SUMMARY
Determining the composition of protein complexes is an essential step towards understanding the cell as an integrated system. Using co-affinity purification coupled to mass spectrometry analysis, we examined protein associations involving nearly five thousand individual, FLAG-HA epitope-tagged Drosophila proteins. Stringent analysis of these data, based on a novel statistical framework to define individual protein-protein interactions, led to the generation of a Drosophila Protein interaction Map (DPiM) encompassing 556 protein complexes. The high quality of DPiM and its usefulness as a paradigm for metazoan proteomes is apparent from the recovery of many known complexes, significant enrichment for shared functional attributes and validation in human cells. DPiM defines potential novel members for several important protein complexes and assigns functional links to 586 protein-coding genes lacking previous experimental annotation. DPiM represents, to our knowledge, the largest metazoan protein complex map and provides a valuable resource for analysis of protein complex evolution.
Keywords: Drosophila, proteome, protein complex map, interactome
INTRODUCTION
The vast majority of proteins work as parts of assemblies composed of several elements, thereby defining protein complexes as essential cellular functional units. The functionality of proteins relies on their ability to interact with one another while pathogenic conditions can reflect the loss of such function. Given the fundamental importance of protein interactions, proteome-wide “interactome” maps based on pairwise protein interactions using the yeast two-hybrid (Y2H) system have been determined for several organisms (Giot et al., 2003; Ito et al., 2001; Li et al., 2004; Rual et al., 2005; Stanyon et al., 2004; Stelzl et al., 2005; Uetz et al., 2000). Alternatively, protein complex isolation based on co-affinity purification combined with tandem mass spectrometry (coAP-MS) has been used to generate protein complex maps at proteome-scale for Saccharomyces cerevisiae (Gavin et al., 2006; Ho et al., 2002; Krogan et al., 2006), Escherichia coli (Hu et al., 2009), and Mycoplasma pneumoniae (Kuhner et al., 2009). This approach has been proven successful in the study of defined metazoan proteomic sub-spaces (Behrends et al., 2010; Bouwmeester et al., 2004; Ewing et al., 2007; Guerrero et al., 2008; Sowa et al., 2009), but there are no large-scale protein complex maps available for metazoans [reviewed in (Gavin et al., 2011)]. Here we present a substantial resource of affinity tagged proteins and generation of a protein complex map of Drosophila that provides a paradigmatic blueprint of interactions in a metazoan proteome.
Extensive genetic analyses in Drosophila have contributed fundamentally to our understanding of metazoan morphogenesis. However, many functional associations defined genetically in the animal lack mechanistic explanations. A comprehensive protein complex map would serve as a powerful resource to uncover the molecular basis of these genetic interactions and provide necessary mechanistic insights. Moreover, despite the success of the extensive molecular genetic studies in Drosophila, one third of (~14,000) predicted Drosophila proteins (Adams et al., 2000) remain without functional annotation (Tweedie et al., 2009). The genetic tools available in Drosophila enable testing of predicted physical interactions in vivo, making it an ideal model organism for the generation of a comprehensive protein complex map. Such a map is a compelling tool for gene annotation, which is also incomplete in mammals, so a Drosophila map will be of considerable value for annotating mammalian proteomes.
Here we describe the generation of a large-scale Drosophila Protein interaction Map (DPiM) by coAP-MS analysis based on ~3,500 affinity purifications. We developed a semi-quantitative statistical approach to score protein interactions and defined a high quality map. The map recovers many known, and hundreds of previously uncharacterized protein complexes, thus providing functional associations and biological context for 586 proteins that previously lacked annotation. To our knowledge, DPiM is the first large-scale metazoan protein complex analysis that is not focused on a specific sub-proteomic space, thereby providing a systems view of a metazoan proteome. The map defines a primary protein interaction landscape for Drosophila cells that allows study of the developmental dynamics and tissue level variation of any protein complex in the map. Finally, DPiM offers a new reference point in the analysis of protein complex evolution.
RESULTS
High-throughput Drosophila Proteomics Platform
To systematically isolate Drosophila protein complexes and determine their composition, we developed a large collection of affinity-tagged clones called the Universal Proteomics Resource [(Yu et al., 2011) http://www.fruitfly.org/EST/proteomics.shtml] as part of the Berkeley Drosophila Genome Project (BDGP; see Methods). From this collection, 4,273 individual clones were transiently transfected into S2R+ cells. Approximately 80% of the clones successfully expressed “bait” protein at detectable levels and associated protein complexes were affinity purified. Purifications that resulted in detection of one or more unique, bait-derived peptides by mass spectrometry were considered for subsequent analysis, with few exceptions (see Methods). This resulted in identification of a total of 4,927 Drosophila proteins (at 0.8% False Discovery Rate) from 3,488 individual affinity purifications (Figure 1A). In general, mass spectrometric analysis of tryptic peptides cannot distinguish a specific protein isoform with confidence. So for this analysis, all the identified isoforms were traced back to the genes encoding them. From hereon, all gene products are referred to as proteins without specifying isoforms. The raw mass spectrometry data are available in Supplemental Table S1 and are accessible through FlyBase Linkouts and the DPiM website (https://interfly.med.harvard.edu/).
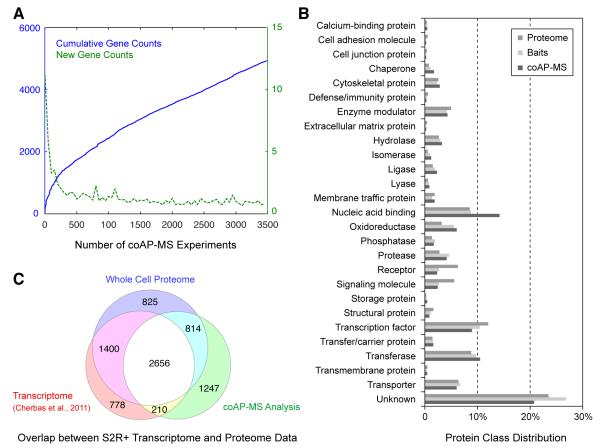
Figure 1. Analysis of proteins identified in the coAP-MS pipeline.
