Abstract
Interactions of microsomal cytochromes P450 (CYPs) with other proteins in the microsomal membrane are important for their function. In addition to their redox partners, CYPs have been reported to interact with other proteins not directly involved in their enzymatic function. In this study, proteins were identified that interact with CYP2C2 in vivo in mouse liver. Flag-tagged CYP2C2 was expressed exogenously in mouse liver and was affinity purified, along with associated proteins which were identified by MS and confirmed by western blotting. Over 20 proteins reproducibly co-purified with CYP2C2. The heterogeneous sedimentation velocity of CYP2C2 and associated proteins by centrifugation in sucrose gradients and sequential immunoprecipitation analysis were consistent with multiple CYP2C2 complexes of differing composition. The abundance of CYPs and other drug metabolizing enzymes and NAD/NADP requiring enzymes associated with CYP2C2 suggest that complexes of these protein may improve enzymatic efficiency or facilitate sequential metabolic steps. Chaperones, which may be important for maintaining CYP function, and reticulons, endoplasmic reticulum proteins that shape the morphology of the endoplasmic reticulum and are potential endoplasmic reticulum retention proteins for CYPs, were also associated with CYP2C2.
Keywords: cytochrome P450, drug metabolizing enzymes, mass spectrometry, protein-protein interactions, reticulons
1 Introduction
Mitochondrial and microsomal cytochromes P450 (CYPs) catalyze the oxidation of a large number of xenobiotics as well as endogenous compounds [1]. Microsomal CYPs are integral membrane proteins that are anchored in the membrane by hydrophobic amino-terminal sequences and interact functionally with other microsomal proteins. CYP reductase (CPR) donates electrons to CYPs during their catalytic cycle [2-4]. While there are multiple forms of CYPs, there is a single CPR. Cytochrome b5 also increases the activities of some CYPs by donating electrons or by allosteric binding [4]. CPR and cytochrome b5, like CYPs are membrane bound ER proteins.
There is substantial evidence that CYPs do not exist as simple monomers in the endoplasmic reticulum (ER) membranes. Homooligomerization of purified CYP3A4 [5], CYP2B4 [6], and CYP1A2 [7] in liposomes and CYP2C2 [8, 9] and CY2C8 [10] and CYP19 and CYP17 [11] in natural membranes has been detected. Heterooligomerization between CYP2C9/CYP3A4 [12], CYP2B4/CYP1A2 [13], CYP2C9/CYP2D6 [14], and CYP2C9/CYP2C19 [15] in reconstituted systems and between CYP1A1/CYP3A4 [16], and CYP2C2/CYP2E1 [8] in mammalian ER membranes has been demonstrated. The presence of one CYP can influence the catalytic characteristics of another CYP [17, 18] so that the physical interactions among CYPs may have functional significance.
CYPs can also interact with other drug metabolism enzymes in the microsomal membrane. Microsomal epoxide hydrolase (mEH) and some forms of UDP-glucurosyltransferase (UGT) have been shown to be associated with CYP1A1 by affinity chromatography [19]. CYP3A2, CYP2B2, CYP2C11/13 and CYP1A2 also co-immunoprecipitated with UGTs [20]. Both activation and suppression of UGT function was observed depending on the specific CYP and UGT that were simultaneously expressed in cultured cells [21]. These observations suggest that the protein-protein interactions between CYPs and other metabolic enzymes may be physiologically relevant, for example to facilitate sequential metabolism of a single substrate by two enzymes.
Interactions with non-metabolic enzymes also may modulate CYP activity or cellular localization. Progesterone receptor membrane component 1 (PGRMC1) has been shown to stimulate the activity of sterol metabolizing CYPs [22] and to inhibit the activity of several drug metabolizing CYPs [23]. B-cell associated protein 31 (BAP31) interacts with CYP2C2 and down regulation of BAP31 results in altered cellular localization of CYP2C2 as well as increased protein levels [24].
Most of the studies described above were done in vitro or involved studying interactions between exogenously expressed proteins in cells, usually nonhepatic cells. The environment of CYPs in vivo in the microsomes of hepatocytes is likely to be different from that in these earlier studies with different concentrations of proteins, different lipid compositions, and the full complement of ER membrane proteins. In this study, we have used a non-biased approach to identify proteins that interact with flag-tagged CYP2C2 in mouse liver in vivo. Flag/CYP2C2 and associated proteins were purified by binding to anti-Flag M2 agarose and the proteins were identified by MS.
2 Materials and Methods
2.1 Materials
Anti-Flag antibody was from Sigma-Aldrich. Antibodies to CYP3A11 and CYP2D1 were from Millipore Corporation, to long chain acyl CoA synthetase (ACSL1) from Cell Signaling, to 11β–hydroxysteroid dehydrogenase (11β-HSD1) from R&D Systems, and to PGRMC1, BAP31, CPR, kinectin 1, mEH and androsterone-UGT from Santa Crutz Biotechnology.
2.2 Cell Culture
Human hepatoma HepG2 (ATCC HB8065), human kidney COS1, and human embryonic cell-derived Ad293 (Cell Biolabs, Inc.) cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 100 units/ml penicillin G-streptomycin sulfate and 10% fetal bovine serum as described [25].
2.3 Generation of Ad2C2/Flag/His Recombinant Adenovirus
CYP2C2 was expressed in mouse liver using recombinant adenovirus [26]. Full length rabbit CYP2C2 cDNA from pC2 [27] was amplified by PCR using forward and reverse primers incorporating Not I and XhoI sites, respectively. The PCR product was ligated with 3XFlag and 6XHis oligonucleotide sequences containing XhoI/XbaI and Xba1/EcoRV sites, respectively, and the combined sequence was inserted into the Adtrack CMV shuttle vector at the EcoRV and NotI sites to form Adtrack-2C2/Flag/His. For construction of the recombinant Ad2C2/Flag/His adenovirus, Escherichia coli BJ5183 cells containing the AdEasy backbone vector were transformed with Ad2C2/Flag/His DNA that had been linearized by PmeI digestion [28]. Ad293 cells were transfected with the resulting recombinant adenoviral DNA, and recombinant adenovirus was then amplified by several rounds of infection. The virus was isolated by CsCl step gradient centrifugation and dialyzed in phosphate-buffered saline-10% glycerol. Total viral particles were determined by absorbance at A260 (1 A260 unit is approximately 1012 particles).
