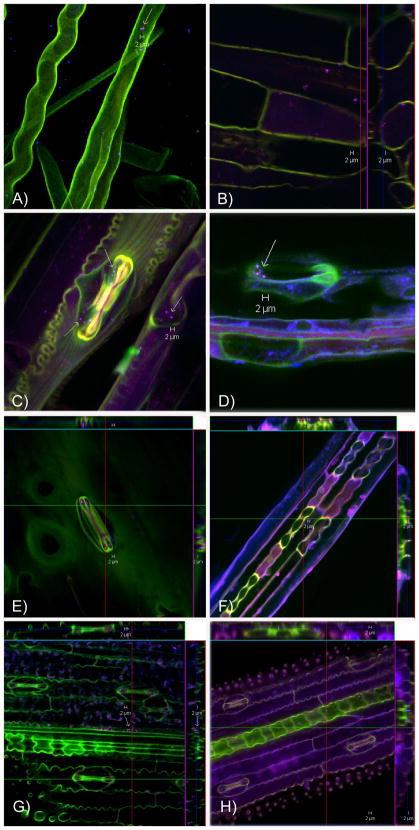
FISH and confocal laser-scanning microscopy (CLSM) analysis were performed to determine the colonization behavior of B. pseudomallei in roots and foliage of grasses collected from the field sites as well as from inoculated plant cultures (see Figure 2). All roots and foliage positive for B. pseudomallei by FISH analysis were also found to be positive for B. pseudomallei by B. pseudomallei specific TTS1 real-time PCR (Kaestli et al., 2007). Microscopy showed that B. pseudomallei cells were smaller in size in grass samples when compared to B. pseudomallei in culture, with an approximate size of 1–2 μm. B. pseudomallei was detected in the rhizosphere of all grass species examined (Tully, Paspalum, Mission Grass collected from field sites as well as inoculated Sorghum intrans and Oryza rufipogon (Wild Rice)). While the bacteria often clustered, they were mainly found around root hairs and in junctions of root branches. Less frequently, B. pseudomallei was also detected inside roots such as of Mission or Tully Grass from field sites, i.e. inside root hairs and root cortex cells as well as in between the cortex cells. A small number of B. pseudomallei were found in the xylem of a field collected Paspalum from field site B. In the foliage of grasses (namely Tully Grass, Paspalum and Mission Grass from field sites), B. pseudomallei were found around the stomata, at the surface of these openings as well as within the stomatal guard cells. In Wild Rice where the rhizosphere was inoculated with B. pseudomallei, the bacterium was detected along the vascular bundles of the foliage, at the surface of the lower side of the leaf in a gully-like structure formed by the vascular bundles.
Figure 2.
FISH and confocal laser-scanning microscopy on roots and leaves using a validated set of fluorescently labeled oligonucleotides targeting the 16S rRNA of B. pseudomallei (red); β-Proteobacteria (blue) as positive control and the probe mix non-EUB-338-I, II, III (green) as negative control for nonspecific probe or dye binding. The composite pictures are shown with B. pseudomallei cells in magenta (combination of red and blue). No unspecific green signals were evident for B. pseudomallei signals (see Supplement Figure 3). The orthogonal views (B, E, F, G, H) depict the intra- or extracellular location of B. pseudomallei by showing an internal of three dimensional z-stacks. A) Two B. pseudomallei are seen within a root hair (inside location confirmed by orthogonal view – see Supplement Figure 3) and further β-Proteobacteria among root hairs of Tully Grass whose rhizosphere was inoculated with B. pseudomallei. B) B. pseudomallei in the rhizosphere and inside root cells of Mission Grass from a highly positive field site. C) Various B. pseudomallei and other β-Proteobacteria are shown close to and within stomatal guard cells of a Mission Grass leaf of a highly positive field site. D) Three B. pseudomallei outside of a stomatal opening of a leaf of an introduced pasture grass (Digitaria milanjiana cultivar Jarra) whose rhizosphere was inoculated with B. pseudomallei. E) B. pseudomallei in stomatal guard cells and on the surface of a stomata of a Paspalum leaf collected at field site B. F)–H) B. pseudomallei are seen along vascular bundles of leaves of grasses whose rhizosphere was inoculated with B. pseudomallei. They are at the surface of the lower side of the leaf in a gully-like structure created by the vascular bundles F) Leaf of Sorghum intrans. G)–H) Leaves of Wild Rice.
B. pseudomallei colonization exerts a pathogenic effect upon exotic Mission Grass in contrast to native Wild Rice
140 seedlings of Mission Grass, Tully Grass and Wild Rice were inoculated with no, low or a high dose of B. pseudomallei (see Supplement Figure 1). After four weeks, there was evidence that B. pseudomallei had a pathogenic effect upon Mission Grass. Seedling mortality increased proportionally from the negative to the low and high dose inoculation group (4% vs. 21% vs. 37% dead seedlings; n=62; Fisher’s Exact, P=0.033). Furthermore, a high inoculation dose was a predictor for death of Mission Grass seedlings with an unadjusted Odds Ratio of 13.4 (95% CI 1.5–122.1, P=0.021). There was also a trend of reduced foliage in Mission Grass seedlings which were inoculated at a high dose and alive after 4 week; however, this trend was not statistically significant and may reflect low numbers (median growth 23.5 cm in negative and low dose, 95% CI 13–38 (n=38) as opposed to 4.5 cm, 95% CI 0–42.7 cm for high dose (n=12), Mann-Whitney, P=0.16). Although not statistically significant, a higher proportion of dead Tully Grass was observed in the high inoculation group as compared to the low inoculation and negative groups (27% dead plants (4/15) vs 9% (3/32), P=0.188) and a decreased leaf growth was found (see Supplement Figure 1).