2.4 CYP2C2 activity assay in mammalian cells
Cells were seeded into six-well plates and transfected with AdEmpty or Ad2C2/Flag/His using LipofectAMINE (Invitrogen). Cell culture media was removed 20 h after transfection and replaced with 500 μl serum free media supplemented with 5 mM Luciferin-ME in each well. CYP2C2 activity was assayed with the P450-Glo™ Assay kit (Promega).
2.5 Adenoviral infection and protein expression in mice
AdEmpty virus or AdC2/Flag/His virus (1010 active viral particles) in 200 μl of phosphate buffered saline was injected in the six to eight week old BALB/c male mice mouse via the tail vein. Mice were sacrificed 4 days later and livers were removed for affinity isolation of CYP2C2 and associated proteins. Efficiency of infection was estimated by the expression of green fluorescent protein (GFP) in frozen sections by detection with a Zeiss LSM confocal microscope. Animal experimentation was approved by the Institutional Animal Care and Use Committee of the University of Illinois at Urbana-Champaign.
2.6 Affinity purification of CYP2C2 protein complexes
Livers from mice infected with AdEmpty or Ad2C2/Flag/His were finely minced and washed with 0.15 M NaCl, 0.015 M sodium citrate buffer. After centrifugation at 2,500 x g for 5 min, the cell pellet was resuspended in hypotonic buffer (10 mM HEPES, pH 7.9; 1.5 mM NaCl; 10 mM KCl; 1 mM EDTA; 0.5 mM sucrose; 0.1 mM PMSF and 1X protease inhibitor cocktail) and lysed by 30 strokes with a Type B pestle in a Dounce homogenizer. After centrifugation at 2,500 x g for 5 min, the supernatant was centrifuged at 113,000 x g for 1 h. The resulting microsomal pellet was resuspended in lysis buffer (20 mM Tris-HCl, pH 7.8, 150 mM NaCl; 0.5% 3-[(3-Cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO); 0.1 mM PMSF and 1X protease inhibitor cocktail solution) at 4°C for 1 h to solubilize the microsomal proteins. Insoluble material was removed by centrifugation at 113,000 g for 1 h. Flag-CYP2C2 was isolated by binding to anti-Flag M2 affinity resin overnight at 4°C, washing 3X with lysis buffer containing 0.1% CHAPSO, and elution with 3X Flag peptide. For subsequent nickel-nitrilotriacetic acid (Ni-NTA) purification, the M2-bound fraction was incubated overnight at 4°C with Ni-NTA beads and the beads were washed 3X with 0.1% CHAPSO, 150 mM imidazole, and bound proteins were eluted with 500 mM imidazole, 0.1% CHAPSO. The affinity purified proteins were separated by SDS-PAGE and proteins were detected by Coomassie Blue staining or western blotting [29].
2.7 LC/MS/MS analysis
MS analysis was conducted using a Waters Q-ToF API-US mass spectrometer. Prior to LC/MS/MS analysis, proteins were digested in-gel with trypsin as described [30]. Digested peptides were extracted with 50% acetonitrile, 5% formic acid. After drying, the peptides were suspended in 5% acetonitrile, 1% formic acid for injection for LC\MS.
HPLC for the trypsin digested peptides was performed with a Waters nanoAcquity UPLC using a Waters Atlantis dC18 nanoAcquity column (3 micron beads, 75 micron inner diameter x 150 mm length), at a flow rate of 250 nl/min. The solvents were water (A) and acetonitrile (B), each containing 0.1% formic acid, and the gradient was from 100% A to 60% B in 60 min. The effluent from the UPLC was infused directly into a Waters Q-ToF using a Waters nano-ESI ion source. After an initial full scan, the top four most intense ions, determined with Waters MassLynx 4.1, were subjected to MS/MS fragmentation by collision induced dissociation. The raw MS results were processed by Waters ProteinLynx Global Server 2.2.5 for background subtraction, and noise filtering and deisotoping. For data analysis, the spectra of all the files for individual analyses for the gel slices were combined and processed using Mascot (Version 2.3, Matrix Science, Boston, MA) and Peaks 4.5 (Bioinformatics Solutions Incorporated, Waterloo, Ontario, Canada). In the analysis, the peptide precursor mass tolerance was 0.5 Da, the ms/ms mass tolerance was 0.5 Da, and cleavage rules were that cleavages occur after Lys and Arg residues but not before a Pro residue. The NCBI NR 20100403 database was searched which contains about 1.1 x 107 sequences and 3.7 X 109 residues. As described for Mascot scoring [31], ion scores were -10*Log(P), where P is the probability that the observed match is a random event. Individual ion scores >42 indicated either identity or extensive homology (p< 0.05) and protein scores were derived from ion scores as a non-probabilistic basis for ranking protein hits [31]. To estimate a false discovery rate, a randomized peptide database derived from all identified peptides in this study was searched resulting in a false discovery rate of about 10%.
2.8 Immunoprecipitation of microsomal proteins and western blotting
Solubilized microsomal membrane proteins were incubated in lysis buffer with antibodies against proteins or rabbit IgG as indicated in the figures at 4°C for 4 h to overnight, and the immune complexes were collected by incubation with a 25% protein A or G agarose slurry for 3 h. Immunoprecipitates were washed with lysis buffer 4X and proteins were separated by SDS-PAGE. CYP2C2/Flag/His was detected by western blotting using Flag antisera.
2.9 Sucrose gradient fractionation
M2 affinity purified protein complexes were analyzed in step sucrose gradients from 5%-45% sucrose in lysis buffer containing 0.5% CHAPSO by centrifugation at 92,000 x g for 14 h at 4°C. Ten fractions were collected and concentrated using a Microcon centrifugal filter (Millipore), and proteins were separated by SDS-PAGE and detected by western analysis. A standard curve for the gradient was generated using protein standards (Sigma-Aldrich) to estimate the sizes of the protein complexes.
2.10 Co-immunoprecipitation of isolated CYP2C2 complexes
Samples that were either bound or unbound to M2 affinity resin, were incubated in lysis buffer with antibodies against proteins as indicated in the figures or rabbit IgG or mouse IgG at 4°C for 4 h to overnight. Immune complexes were collected by binding to protein A or G beads. The samples were centrifuged and the beads washed lysis buffer 4X. Bound proteins were eluted with SDS-PAGE sample buffer equal to 1/10 that of the supernatant. Equal volumes of the supernatant (unbound fraction) and eluted proteins (bound fraction) were analyzed by SDS-PAGE and western blotting.
3 Results
3.1 CYP2C2 expression in mouse liver
Rabbit CYP2C2 is used as a representative of this subfamily of CYPs. The members of this subfamily have comparable sequence similarities within and between species so it is not possible to identify orthologous CYP2C members among mammalian species [32]. The general structures, functions as broad spectrum xenobiotic metabolizing enzymes, and cellular localizations are the same among the mammalian CYP2C’s so similar protein interactions would be expected for the mouse and rabbit CYP’s. To establish that 2C2/Flag/His was active, pAdTrack-2C2/Flag/His was transfected into mammalian Ad-293, HepG2, and COS1 cells and activity was measured after 20 h. Robust activity was observed in each cell type infected with Ad2C2/Flag/His compared to controls (Supplemental Information, Fig. 1S-A). For expression in mice, adenoviral vectors or saline was injected into mice via tail veins and efficiency of infection was estimated by GFP expression which was similarly detected for both AdEmpty and Ad2C2/Flag/His (Supplemental Information, Fig. 1S-B). Expression of 2C2/Flag/His was also detected by western analysis with either flag or CYP2C2 antisera only in extracts of liver from mice infected with Ad2C2/Flag/His (Supplemental Information, Fig. 1S-C).
3.2 Purification of the CYP2C2 protein complexes
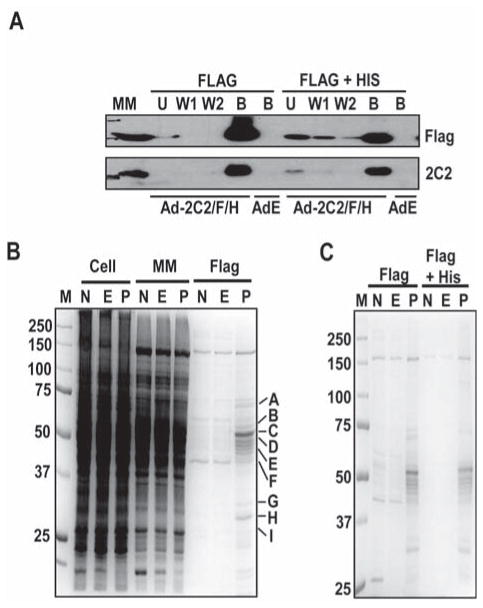
For expression of 2C2/Flag/His in liver and isolation of proteins complexed to 2C2/Flag/His, mice were injected with Ad2C2/Flag/His or with AdEmpty or saline as controls. Four days after infection, liver microsomal membrane fractions were prepared and 2C2/Flag/His was isolated as described in Materials and Methods. In preliminary studies, incubation of liver microsomal membrane with 0.5% CHAPSO, which has been used to solubilize other membrane protein complexes [33], efficiently solubilized the microsomal membrane proteins (not shown). 2C2/Flag/His, detected by western analysis, bound efficiently to the anti-Flag M2 agarose beads with little protein in flow through and wash fractions (Fig. 1A). The M2-bound sample was then further purified by binding to Ni-NTA beads (Fig. 1A). A number of proteins in the samples from Ad2C2/Flag/His infected mice were enriched in the M2-bound fraction compared with those from the control mice injected with saline or AdEmpty (Fig. 1B) and the pattern of enriched proteins was reproducible in a second independent experiment (Fig. 1C). A very similar profile of enriched proteins was observed after further purification by affinity to Ni-NTA beads but “nonspecific” proteins in the control samples were substantially decreased (Fig. 1C). These results suggest that the enriched proteins are in complexes with 2C2/Flag/His that are stable through two different purification procedures.
Figure 1.
Purification of Ad2C2/Flag/His protein complexes from mouse liver. (A) Binding of 2C2/Flag/His to M2 agarose and nickel beads. 2C2/Flag/His was purified by affinity to M2 agarose (Flag) and then further purified by affinity to Ni-NTA (Flag + His) as indicated. As a control, proteins bound to the M2 agarose or nickel beads from livers infected with Ad-empty (AdE) were also analyzed. MM, solubilized microsomal membranes; U, unbound fractions; W1 and W2, wash fractions 1 and 2; B, bound fractions. (B, C). SDS-PAGE analysis of proteins associated with M2 agarose-purified 2C2/Flag/His (B) or proteins associated with M2 agarose-purified 2C2/Flag/His followed by purification with nickel beads (C). Whole cell lysates (Cell), solubilized microsome membrane proteins (MM), proteins bound to M2 agarose beads (Flag), or proteins further purified by binding to nickel beads (Flag + His) from livers of uninfected mice (N) or mice infected with Ad-Empty (E) or Ad-2C2/Flag/His (P) were analyzed by SDS-PAGE. Proteins were visualized by Coomassie Blue staining. Visible bands in sections of the gel from the Ad-2C2/Flag/His bound sample were excised as indicated by the letters on the right of panel B and analyzed by MS. M = markers
3.3 MS identification of CYP2C2 associated proteins from mouse liver
To identify the proteins that copurified with CYP2C2, gel segments as marked in Fig. 1B were excised and analyzed by in-gel tryptic digestion followed by MS and identification of proteins as described in Materials and Methods. CYP2C2 was detected in the 55-kDa region (Segment C). Using a cutoff score of 42, which is above the score for keratin, a common contaminate in these analyses, multiple proteins were identified in each gel segment so that more than 20 peptides were detected in the M2 bound fraction (Table 1, Supplemental Information, Table 1S). Interestingly, many of the proteins identified by MS are drug metabolizing enzymes. Other proteins included chaperone proteins, like Hsc70 and GP78, and a number of dehydrogenases. Intriguingly, reticulon-3, isoform 4 scored very highly. The reticulons are integral ER proteins that are responsible for determining the morphology of the ER membrane [34] and, therefore, are candidates for ER retention “receptors” of CYPs. The identified proteins included both known and novel CYP binding proteins (see Discussion Section), but other proteins that are known to interact with CYPs were not detected, for example CPR, so that the identified proteins are a subset of all the CYP interacting proteins.