No decrease in foliage or root growth was evident in each of the three grass species when inoculated with a low amount of B. pseudomallei.
B. pseudomallei did not exert any obvious negative effects on Wild Rice. There was no reduction in foliage, root growth or seedling mortality upon increased dose of inoculation. Interestingly, there was evidence for increased root growth in the high inoculation dose (median growth 4 cm in negative and low dose, 95% CI 3.4–7.6 (n=13) as opposed to 8 cm, 95% CI 6.3–9.7 cm for high dose (n=7), Mann-Whitney, P=0.038).
Semi-quantitative analysis of Burkholderia and B. pseudomallei load in grasses
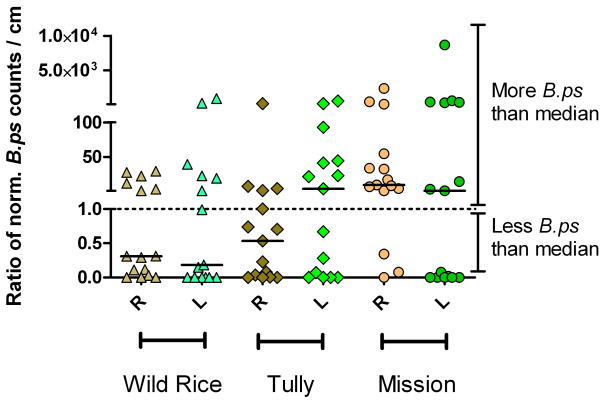
Semi-quantitative analysis of B. pseudomallei load was carried out in 90 root and aerial samples of plants from the above plant growth and time series experiments (see Figure 3 and Supplement Figure 2) restricting the analysis to those plants which were inoculated at least three days prior to harvesting. There was evidence for the median B. pseudomallei load being more than 100 times higher in Mission Grass roots as compared to Wild Rice roots (bootstrap estimate 95% CI 18–205, Mann-Whitney Test, P=0.009)(see Figure 3). A positive correlation was found between B. pseudomallei counts in roots and leaves (Spearman’s rho 0.44, P=0.008).
Figure 3.
Semi-quantitative analysis of B. pseudomallei load from day three after inoculation in 90 root or leaf samples with 15 samples per group. Samples were of the time series and plant growth comparison experiments. Load comparison was by comparison of ratios with the denominator being the median growth in all root or leaf samples, respectively. B. pseudomallei specific TTS1 counts were normalized with an internal plasmid control adjusting for differences in DNA extraction and PCR efficiency. R=roots; L=leaves.
A comparison of semi-quantitative load of bacteria of the genus Burkholderia in grasses of the plant growth experiment revealed evidence for 19 times less Burkholderia in Mission Grass leaves than in leaves of Tully Grass (median bootstrap estimate 95% CI 1.3–37.6, Mann-Whitney Test, P=0.010) or 7 times less than in leaves of Wild Rice (95% CI −16.8–31.8, P=0.055) though the latter was not statistically significant.
Discussion
To date, the literature has mainly suggested soil and surface water as the natural habitat of B. pseudomallei (Cheng et al., 2005). To our knowledge, only two reports showed an association with the rhizosphere of plants (Inglis et al., 2000; Levy et al., 2003). Using real-time PCR and FISH, we found B. pseudomallei not only in the rhizosphere of various grasses in the Top End of Australia but also endophytically, colonizing both roots and aerial parts.
The rhizosphere and particularly areas around root hairs and lateral root emergence are rich in root exudates and nutrients and an ideal habitat for a saprophytic bacterium such as B. pseudomallei. This prime ecological niche was described as a “playground and battlefield” for microbes (Raaijmakers et al., 2009). The fierce competition among the microbial community results in selection pressure for developing traits such as improved iron scavenging by siderophore production (Alice et al., 2006; Lemanceau et al., 2009), exhibiting antagonistic activities against other microbes and optimizing survival mechanisms in the eukaryotic host, be it a plant or root-associated mycorrhizal fungi (Berg et al., 2005; Levy et al., 2003; Raaijmakers et al., 2009). Some of these traits are likely to also aid in successful colonization of human tissues and organs and thus, the rhizosphere is a known source of versatile human opportunistic pathogens such as bacteria of the B. cepacia complex (Berg et al., 2005; Fiore et al., 2001). Other bacteria with human-pathogenic traits and previously associated with plants include common vegetable contaminants such as bacteria of the Enterobacteriaceae family, Listeria spp. and Pseudomonas spp. (Schwaiger et al., 2011)
FISH microscopy also suggested the presence of B. pseudomallei inside and in between root cortex cells. Entry of B. pseudomallei into roots might be similar to that of some rhizobial bacteria and occur via natural openings or wounds in the epidermis such as caused by lateral root emergence (Roberts K ed., 2007). A few B. pseudomallei were also found in the xylem, which transports dissolved mineral nutrients and water from the roots into the aerial parts of the plant.
We detected various B. pseudomallei at the surface of and inside stomata and veins of leaves. It is unclear whether the bacteria invaded the leaf internal tissue via roots, xylem and vascular bundles and/or via stomata after leaf surface contamination by B. pseudomallei contaminated water droplets from wind/rain, or, in the plant culture experiments by condensation in the culture container. Stomatal openings promote gas exchange and transpiration. They are a common entry point for bacteria into leaves and some plants have developed molecular defense mechanisms such as bacterial-signal induced stomatal closure (Melotto et al., 2008). Reduced stomatal size has also been associated with resistance to disease (Riikonen et al., 2008). We indeed found the stomata of more resistant Wild Rice to be smaller than the ones of Mission Grass. However, this needs formal study.