Table 1.
Proteins identified by MS that were associated with M2 affinity-purified 2C2/Flag/His. The lettered bands correspond to the excised gel sections as labeled in Fig. 2B.
| Band | Accession # | Protein a | Mass (KDa) | Scoreb |
# Scored Peptidesc |
|
|---|---|---|---|---|---|---|
| Total | E<0.05 | |||||
| A | gi|309319 | Hsc70 | 70 | 524 | 12 | 9 |
| gi|729927 | ACSL1 | 78 | 205 | 8 | 5 | |
| gi|1304157 | GP78(BIP) | 72 | 127 | 4 | 3 | |
| B | gi|3334185 | FMO5 | 60 | 137 | 7 | 5 |
| C | gi|117219 | rabbit CYP2C2 | 55 | |||
| D | gi|6681113 | Cyp3a11 | 58 | 170 | 7 | 5 |
| gi|13386414 | Cyp2d26 | 57 | 190 | 7 | 5 | |
| gi|436187 | UGT1 | 57 | 64 | 1 | 1 | |
| E | gi|387141 | Cyp2d10 | 57 | 530 | 8 | 5 |
| gi|74180862 | Cyp2d9 | 55 | 235 | 8 | 5 | |
| F | gi|6753762 | mEH | 50 | 188 | 9 | 4 |
| gi|28261389 | IFN GTPase | 47 | 152 | 5 | 3 | |
| G | gi|148688291 | 17β-HSD13 | 32 | 54 | 3 | 2 |
| gi|633242 | 11β-HSD1 | 33 | 58 | 2 | 2 | |
| H | gi|8567342 | RDH7 | 35 | 232 | 10 | 6 |
| gi|20071598 | Bdh1 | 29 | 81 | 2 | 2 | |
| gi|192005 | ApoE | 33 | 74 | 3 | 2 | |
| I | gi|2801793 | PGRMC11 | 21 | 283 | 7 | 4 |
| gi|16716353 | Rtn3-4 | 25 | 176 | 6 | 4 | |
| gi|1576133 | GST Pi | 23 | 49 | 2 | 1 | |
| gi|19354269 | Mettl7b | 22 | 42 | 5 | 0 | |
Hsc70, Heat Shock Protein 70 cognate; ACSL5, Long-chain fatty acid-CoA ligase 1; GP78, 78 kDa glucose-regulated protein; FMO5, Dimethylaniline monooxygenase 5; Cyp, cytochrome P450; UGT1, UDP-glucuronosyltransferase, family 1; mEH, Epoxide hydrolase 1, microsomal; IFN GTPase, Interferon-γ-induced GTPase; 17β-HSD1, 17β-hydroxysteroid dehydrogenase 13; 11β-HSD, 11β-hydroxysteroid dehydrogenase; RDH7, Retinol Dehydrogenasae 7; bdh1, 3-hydroxybutyrate dehydrogenase, type 1; ApoE, Apolipoprotein E; PGRMC1, Progesterone Receptor Membrane Component; RTN3-4; reticulon-3 isoform 4; GST Pi, Glutathione S-transferase, Class Pi; Mettl7b, methyltransferase-like 7b.
Standard Mascot score
(Total) = total number of scored peptides, E<0.05, number of peptides with an ion expectation value (E) as calculated by Mascot of less than 0.05.
3.4 Characterization of the CYP2C2 protein complexes
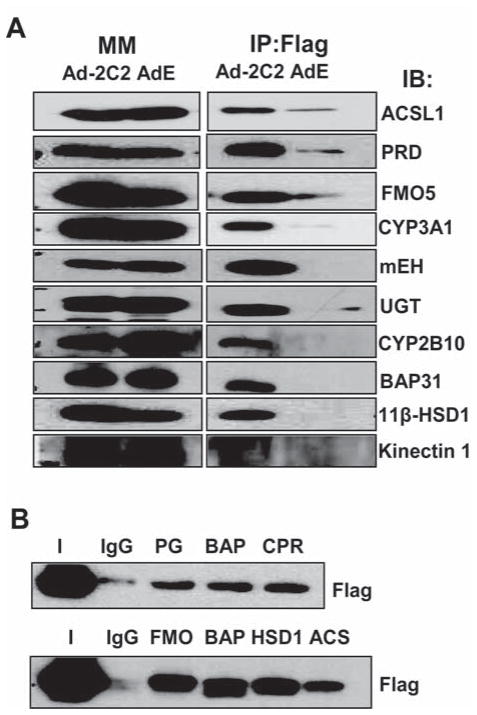
To test whether the proteins detected by MS were actually enriched in the affinity purified sample, we selected one protein that was identified in each of 6 gel segments for analysis by western blotting. As expected, similar levels of protein were present in the input microsomal membrane fractions of the AdEmpty and Ad2C2/Flag/His samples, however, after M2-agarose purification, each of the proteins was substantially enriched in the Ad2C2/Flag/His samples compared to the AdEmpty samples (Fig. 2A). The enrichment was similar to that of two known CYP binding proteins, CPR and BAP31 [24]. In contrast, the microsomal protein kinectin was not enriched in either of the two Ad samples and served as a negative control (Fig. 2A). These results demonstrate that the binding of these proteins to the M2-agarose beads is dependent on the presence of 2C2/Flag/His in the extracts. Since all 6 proteins in this sample of the proteins identified by MS were enriched with 2C2/Flag/His, it is likely that the remaining identified proteins are also in 2C2/Flag/His-containing complexes.
Figure 2.
Analysis of proteins associated with M2 agarose-purified 2C2/Flag/His by western blotting. (A) Proteins present in solubilized microsomal membranes (MM) or associated with purified 2C2/Flag/His (Flag) from livers infected with either Ad-Empty (E) or Ad-2C2/Flag/His (Ad-2C2) were detected by western blotting with antibodies for the indicated proteins. (B) Solubilized microsomal membrane proteins from mouse liver were immunoprecipitated with IgG as control or with antibodies to proteins as indicated, and 2C2/Flag/His in the immunoprecipitates was detected by western blotting using anti-Flag antibody.