Leaves of perennial grasses might also serve as a “refuge” for B. pseudomallei in the dry season when surface soil moisture is commonly too low for growth. We found a significantly higher proportion of leaves to be positive for B. pseudomallei in the dry as opposed to the wet season. Stomata provide a protected hydrophilic environment and bacteria of the genus Burkholderia have previously been found inside stomatal chambers (Compant et al., 2008). We also detected B. pseudomallei in natural pockets along vascular leaf bundles which are known to be a protected niche for bacterial epiphytic survival (Melotto et al., 2008).
The fact that B. pseudomallei colonizes aerial parts of grasses raises the question whether this also facilitates spread of these bacteria by animals traversing grass fields and/or by oral-fecal transmission by animals such as grazing macropods which feed on grasses. Mission Grass is a preferred diet of wallabies (Phil Hickey, pers. comm.) and we have previously found a significant association of B. pseudomallei occurrence with sites containing animal waste including of macropods (Kaestli et al., 2009) and we have detected B. pseudomallei in macropod droppings in rural Darwin by molecular detection as described in (Kaestli et al., 2007).
In a survey on B. pseudomallei occurrence in rural Darwin (Kaestli et al., 2009), we found that anthropogenic manipulations of the environment such as irrigation or keeping animals have an impact upon B. pseudomallei occurrence. We also observed that some areas with seemingly less signs of anthropogenic environmental disturbance but with repeatedly high counts of B. pseudomallei showed a dense grass growth, even in the dry season. Work described in this report provides evidence that the major contributor to the high B. pseudomallei load at these less disturbed sites are introduced grass species. Given that much of northern Australia is free from other anthropogenic environmental disturbances such as construction and farm animals, the magnitude of contribution to overall B. pseudomallei load from imported grasses in these sites is likely to be substantial. These grasses were introduced to Australia in the 20th century as pasture grass, including Tully Grass, Mission Grass, both native to Africa and Paspalum, native to the Americas. Melioidosis case reports from these continents are rare (Currie et al., 2008) and B. pseudomallei might only occur in localized foci in these regions. Upon introduction of these grasses to Australia, they have since spread to non-targeted areas and Mission Grass has been declared a weed (Walden et al., 2008). We chose four field sites with a varying degree of exotic grasses and using a grid-like sampling approach (Limmathurotsakul et al., 2010b) and multivariable analysis, exotic grasses were indeed the most dominant predictor for presence of B. pseudomallei in the dry season. Invasive plants are known to change the microbial community of the soil and rhizosphere (Batten et al., 2006; Kourtev et al., 2002) such as by plant exudates or by impacting upon the soil composition through changing soil biomass, nutrient status, pH or moisture (Ehrenfeld et al., 2001). Various negative or positive feedback loops between invasive plants and soil microbiota have been shown (Callaway et al., 2004; Reinhart et al., 2006). Invasive grass species in Northern Australia increase biomass and persistence of annual soil wetting (Wurm et al., 2006) and impact upon soil nitrogen cycles (Rossiter et al., 2006) and thus, might also indirectly influence the occurrence of B. pseudomallei. Moreover, in contrast to many native grasses which only grow during the wet season, the perennial nature of Paspalum, Tully and Mission Grass allows B. pseudomallei a year-round, stable habitat which results in a high prevalence of these bacteria in the dry season when B. pseudomallei occurrence is otherwise usually low (Thomas et al., 1979).
Differences in plant immunity as well as microbial competition between established native and introduced plants might also influence the level of colonization success by B. pseudomallei. In plant inoculation experiments using molecular semi-quantification, we found that Wild Rice which is native to melioidosis-endemic northern Australia harbored less B. pseudomallei per cm leaf or root compared with Mission Grass. The opposite was evident when comparing the load of bacteria of the whole genus Burkholderia, with more Burkholderia detected in Wild Rice than in Mission Grass. This was also evident when culturing bacteria from these plants using selective Ashdown’s media. The most dominant species cultured from Mission Grass was B. pseudomallei while primarily other bacteria such as those of the B. cepacia complex were recovered from Wild Rice.
Moreover, we consistently found B. pseudomallei recovered from Mission Grass roots or leaves to be of common rough colony morphology type I (Chantratita et al., 2007) which corresponded to the inoculation type. This was in stark contrast to Wild Rice, from which B. pseudomallei was mainly retrieved as smooth colony morphology type III. This smooth type was previously associated with an environmental stress response and increased flagella expression which was suggested to assist the bacteria in moving towards more favorable conditions (Chantratita et al., 2007; Tandhavanant et al., 2010).
These observations suggest that B. pseudomallei encounters a reduced level of microbial competition and likely, reduced specific plant defense mechanisms in exotic Mission Grass as opposed to in native Wild Rice. Our findings of increased resistance of Wild Rice to B. pseudomallei colonization match results by Lee et al. (Lee et al., 2010) who showed that rice plants were resistant to B. pseudomallei infection. B. pseudomallei is now thought to have originated within Australia (Pearson et al., 2009) and Wild Rice and B. pseudomallei possibly shared the same habitat for many millennia, suggesting co-evolution resulting in balanced bacterial-host interactions after the likely development of some plant immunity against these opportunistic pathogens.
While high loads of B. pseudomallei exerted a pathogenic effect upon Tully or Mission Grass in plant inoculation experiments, plants of these species showed no signs of disease in situ at the field sites despite being colonized by B. pseudomallei. In native wild rice, there was even evidence for increased root growth upon high dose inoculation of B. pseudomallei in plant inoculation experiments. The ecological role of B. pseudomallei in plants needs more formal study; we did not detect noteworthy production of indole derivatives such as auxins by B. pseudomallei and neither any cellulose degradation (see supplement). While auxin is a plant hormone often produced by plant associated bacteria and found in both, plant growth promoting bacteria and plant pathogens (Patten et al., 1996), the ability to degrade cellulose is common to plant pathogens (Compant et al., 2008).
More studies are needed to explore future implications of our findings of a preferential colonisation of some introduced grass species by B. pseudomallei in a melioidosis endemic area. As these grass species are now commonly found in rural areas in Northern Australia, an increased risk of exposure to B. pseudomallei for the rural population is possible as a consequence of this spreading. In contrast, while it is to date unknown what interactions occur between B. pseudomallei and wild rice in situ, due to the main distributions of wild rice being in remote wetlands, such interactions do not currently directly impact upon the risk of exposure to the human population. In terms of global implications and following potential dispersal of B. pseudomallei to a non-endemic area, B. pseudomallei survival in a new environment not only depends on local climate, physico-chemical soil conditions, soil disturbance and type of vegetation but as indicated by antibiosis studies (Lin et al., 2011; Marshall et al., 2010) and our data, also on the level of competition from the local established microbial community. The ongoing worldwide spread and use of these grasses for pasture might be contributing to global dispersal of B. pseudomallei. Nevertheless, although these grass species now occur worldwide it remains to be seen whether the association with B. pseudomallei found in northern Australia is also evident in other parts of the world. The confirmation of colonization by B. pseudomallei of invasive grasses beyond Australia would assist in predicting the distribution of B. pseudomallei in the environment globally.
To our knowledge, this is the first report on the widespread occurrence of B. pseudomallei in grasses in a melioidosis endemic area. Our data indicate that B. pseudomallei readily colonizes the rhizosphere of various grasses and is also an endophyte – likely of opportunistic nature - profiting from a protected, hydrophilic and nutrient-rich environment. A habitat in aerial parts of grasses also raises questions about potential dispersal mechanisms involving grazing animals. The ongoing spread of some invasive grass species both within Australia and globally contributes to changes in landscape ecology with new habitats suitable for B. pseudomallei survival, with public health implications from exposure of populations to this potentially deadly bacterium.
Experimental Procedures
Field studies and description of field sites and soils
Four field study sites were chosen in rural Darwin (12°S) in the tropical Top End of the Northern Territory of Australia. This location has the highest reported incidence of melioidosis in the world (Currie et al., 2010; Limmathurotsakul et al., 2010a). These field sites were previously found to be repeatedly positive for B. pseudomallei and were in riparian zones with a stream marking one side, a drain of a road another side and open woodland and open forest the other two sides. The vegetation of the sites consisted of an open forest of Melaleuca (Paperbarks), Eucalyptus and Pandanus species with a grass understory of native (such as Sorghum spp. or Whiteochloa spp) and at three sites, introduced grasses (mainly Brachiaria humidicola cv Tully (Tully Grass), Pennisetum pedicellatum and polystachion (Mission Grass) and Paspalum plicatulum (Paspalum)). Following the “Australian Soil and Land Survey” Field Handbook (McDonald et al., 1998), soil profiles were classified as redoxic Hydrosols for sites B, P and S and as Anthroposols (in a Redoxic Hydrosol environment) for site E as according to the Australian Soil Classification (Isbell, 2002)(see Supplement Table 1). The topsoil of these sites was generally consistent both within and between sites and consisted of sandy loam to sandy clay, with the predominance of sandy loam followed by sandy clay loam. Sites B and E also had a layer of peat indicating decomposition of plant remains but the organic carbon content of the topsoil was low with a mean of 1.1 % (see Supplement Table 2). An area covered approximately 100 by 60 meters. Per area and time point, 30 samples at a depth of 30 cm were collected using a grid-like sampling along three transects. Each transect was approx. 20 metres apart and with 10 samples collected from each at 10 meter intervals. Soil sampling was as described previously (Kaestli et al., 2009). If grasses were present at the soil sampling points, these were collected in plastic bags and processed within 24 hours. Various abiotic factors were recorded such as soil pH (soil pH field kit, Inoculo, Australia), presence of water logging; distance to closest stream and road drainage, soil water status and soil texture following the “Australian Soil and Land Survey” Field Handbook (McDonald et al., 1998) and a common soil texture flowchart (http://www.h2ou.com/h2twss96.htm) as well as soil color using the Munsell Soil Color Chart (ed. 2009). Using a soil moisture field meter (MPM160 meter, ICT International Pty Ltd, Australia) on a selection of soil samples “moist to wet soil” corresponded to approx. >200 mV or >4% vol soil water content. Soil chemistry analysis was performed on 18 key soil samples collected in the dry season from sites B, E and P (CSBP Soil and Plant Analysis Laboratory, Bibra Lake, Australia)(see Supplement Table 2).
Sampling started in the dry season, in September 2009 and was repeated three times at 6 month intervals, this covered two consecutive dry seasons and wet seasons. In the wet season, the sampling points closest to the stream at each site were inaccessible due to flooding. In summary, 390 samples were collected from four sites at four time points over 1.5 years.
Inoculation of grasses with B. pseudomallei
Seeds of Pennisetum pedicellatum were collected from seeding Annual Mission Grass in rural Darwin. Seeds of Sorghum intrans and Oryza rufipogon were obtained from Greening Australia and of Brachiaria humidicola cv Tully from Southedge Seeds (Australia). No B. pseudomallei were detected in these seeds by direct B. pseudomallei molecular detection (Kaestli et al., 2007). The seeds were germinated on moist sterile cotton or in commercially available potting mix. Once germinated, the seedlings were inoculated with B. pseudomallei by soaking only the roots in 2×10e7 CFU B. pseudomallei/mL sterile water for one hour after which seedlings were transferred into soil with a soil moisture content of 15 %. The soil represented a soil type common to the Top End, namely sandy clay loam collected from rural Darwin and negative for B. pseudomallei by culture and direct molecular detection (Kaestli et al., 2007). The seedlings were grown in closed containers (phytatrays, Sigma-Aldrich Australia) under fluorescent light at 35°C for 12 hours and in darkness for 12 hours at 30°C. Inoculation experiments were conducted with B. pseudomallei strain MSHR2817 which was isolated from the rhizosphere of Tully Grass in rural Darwin (ST 144). B. pseudomallei strains with ST144 were previously isolated from environmental and clinical sources in the Darwin area.