To further confirm the interaction of 2C2/Flag/His with the identified proteins, selected proteins were first immunoprecipitated and 2C2/Flag/His in the immunoprecipitate was detected by western analysis. In each case 2C2/Flag/His co-immunoprecipitated while little 2C2/Flag/His was precipitated nonspecifically by control IgG (Fig. 2B), which provides further evidence that the proteins identified by MS are present in a complex with 2C2/Flag/His.
3.5 Sucrose gradient analysis of CYP2C2 protein complexes
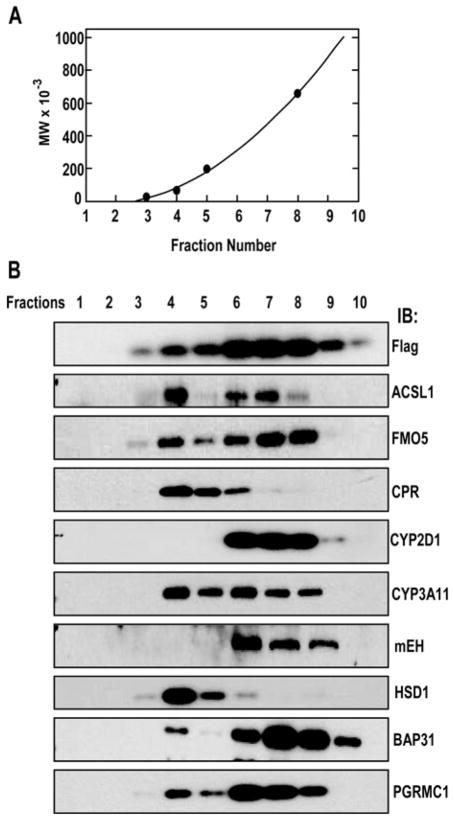
To determine whether there is a single complex or multiple complexes of 2C2/Flag/His with the identified proteins, we separated the 2C2/Flag/His protein complexes by sucrose density gradient centrifugation in the presence of 0.5% CHAPSO to determine the sizes of the complexes (Fig. 3). 2C2/Flag/His, detected with M2 antibody, was distributed broadly in the gradient, consistent with multiple heterogeneous complexes Fig. 3B). Proteins in fraction 4 corresponded to a molecular weight of 66,000 and probably represents 2C2/Flag/His monomer while protein in fractions 5-10 correspond to molecule weights of 200,000 to >600,000. Most other proteins were also detected in fraction 4 which also likely correspond to monomers of these proteins. However, different distribution patterns at higher molecular weights were observed for different proteins indicating multiple complexes with 2C2/Flag/His. CPR and 11β-HSD were present at highest amounts in fraction 4, presumed monomers, with tailing into fractions 5 and 6 suggesting either dimer formation with 2C2/Flag/His or unstable complexes that dissociated during the centrifugation. The other proteins cosedimented with 2C2/Flag/His in fractions 6 to 8, but with subtle differences. The maximum concentrations of BAP31 and FMO5 were in fractions 7 and 8, maximum concentration of CYP3A11, mEH, and PGRMC1 were in fraction 6, while long-chain fatty acid-CoA ligase 1 (ACSL1) and CYP2D1, like 2C2/Flag/His were symmetrically distributed across fractions 6-8. This complex sedimentation pattern indicates that CYP2C2 is present in a diverse set of heterogeneous protein complexes.
Figure 3.
Fractionation of 2C2/Flag/His protein complexes by sedimentation in a sucrose gradient. Protein complexes purified from anti-Flag M2 affinity resins were separated by centrifugation in a 5% to 40% sucrose gradient. A. Proteins of known molecular weight were analyzed and a standard curve plotted. B. In parallel gradients, fractions were obtained from the top (1) to the bottom (10) of the gradient, and proteins were precipitated and analyzed by SDS-PAGE and western blotting for the indicated proteins as described in Materials and Methods. Protein abbreviations are defined in Table 1.
3.6 Co-immunoprecipitation of 2C2/Flag/His-associated protein complexes
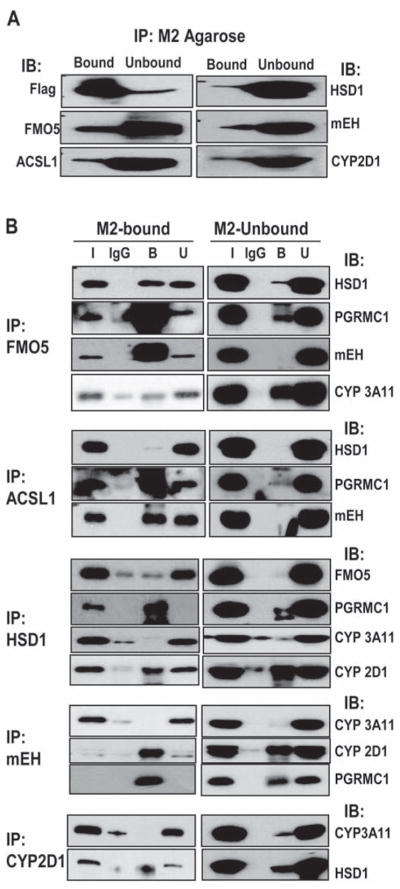
We used co-immunoprecipitation of the 2C2/Flag/His associated proteins with each other to further determine if multiple 2C2/Flag/His complexes existed. As a control, the proteins not binding to the M2-agarose (M2-Unbound) were also analyzed. As expected, 2C2/Flag/His was predominantly present in the M2-Bound fraction, while the other proteins were predominantly present in the M2-Unbound fraction indicating that only a fraction of the total amount of these proteins was complexed with 2C2/Flag/His (Fig. 4A).
Figure 4.