Comparison of plant growth upon inoculation with B. pseudomallei
Seedlings of Tully Grass and annual Mission Grass as well as Wild Rice were divided into three groups; these are negative controls without B. pseudomallei inoculation, low (2×10e4 CFU B. pseudomallei/mL sterile water) and high (2×10e7 CFU B. pseudomallei/mL sterile water) inoculum. For Tully Grass, these groups contained 17, 15 and 15 seedlings (for negative control, low and high inocula), for Mission Grass 24, 19 and 19 and Wild Rice 9, 12 and 10 seedlings. Seedlings were grown in sandy clay loam with a soil moisture content of 15 %. Length of roots and leaves of every plant was recorded before and four weeks after inoculation as well as the appearance of the plant. There was no significant difference between the length of roots or leaves in the different groups at start of experiment. Four weeks after inoculation, 3 plants were combined per group and Burkholderia and B. pseudomallei load was measured in surface sterilized roots and leaves.
Surface sterilization of leaves and roots
Leaves and roots were detached from the plants, washed in sterile water and surface sterilized by 3 min incubation in 70 % ethanol followed by 1 min in 1 % commercial bleach containing 0.01 % Tween and three washing steps in sterile water (Compant et al., 2008). Water of the initial washing step before sterilization and the last washing step after sterilization was cultured on Ashdown’s plates to check for bacterial growth (Ashdown, 1979).
Surface sterilization of leaves was successful with no B. pseudomallei colonies detectable in the last washing step while root surface sterilization was successful or resulted in a 10-to 100-fold reduction of B. pseudomallei colonies.
Soil and plant DNA extraction
Soil DNA extraction was done as previously described (Kaestli et al., 2009). Briefly, 20 g of soil was incubated with 20 mL of Ashdown’s Broth for 39 hours shaking at 37°C, the soil supernatant was centrifuged twice and the pellet processed using the PowerSoil Kit (MoBio Laboratories, USA). Modifications included the addition of 0.8 mg of aurintricarboxylic acid (ATA) and 20 μL of proteinase K (20 mg/mL).
Leaves and roots from field plants were washed with sterile water and 70 % ethanol, cut into fine pieces and processed as above. Leaves and roots from culture plants inoculated with B. pseudomallei were surface sterilized as described in previous section, cut into fine pieces and incubated overnight in 10 mL of sterile water, shaking at 37°C. The supernatant was processed as above.
Detection of B. pseudomallei DNA by TTS1 real-time PCR
B. pseudomallei DNA was targeted by the well validated B. pseudomallei specific TTS1 real-time PCR as described previously (Kaestli et al., 2007; Novak et al., 2006). The limit of detection of this PCR was 15 fg of genomic DNA of B. pseudomallei isolate MSHR186 and sensitivity and specificity were found to be 100% based upon screening 224 B. pseudomallei and 151 non-B. pseudomallei culture isolates. A subset of soil samples was also cultured (Ashdown et al., 1992) and subject to MLST (Godoy et al., 2003).
Semi-Quantification of B. pseudomallei load
In order to semi-quantify and compare B. pseudomallei load between different leaf or root samples, an internal control plasmid (pT7SsCl) was included into the DNA extraction step to check for differences in DNA extraction efficiency and presence of PCR inhibitors. 2.9 pg of a pT7TS vector with a 1,470 bp insert of a chloride channel of Sarcoptes scabiei (kindly provided by Kate Mounsey and Deborah Holt (Mounsey et al., 2007)) were added to the 24 hours enriched plant pellets prior to DNA extraction. In a multiplex PCR targeting TTS1 and pT7SsCl, primers pT7SsCl_forw (final 80 nM; 5′-GTTTTTGACACCATCGCAAGATCG-3′), pT7SsCl_rev (final 100nM; 5′-GTTGTGGTTGATGAGAGATCAGAGA-3′) and the probe pT7SsCl_probe (final 256 nM; 5′-TCGCTCCCAGACGTTTCAGCAGTAACT-3′ labeled with CAL Fluor Red and BHQ-2 (Biosearch Technologies)) were used targeting a 135 bp fragment of the internal control plasmid pT7SsCl. Primer concentrations for pT7SsCl amplification were determined by primer matrix assays and no interference with the TTS1 amplification was found. pT7SsCl standard curves were linear over a dilution series of 6 logs with R2 of 0.996 and a detection limit of less than 10 plasmid copies. No significant difference was found in recovery rates of pT7SsCl between different grass species nor between roots or aerial parts. TTS1 copy numbers were divided by corresponding pT7SsCl copy numbers to normalize samples for potential differences in extraction efficiency or presence of PCR inhibitors.
Semi-quantification of Burkholderia by recA SYBR assay
Bacteria belonging to the genus Burkholderia were semi-quantified using a SYBR assay based on a previously published PCR targeting 385 bp of the recA gene (Payne et al., 2005). 4 uL of root- and foliage- extracted DNA were added to final 10 uL PCR mix. 1x TaqMan® Environmental Master Mix 2.0 (Applied Biosystems), 1x SYBR Green 1 solution (Quantace), 400 ng/μL non-acetylated BSA and 900 nM concentrations of primers Burk3 and Burk4 were used. PCR conditions in a Applied Biosystems 7900 system were 50°C 2 min, 95 °C 10 min, followed by 45 cycles of 95 °C 30 sec, 48 °C 1 min, 72 °C 1 min and a final extension step of 72 °C for 10 min. Dissociation curve analysis was from 60°C – 95 °C, increasing 2 °C/sec. The assay was validated using a standard curve with a dilution series of genomic DNA of B. pseudomallei strain MSHR305 over 6 logs and a limit of detection of 475 fg DNA was found and a R2 of 0.996.
Fluorescent In Situ Hybridization (FISH) and Confocal Laser Scanning Microscopy(CSLM)
Leaves and roots were dispatched from plants, fixed overnight in 3% paraformaldehyde at 4°C and stored at −20°C in 50% ethanol as described previously (Amann et al., 1990b). For FISH analysis, a mix of three 16S-rRNA targeted probes (Eurofins MWG Operon, Germany) were used, labeled at the 5′ end with the fluorescent dyes Fluorescein (green), Cy3 (red) or Cy5 (blue): the B. pseudomallei specific probe BmBpm-463 (5′-ATCCACTCCGGGTATTAG-3′) [together with the unlabeled competitor Bt-463 specific for B. thailandensis (5P′-ATCCACCCCGGTTATTAG-3′)], BET-42a targeting β-Proteobacteria (Manz et al., 1992) with unlabeled competitor GAM-42a targeting γ-Proteobacteria and as negative control for nonspecific probe or dye binding, the probe Non-EUB-338-I which is reverse complementary to the universal bacterial probe EUB338-I (Amann et al., 1990a). Validation of BmBpm-463 in conjunction with Bt-463 was performed in silico using the tool probe match implemented in the software package ARB (Ludwig e t a l., 2004) (www.arb-home.de) and the SILVA database SSURef_100_SILVA_02_08_09_opt.arb (Pruesse et al., 2007) (www.arb-silva.de) and by performing FISH with cultured and fixed strains using 10 different B. pseudomallei strains of clinical and environmental origin as well as 2 B. thailandensis, 1 B. thailandensis-like (MSMB43), 1 B. cepacia, 1 B. pyrrocinia, 2 Burkholderia cluster A and B spp., 1 Cupriavidus spp. and 1 Ralstonia spp. strain. Sensitivity and specificity were 100%. BmBpm-463 is not able to distinguish between B. pseudomallei and B. mallei 16S rRNA; however, as B. mallei did not occur in our target environment, this did not impact on the study.
Hybridizations were carried out on epoxy resin coated glass slides as described previously (Stoffels et al., 2001). Briefly, roots and leaves were cut into fine pieces, dehydrated in ethanol series, air dried and incubated at 46°C in a hybridization solution containing 35% formamide, 30 ng/μl Cy3- and Cy5- and 50 ng/μl Fluorescein-labeled probes and 30 ng/μl of the respective competitors. Washing and mounting of samples was as per (Stoffels et al., 2001).
For microscopy, a Zeiss LSM-510-META confocal laser-scanning microscope (Zeiss, Germany) was used (Stoffels et al., 2001). An argon ion laser supplied a wavelength of 488 nm to excite fluorescein, and two helium neon lasers provided the wavelengths of 543 nm for Cy3 and 633 nm for Cy5.
Statistical Analysis
Statistical analysis was carried out using Stata (Intercooled Stata, version 8.2, USA). For bivariate analysis, Fisher’s Exact test and Mann-Whitney U test were used. For multivariable analysis, odds ratios were calculated in stepwise multivariable logistic regression analyses clustered by site. The specification of the models was assessed using a link test and model performance by a Pearson goodness-of-fit test. For spatial autocorrelation analysis, Geary’s c statistics were used. All tests were 2-tailed, significant results indicated a P value of <0.01.
Probabilities for exotic grasses and moist soil at field site E were mapped using the geo-statistical interpolation technique “indicator kriging” in ArcGIS9 (ESRI, 2008) with a square grid of 10×10 metres. For the exotic grass data, an anisotropic spherical model was fitted resulting in a semi-variogram with a minor correlation range of 66 metres, a nugget size of 0, a mean prediction error of 0.0034 and average standard error of 0.174. For soil moisture, an anisotropic exponential model was fitted with a minor correlation range of 59 metres, a nugget size of 0.031, a mean prediction error of −0.0006 and average standard error of 0.279.
Supplementary Material
Acknowledgments
We would like to thank Ian Harrington (Menzies School of Health Research), Christopher Mangion and Andrew Owens (Dept of Natural Resources, Environment and the Arts) for assistance in field work. We are thankful to Deborah Holt and Kate Mounsey for kindly providing the plasmid pT7SsCl and to Arthur Cameron for providing Jarra grass seeds. We are grateful to Penny Wurm, Sean Bellairs and Phil Hickey for advice on grass ecology and weeds in the Northern Territory and to anonymous reviewers whose valuable comments significantly improved the manuscript. This work was supported by grants from the Swiss National Science Foundation, the Australian National Health and Medical Research Council (NHMRC), the Human Frontier Science Program and the US National Institutes of Health (NIH).