Co-immunoprecipitation of proteins associated with 2C2/Flag/His. (A) Proteins either unbound (M2-Unbound) or bound to M2 agarose (M2-bound) were separated by SDS-PAGE and the indicated proteins were detected by western blotting. (B) Proteins in either the M2-bound or M2-unbound fractions were immunoprecipitated with antisera against the indicated proteins (left side), and co-immunoprecipitated proteins were then detected by western blotting in both the bound (B) and unbound (U) fractions using antibodies against the proteins indicated (right side). The input (I) represents 1/10 of the total proteins in the sample before immunoprecipitation and proteins bound to IgG were detected as a control.
Proteins were precipitated with antibodies to either FMO5, ACSL1, 11β-HSD1, mEH or CYP2D1 and then co-immunoprecipitated proteins were detected by western analysis using an antibody from a species other than that of the initial precipitating antibody (Fig. 4B). In the FMO5 immunoprecipitation, the fraction of PGRMC1 and mEH coprecipitating from the M2-bound fractions was much greater than from the M2-unbound fraction (Fig. 4B). In contrast, much smaller fractions of 11β-HSD1 and Cyp3a11 in the M2-bound fraction co-immunoprecipitated with FMO5. These results suggest that FMO5, PGRMC1 and mEH are in the same complex with 2C2/Flag/His while 11β-HSD1 and Cyp3a11 may be in different complexes. In addition, the small amounts of these proteins that co-immunoprecipitate in the M2-unbound fraction indicate that these proteins do not directly interact with FMO5 in the absence of 2C2/Flag/His with the possible exception of Cyp3a11. Similarly, co-immunoprecipitation with ACSL1 of PGRMC1 and mEH, but not 11β-HSD1 in the M2-bound fraction with little co-immunoprecipitation in the M2-unbound fraction suggest that ACSL1, PGRMC1, and mEH are in a complex dependent on the presence of 2C2/Flag/His. In 11β-HSD1 immunoprecipitates, PGRMC1 and CYP2D1 selectively co-immunoprecipitated in the M2-bound fraction compared to the M2-unbound fraction, while FMO5 and Cyp3a11 did not co-immunoprecipitate efficiently in the M2-bound fraction. The lack of interaction of 11β-HSD1 with FMO5 is consistent with the FMO5 immunoprecipitation studies. Substantial amounts of Cyp2d1 co-immunoprecipitated with 11β-HSD1 in both the M2-bound and M2-bound fractions which suggested a direct interaction between 11β-HSD1 and Cyp2d1. Similar studies with mEH suggested that mEH was in a 2C2/Flag/His-dependent complex with PGRMC1 and Cyp2d1, but not with Cyp3a11 and that Cyp2d1 was in a complex with 11β-HSD1, but not Cyp3a11. These results suggest that 2C2/Flag/His is present in multiple protein complexes with different compositions. The results are consistent with a complex containing 2C2/Flag/His, FMO5, PGRMC1, mEH, ACSL1, and possibly Cyp2d1, which is consistent with the results from sucrose gradient fragmentation analysis in which these proteins sedimented predominantly in fractions 6-8. A second complex containing 2C2/Flag/His, 11β-HSD1, PGRMC1 and Cyp2d1 may also exist as well as complexes with Cyp3a11, which did not co-immunoprecipitate with any of the tested proteins. These results, with the sucrose gradient analysis demonstrate that there are multiple heterogeneous 2C2/Flag/His complexes.
4 Discussion
We have identified about 25 proteins that interact with exogenously expressed CYP2C2 in mouse liver by copurification and analysis by MS or western blotting. A concern is that these hydrophobic proteins may nonspecifically interact after solubilization of the membranes even in the presence 0.5% CHAPSO. Several lines of evidence suggest that most of the proteins complexed with CYP2C2 are specific interacting proteins. First, the identified proteins are substantially enriched in the 2C2/Flag/His samples compared to Ad-empty or uninfected negative controls and these proteins represent only a very small subset of the total ER membrane proteins. Second, impressively, reisolation of M2-affinity purified CYP2C2 by binding to nickel beads resulted in a similar pattern of associated proteins. The consistent pattern through two purification steps suggests that the isolated proteins are binding with high affinity which would be consistent with specific binding. Third, some of the identified proteins had been reported to interact with CYPs, including other CYPs, PGRMC1 [22], retinol dehydrogenase [35], and mEH, and UGT [19]. Fourth, the enrichment of the identified proteins in the 2C2/Flag/His samples was similar to that of known CYP binding proteins. The constancy of the co-purified protein pattern through two steps of purification and the identification of proteins known to interact with CYPs provide confidence that proteins identified interact specifically with CYP2C2 in the microsomal membrane.
Analysis of the size distribution of the 2C2/Flag/His complexes by sucrose gradient centrifugation indicated that CYP2C2 was broadly distributed in the gradient with estimated molecular weights from about 60,000 to >600,000 and different distribution patterns were observed for the interacting proteins suggesting that multiple heterogeneous complexes are present. Co-immunoprecipitation analysis of the proteins associated with 2C2/Flag/His supported this idea since different subsets of the proteins were co-immunoprecipitated by antisera to individual associated proteins. In most cases, such co-immunoprecipitation was not observed with the M2-unbound fraction, indicating that the interaction between the proteins was mediated by 2C2/Flag/His in a protein complex. These results indicate that there are multiple specific CYP2C2 complexes.
The nature of the proteins in the complex provides insight about the possible functional significance of the complexes. One striking observation is that a large fraction of the identified proteins either require NAD or NADPH or have redox partners that do. Several CYPs copurified with CYP2C2 and while CYPs do not require NADPH directly, their redox partner, CPR does. Other associated proteins that require NADPH are FMO5, 11β-HSD1, and 17β-hydroxysteroid dehydrogenase. NADH is required by retinol dehydrogenase and 3-hydroxybutyrate dehydrogenase. FMO5, like the CYPs, is a monooxygenase. The association of these enzymes suggests that complexes of enzymes that catalyze oxidative reactions requiring NAD or NADH may be functionally beneficial.