References
- Alice AF, Lopez CS, Lowe CA, Ledesma MA, Crosa JH. Genetic and transcriptional analysis of the siderophore malleobactin biosynthesis and transport genes in the human pathogen Burkholderia pseudomallei K96243. J Bacteriol. 2006;188:1551–1566. doi: 10.1128/JB.188.4.1551-1566.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Envir Microbiol. 1990a;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990b;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashdown LR. An improved screening technique for isolation of Pseudomonas pseudomallei from clinical specimens. Pathology. 1979;11:293–297. doi: 10.3109/00313027909061954. [DOI] [PubMed] [Google Scholar]
- Ashdown LR, Clarke SG. Evaluation of culture techniques for isolation of Pseudomonas pseudomallei from soil. Appl Environ Microbiol. 1992;58:4011–4015. doi: 10.1128/aem.58.12.4011-4015.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten K, Scow K, Davies K, Harrison S. Two invasive plants alter soil microbial community composition in serpentine grasslands. Biological Invasions. 2006;8:217–230. [Google Scholar]
- Berg G, Eberl L, Hartmann A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environmental Microbiology. 2005;7:1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x. [DOI] [PubMed] [Google Scholar]
- Callaway RM, Thelen GC, Rodriguez A, Holben WE. Soil biota and exotic plant invasion. Nature. 2004;427:731–733. doi: 10.1038/nature02322. [DOI] [PubMed] [Google Scholar]
- Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, Limmathurotsakul D, et al. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol. 2007;189:807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compant S, Kaplan H, Sessitsch A, Nowak J, it Barka E, Clement C. Endophytic colonization of Vitis vinifera L. by Burkholderia phytofirmans strain PsJN: from the rhizosphere to inflorescence tissues. FEMS Microbiology Ecology. 2008;63:84–93. doi: 10.1111/j.1574-6941.2007.00410.x. [DOI] [PubMed] [Google Scholar]
- Currie BJ, Dance DA, Cheng AC. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg. 2008;102(Suppl 1):S1–S4. doi: 10.1016/S0035-9203(08)70002-6. [DOI] [PubMed] [Google Scholar]
- Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin Prospective Study. PLoS Negl Trop Dis. 2010;4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper AD, Mayo M, Harrington G, Karp D, Yinfoo D, Ward L, et al. Association of the melioidosis agent Burkholderia pseudomallei with water parameters in rural water supplies in Northern Australia. Appl Environ Microbiol. 2010;76:5305–5307. doi: 10.1128/AEM.00287-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenfeld JG, Kourtev P, Huang W. Changes in soil functions following invasions of exotic understory plants in deciduous forests. Ecological Applications. 2001;11:1287–1300. [Google Scholar]
- Fiore A, Laevens S, Bevivino A, Dalmastri C, Tabacchioni S, Vandamme P, et al. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environmental Microbiology. 2001;3:137–143. doi: 10.1046/j.1462-2920.2001.00175.x. [DOI] [PubMed] [Google Scholar]
- French CT, Toesca IJ, Wu TH, Teslaa T, Beaty SM, Wong W, et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. PNAS. 2011;108:12095–12100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galyov EE, Brett PJ, DeShazer D. Molecular Insights into Burkholderia pseudomallei and Burkholderia mallei Pathogenesis. Annu Rev Microbiol. 2010;64:495–517. doi: 10.1146/annurev.micro.112408.134030. [DOI] [PubMed] [Google Scholar]
- Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, et al. Multilocus Sequence Typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis TJJ, Garrow SC, Henderson M, Clair A, Sampson J, O’Reilly L, et al. Burkholderia pseudomallei traced to water treatment plant in Australia. Emerging Infectious Diseases. 2000;6:56–59. doi: 10.3201/eid0601.000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis TJJ, Sagripanti JL. Environmental factors that affect the survival and persistence of Burkholderia pseudomallei? Appl Environ Microbiol. 2006;72:6865–6875. doi: 10.1128/AEM.01036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbell R. The Australian Soil Classification. CSIRO PUBLISHING; 2002. [Google Scholar]
- Kaestli M, Mayo M, Harrington G, Ward L, Watt F, Hill JV, et al. Landscape changes influence the occurrence of the melioidosis bacterium Burkholderia pseudomallei in soil in northern Australia. PLoS Negl Trop Dis. 2009;3:e364. doi: 10.1371/journal.pntd.0000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestli M, Mayo M, Harrington G, Watt F, Hill J, Gal D, et al. Sensitive and Specific Molecular Detection of Burkholderia pseudomallei, the Causative Agent of Melioidosis, in the Soil of Tropical Northern Australia. Appl Environ Microbiol. 2007;73:6891–6897. doi: 10.1128/AEM.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtev PS, Ehrenfeld JG, Haeggblom M. Exotic plant species alter the microbial community structure and function of the soil. Ecology. 2002;83:3152–3166. [Google Scholar]
- Lee YH, Chen Y, Ouyang X, Gan YH. Identification of tomato plant as a novel host model for Burkholderia pseudomallei. BMC Microbiol. 2010;10:28. doi: 10.1186/1471-2180-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanceau P, Bauer P, Kraemer S, Briat JF. Iron dynamics in the rhizosphere as a case study for analyzing interactions between soils, plants and microbes. Plant and Soil. 2009;321:513–535. [Google Scholar]
- Levy A, Chang BJ, Abbott LK, Kuo J, Harnett G, Inglis TJJ. Invasion of Spores of the Arbuscular Mycorrhizal Fungus Gigaspora decipiens by Burkholderia spp. Appl Envir Microbiol. 2003;69:6250–6256. doi: 10.1128/AEM.69.10.6250-6256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, Chetchotisakd P, et al. Increasing incidence of human melioidosis in Northeast Thailand. Am J Trop Med Hyg. 2010a;82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limmathurotsakul D, Wuthiekanun V, Chantratita N, Wongsuvan G, Amornchai P, Day NPJ, et al. Burkholderia pseudomallei is spatially distributed in soil in Northeast Thailand. PLoS Negl Trop Dis. 2010b;4:e694. doi: 10.1371/journal.pntd.0000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Chen YS, Li YC, Tseng IL, Hsieh TH, Buu LM, et al. Burkholderia multivorans acts as an antagonist against the growth of Burkholderia pseudomallei in soil. Microbiology and Immunology. 2011;55:616–624. doi: 10.1111/j.1348-0421.2011.00365.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Research. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz W, Amann RI, Ludwig W, Wagner M, Schleifer KH. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Systematic and Applied Microbiology. 1992;25:593–600. [Google Scholar]