A second striking observation is the large number of enzymes involved in drug metabolism in the CYP2C2 complexes. In addition to NAD/NADH related drug metabolizing enzymes, CYPs, mEH, and FMO5, discussed above, UGTs and glutathionine S-transferases were detected. This suggests the presence of drug metabolizing complexes which might facilitate sequential metabolism of a substrate, redundant metabolic functions, or protective mechanisms. FMO5 and CYPs have unique substrates, but also common substrates. FMO5 differs from CYPs in being resistant to inactivation by reactive metabolites that are generated by the two enzymes [36]. FMO5 metabolites may, therefore, affect CYP activity and FMO5 also provides a mechanism for continued metabolism of substrates with metabolites that inactivate CYP Association of mEH is interesting since mEH would be protective against epoxides formed by CYP mediated metabolism. Sequential modification of the same substrate would be facilitated by association of CYPs with enzymes like UGT which catalyze conjugations to functional groups introduced into substrates by CYPs. Complexes of drug-metabolizing enzymes, therefore, are potentially functionally significant.
Functional grouping of the other proteins that are associated with CYP2C2 is less clear. Interferon-γ-induced GTPase, Mettl7b, and long-chain fatty acid-CoA ligase have no obvious role in CYP function. Hsc70 and PGRMC1 are chaperone-like proteins which may be important for CYP activity. For example, PGRMC1 activates CYP51 and other sterol metabolizing CYPs [22] while it inhibits the drug metabolizing forms, CYP2C2, CYP2E1, and CYP3A4 [23]. The reticulons are particularly interesting ER structural proteins that are important in shaping the curvature of the ER and, thus, are present in the tubular components of the smooth ER [37]. Since CYPs are localized to the smooth ER, the reticulons are well positioned to retain the CYPs in the ER. Further analysis will be required to determine the role of these proteins in ER retention of CYPs.
These experiments provide strong evidence that CYP2C2 interacts with a wide range of ER membrane proteins in multiple heterocomplexes in vivo. While the present data do not address the functional significance of the interacting proteins, the striking overabundance of NAD/NADP dependent enzymes and drug metabolizing enzymes suggest that the complexes may increase the efficiency of these classes of enzymes. In addition, interaction with chaperone-like proteins may be important for CYP function and the reticulons may be important determinants of CYP localization in the cell.
Supplementary Material
Acknowledgments
This study was supported by a grant from the NIH GM35897. MS analysis was conducted at the Protein Sciences Facility, Carver Biotechnology Center, University of Illinois, Urbana, IL 61801. We thank Brian Imai of the Protein Sciences Facility for helpful discussions and data anlaysis.
Abbreviations
- ACSL1
long-chain fatty acid-CoA ligase 1
- BAP31
B-cell associated protein 31
- CHAPSO
3-[(3-Cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate
- CPR
CYP reductase
- CYP
cytochrome P450
- ER
endoplasmic reticulum
- 11β-HSD1
11β–hydroxysteroid dehydrogenase-1
- FMO5
flavin monooxygenase 5
- GFP
green fluorescent protein
- mEH
microsomal epoxide hydrolase
- Ni-NTA
nickel-nitrilotriacetic acid
- PGRMC1
Progesterone receptor membrane component 1
- UGT
UDP-glucurosyltransferase (UGT)
Footnotes
Conflict of Interest Statement
The authors have no financial/commercial conflicts of interest.
References
- 1.Hrycay EG, Bandiera SM. Expression, function and regulation of mouse cytochrome P450 enzymes: comparison with human P450 enzymes. Curr Drug Metab. 2009;10:1151–1183. doi: 10.2174/138920009790820138. [DOI] [PubMed] [Google Scholar]
- 2.Gut J, Richter C, Cherry RJ, Winterhalter KH, Kawato S. Rotation of cytochrome P-450. Complex formation of cytochrome P-450 with NADPH-cytochrome P-450 reductase in liposomes demonstrated by combining protein rotation with antibody-induced cross-linking. J Biol Chem. 1983;258:8588–8594. [PubMed] [Google Scholar]
- 3.Iwase T, Sakaki T, Yabusaki Y, Ohkawa H, et al. Rotation and interactions of genetically expressed cytochrome P-450IA1 and NADPH-cytochrome P-450 reductase in yeast microsomes. Biochemistry. 1991;30:8347–8351. doi: 10.1021/bi00098a010. [DOI] [PubMed] [Google Scholar]
- 4.Yamada M, Ohta Y, Bachmanova GI, Nishimoto Y, et al. Dynamic interactions of rabbit liver cytochromes P450IA2 and P450IIB4 with cytochrome b5 and NADPH-cytochrome P450 reductase in proteoliposomes. Biochemistry. 1995;34:10113–10119. doi: 10.1021/bi00032a003. [DOI] [PubMed] [Google Scholar]
- 5.Davydov DR, Sineva EV, Sistla S, Davydova NY, et al. Electron transfer in the complex of membrane-bound human cytochrome P450 3A4 with the flavin domain of P450BM-3: the effect of oligomerization of the heme protein and intermittent modulation of the spin equilibrium. Biochim Biophys Acta. 2010;1797:378–390. doi: 10.1016/j.bbabio.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Myasoedova KN, Berndt P. Cytochrome P-450LM2 ologomers in proteoliposomes. FEBS Lett. 1990;275:235–238. doi: 10.1016/0014-5793(90)81479-8. [DOI] [PubMed] [Google Scholar]
- 7.Myasoedova KN, Magretova NN. Cross-Linking study of cytochrome P450 1A2 in proteoliposomes. Biosci Rep. 2001;21:63–72. doi: 10.1023/a:1010486118448. [DOI] [PubMed] [Google Scholar]
- 8.Ozalp C, Szczesna-Skorupa E, Kemper B. Bimolecular fluorescence complementation analysis of cytochrome P450 2C2, 2E1 and NADPH-cytochrome P450 reductase molecular interactions in living cells. Drug Metab Dispos. 2005;33:1382–1390. doi: 10.1124/dmd.105.005538. [DOI] [PubMed] [Google Scholar]
- 9.Szczesna-Skorupa E, Mallah B, Kemper B. Fluorescence resonance energy transfer analysis of cytochromes P450 2C2 and 2E1 molecular interactions in living cells. J Biol Chem. 2003;278:31269–31276. doi: 10.1074/jbc.M301489200. [DOI] [PubMed] [Google Scholar]