- Marshall K, Shakya S, Greenhill AR, Padill G, Baker A, Warner JM. Antibiosis of Burkholderia ubonensis againist Burkholderia pseudomallei, the causative agent for melioidosis. Southeast Asian J Trop Med Public Health. 2010;41:904–912. [PubMed] [Google Scholar]
- Mayo M, Kaestli M, Harrington G, Cheng AC, Ward L, Karp D, et al. Burkholderia pseudomallei in un-chlorinated domestic bore water in Tropical Northern Australia; distribution, diversity and relationships to human melioidosis cases. Emerg Infect Dis. 2011;17 doi: 10.3201/eid1707.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald R, Isbell RF, Speight JG, Walker J, Hopkins MS. Australian Soil and Land Survey Field Handbook. 2. Dept of Primary Industries and Energy and CSIRO Australia; 1998. [Google Scholar]
- Melotto M, Underwood W, He SY. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu Rev Phytopathol. 2008;46:101–122. doi: 10.1146/annurev.phyto.121107.104959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounsey KE, Dent JA, Holt DC, McCarthy J, Currie BJ, Walton SF. Molecular characterisation of a pH-gated chloride channel from Sarcoptes scabiei. Invertebrate Neuroscience. 2007;7:149–156. doi: 10.1007/s10158-007-0050-6. [DOI] [PubMed] [Google Scholar]
- Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, et al. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J Clin Microbiol. 2006;44:85–90. doi: 10.1128/JCM.44.1.85-90.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palasatien S, Lertsirivorakul R, Royros P, Wongratanacheewin S, Sermswan RW. Soil physicochemical properties related to the presence of Burkholderia pseudomallei. Trans RSoc Trop Med Hyg. 2008;102(Suppl 1):S5–S9. doi: 10.1016/S0035-9203(08)70003-8. [DOI] [PubMed] [Google Scholar]
- Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Can J Microbiol. 1996;42:207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- Payne GW, Vandamme P, Morgan SH, LiPuma JJ, Coenye T, Weightman AJ, et al. Development of a recA Gene-Based Identification Approach for the Entire Burkholderia Genus. Appl Envir Microbiol. 2005;71:3917–3927. doi: 10.1128/AEM.71.7.3917-3927.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 2009;7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Research. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J, Paulitz T, Steinberg C, Alabouvette C, Moenne-Loccoz Y. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant and Soil. 2009;321:341–361. [Google Scholar]
- Reinhart KO, Callaway RM. Soil biota and invasive plants. New Phytologist. 2006;170:445–457. doi: 10.1111/j.1469-8137.2006.01715.x. [DOI] [PubMed] [Google Scholar]
- Riikonen J, Syrjaelae L, Tulva I, Maend P, Oksanen E, Poteri M, et al. Stomatal characteristics and infection biology of Pyrenopeziza betulicola in Betula pendula trees grown under elevated CO2 and O3. Environmental Pollution. 2008;156:536–543. doi: 10.1016/j.envpol.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Roberts K, editor. Handbook of Plant Science. John Wiley & Sons Ltd; 2007. [Google Scholar]
- Rossiter N, Setterfield S, Douglas M, Hutley L, Cook G. The impact of exotic grass invasions on nitrogen cycling: a mini-review. In: Preston C, Watts JH, Crossman ND, editors. Managing Weeds in a Changing Climate; Proceedings of the 15th Australian Weeds Conference; Adelaide, SA: 2006. p. 815. [Google Scholar]
- Rotz LD, Khan AS, Lillibridge SR, Ostroff SM, Hughes JM. Public health assessment of potential biological terrorism agents. Emerg Infect Dis. 2002;8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaiger K, Helmke K, Hoelzel CS, Bauer J. Comparative analysis of the bacterial flora of vegetables collected directly from farms and from supermarkets in Germany. International Journal of Environmental Health Research. 2011 doi: 10.1080/09603123.2010.515672. [DOI] [PubMed] [Google Scholar]
- Stace HCT, Hubble GD, Brewer R, Northcote KH, Sleeman JR, Mulchay MJ, Halsworth EG. A Handbook of Australian soils. Rellim Technical Publications for the Commonwealth Scientific and Industrial Research Organisation and the International Society of Soil Science. 1968. [Google Scholar]
- Stoffels M, Castellanos T, Hartmann A. Design and Application of New 16S rRNA-targeted Oligonucleotide Probes for the Azospirillum-Skermanella-Rhodocista-Cluster. Systematic and Applied Microbiology. 2001;24:83–97. doi: 10.1078/0723-2020-00011. [DOI] [PubMed] [Google Scholar]
- Tandhavanant S, Thanwisai A, Limmathurotsakul D, Korbsrisate S, Day N, Peacock S, et al. Effect of colony morphology variation of Burkholderia pseudomallei on intracellular survival and resistance to antimicrobial environments in human macrophages in vitro. BMC Microbiology. 2010;10:303. doi: 10.1186/1471-2180-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Forbes FJ, Parker M. Isolation of Pseudomonas pseudomallei from clay layers at defined depths. Am J Epidemiol. 1979 doi: 10.1093/oxfordjournals.aje.a112832. [DOI] [PubMed] [Google Scholar]
- Walden D, Gardener M. Weed management in Kakadu National Park. 2008. Invasive species. [Google Scholar]
- White NJ. Melioidosis. Lancet. 2003;361:1715–1722. doi: 10.1016/s0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- Wurm PAS, Bellairs S, Kernich B. Suppression of native wild rice germination by exotic para grass. In: Preston C, Watts JH, Crossman ND, editors. Managing Weeds in a Changing Climate; Proceedings of the 15th Australian Weeds Conference; Adelaide: 2006. [Google Scholar]
- Wuthiekanun V, Smith MD, Dance DA, White NJ. Isolation of Pseudomonas pseudomallei from soil in north-eastern Thailand. Trans R Soc Trop Med Hyg. 1995;89:41–43. doi: 10.1016/0035-9203(95)90651-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.