- 10.Hu G, Johnson EF, Kemper B. CYP2C8 exists as a dimer in natural membranes. Drug Metab Dispos. 2010;38:1976–1983. doi: 10.1124/dmd.110.034942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Praporski S, Ng SM, Nguyen AD, Corbin CJ, et al. Organization of cytochrome P450 enzymes involved in sex steroid synthesis: Protein-protein interactions in lipid membranes. J Biol Chem. 2009;284:33224–33232. doi: 10.1074/jbc.M109.006064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian M, Tam H, Zheng H, Tracy TS. CYP2C9-CYP3A4 protein-protein interactions: role of the hydrophobic N terminus. Drug Metab Dispos. 2010;38:1003–1009. doi: 10.1124/dmd.109.030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed JR, Eyer M, Backes WL. Functional interactions between cytochromes P450 1A2 and 2B4 require both enzymes to reside in the same phospholipid vesicle: evidence for physical complex formation. J Biol Chem. 2010;285:8942–8952. doi: 10.1074/jbc.M109.076885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian M, Low M, Locuson CW, Tracy TS. CYP2D6-CYP2C9 protein-protein interactions and isoform-selective effects on substrate binding and catalysis. Drug Metab Dispos. 2009;37:1682–1689. doi: 10.1124/dmd.109.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazai E, Kupfer D. Interactions between CYP2C9 and CYP2C19 in reconstituted binary systems influence their catalytic activity: possible rationale for the inability of CYP2C19 to catalyze methoxychlor demethylation in human liver microsomes. Drug Metab Dispos. 2005;33:157–164. doi: 10.1124/dmd.104.001578. [DOI] [PubMed] [Google Scholar]
- 16.Alston K, Robinson RC, Park SS, Gelboin HV, Friedman FK. Interactions among cytochromes P-450 in the endoplasmic reticulum. Detection of chemically cross-linked complexes with monoclonal antibodies. J Biol Chem. 1991;266:735–739. [PubMed] [Google Scholar]
- 17.Hazai E, Bikadi Z, Simonyi M, Kupfer D. Association of cytochrome P450 enzymes is a determining factor in their catalytic activity. J Comput Aided Mol Des. 2005;19:271–285. doi: 10.1007/s10822-005-4995-4. [DOI] [PubMed] [Google Scholar]
- 18.Cawley GF, Zhang S, Kelley RW, Backes WL. Evidence supporting the interaction of CYP2B4 and CYP1A2 in microsomal preparations. Drug Metab Dispos. 2001;29:1529–1534. [PubMed] [Google Scholar]
- 19.Taura KI, Yamada H, Hagino Y, Ishii Y, et al. Interaction between cytochrome P450 and other drug-metabolizing enzymes: evidence for an association of CYP1A1 with microsomal epoxide hydrolase and UDP-glucuronosyltransferase. Biochem Biophys Res Commun. 2000;273:1048–1052. doi: 10.1006/bbrc.2000.3076. [DOI] [PubMed] [Google Scholar]
- 20.Ishii Y, Iwanaga M, Nishimura Y, Takeda S, et al. Protein-protein interactions between rat hepatic cytochromes P450 (P450s) and UDP-glucuronosyltransferases (UGTs): evidence for the functionally active UGT in P450-UGT complex. Drug Metab Pharmacokinet. 2007;22:367–376. doi: 10.2133/dmpk.22.367. [DOI] [PubMed] [Google Scholar]
- 21.Ishii Y, Takeda S, Yamada H, Oguri K. Functional protein-protein interaction of drug metabolizing enzymes. Front Biosci. 2005;10:887–895. doi: 10.2741/1583. [DOI] [PubMed] [Google Scholar]
- 22.Hughes AL, Powell DW, Bard M, Eckstein J, et al. Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 2007;5:143–149. doi: 10.1016/j.cmet.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Szczesna-Skorupa E, Kemper B. Progesterone Receptor Membrane Component 1 (Pgrmc1) Inhibits the Activity of Drug-Metabolizing Cytochromes P450 and Binds to Cytochrome P450 Reductase. Mol Pharmacol. 2011;79:340–350. doi: 10.1124/mol.110.068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szczesna-Skorupa E, Kemper B. BAP31 is involved in the retention of cytochrome P450 2C2 in the endoplasmic reticulum. J Biol Chem. 2006;281:4142–4148. doi: 10.1074/jbc.M509522200. [DOI] [PubMed] [Google Scholar]
- 25.Fang S, Miao J, Xiang L, Ponugoti B, et al. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He TC, Zhou S, da Costa LT, Yu J, et al. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szczesna-Skorupa E, Kemper B. N-terminal substitutions of basic amino acids induce translocation across the microsomal membranes and glycosylation of rabbit cytochrome P450IIC2. J Cell Biol. 1989;108:1237–1243. doi: 10.1083/jcb.108.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo J, Deng ZL, Luo X, Tang N, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 29.Ahn K, Szczesna-Skorupa E, Kemper B. The amino-terminal 29 amino acids of cytochrome P450 2C1 are sufficient for retention in the endoplasmmic reticulum. J Biol Chem. 1993;268:18726–18733. [PubMed] [Google Scholar]
- 30.Kang YJ, Stevenson AK, Yau PM, Kollmar R. Sparc protein is required for normal growth of zebrafish otoliths. J Assoc Res Otolaryngol. 2008;9:436–451. doi: 10.1007/s10162-008-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Nebert DW, Nelson DR, Coon MJ, Estabrook RW, et al. The P450 superfamily: update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 1991;10:1–14. doi: 10.1089/dna.1991.10.1. [DOI] [PubMed] [Google Scholar]
- 33.Yang T, Espenshade PJ, Wright ME, Yabe D, et al. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 34.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, et al. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imaoka S, Wan J, Chow T, Hiroi T, et al. Cloning and characterization of the CYP2D1-binding protein, retinol dehydrogenase. Arch Biochem Biophys. 1998;353:331–336. doi: 10.1006/abbi.1998.0644. [DOI] [PubMed] [Google Scholar]
- 36.Cashman JR. Some distinctions between flavin-containing and cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:599–604. doi: 10.1016/j.bbrc.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 37.Shibata Y, Voss C, Rist JM, Hu J, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.