(A) Cumulative number of gene counts (blue) and unique gene counts (green) detected as a function of the number of high quality affinity purification experiments. (B) Comparison of protein class distribution between Drosophila proteome, baits used and proteins identified in DPiM analysis (coAP-MS) using PANTHER (Thomas et al., 2003). (C) A conservative estimate of overlap between the S2R+ cell transcriptome [5,044 protein coding genes with gene score ≥ 300; (Cherbas et al., 2011)], S2R+ proteome whole cell lysate MS analysis (5,695 proteins) and the proteins identified in coAP-MS analysis (4,927 proteins). The intersections of the data sets are as follows: 4,056 (Transcriptome and Whole Cell Proteome), 3,470 (coAP-MS and Whole Cell Proteome) and 2,866 (Transcriptome and coAP-MS). See also Supplemental Figure S1.
Comparison of protein functional class distribution using the PANTHER classification system (Thomas et al., 2003) indicates that the distribution of protein categories of baits used and proteins identified in coAP-MS is very similar to the overall distribution of the Drosophila proteome, much of which remains unannotated (Figure 1B). A few minor differences are noted: nucleic acid binding proteins and oxidoreductases are overrepresented while receptor and signaling molecules are underrepresented in the coAP-MS data set (Figure 1B).
We determined the proteome composition of the S2R+ cell by high-resolution mass spectrometry, resulting in the identification of 6,081 proteins corresponding to 5,695 genes (1% FDR) in S2R+ cells (Figure 1C) (see Methods, Supplemental Figure S1 and Table S2). The transcriptome data (Cherbas et al., 2011) and whole cell proteome analyses indicate that more than one third of the predicted Drosophila proteome is expressed in these cells. A large fraction of baits used for generating this map is expressed in S2R+ cells (61%) and 75% of proteins identified by coAP-MS were found in either transcriptome or whole cell proteome analysis. Our analysis has interrogated a large portion of the S2R+ cell proteome but not saturated it. These are conservative estimates as strict comparisons with the transcriptome data are not possible given the methodological differences and absence of a rigorously defined false discovery rate for the transcriptome data.
A Drosophila Protein Interaction Map
Proteins identified by coAP-MS represent a mixture of genuine direct or indirect interactors and non-specific interactors (Ewing et al., 2007; Rees et al., 2011). The non-specific interactors are present in a large number of data sets independent of the bait used while genuine interactors tend to co-occur across relevant experiments. We developed a scoring system based on the hypergeometric probability distribution (Hart et al., 2007) to calculate the significance of co-occurrence of protein pairs by incorporating the Total Spectral Counts (TSCs) for each protein. The number of TSCs correlates roughly with protein abundance in a sample (Liu et al., 2004) and thus increases the sensitivity of our approach by providing a semi-quantitative dimension to the score. We refer to this scoring system as the HGSCore (HyperGeometric Spectral Counts score; see Methods). A matrix model was used for both bait-prey and prey-prey interactions, and a total of 209,912 potential protein-protein interactions were scored among 4,927 Drosophila proteins (Supplemental Table S3).
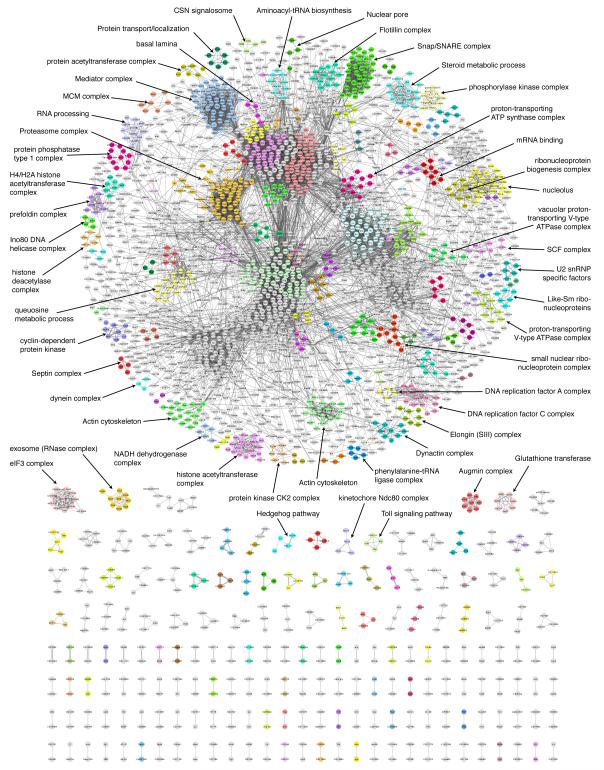
This statistical analysis led to the prediction of 10,969 high-confidence co-complex membership interactions (0.05% FDR) involving 2,297 Drosophila proteins, which are visualized as a network (Figure 2). Further analyses of these high confidence co-complex membership interactions based on the Markov clustering algorithm (MCL) (Enright et al., 2002) defined 556 putative complexes encompassing 2,240 proteins (Supplemental Table S4). We use the term Drosophila Protein interaction Map (DPiM) to refer to the composite data set and the resulting network. The map shows a distinct grouping of 1,817 proteins (80% of total) as the giant component of the network encompassing 377 putative complexes (68%) with a high degree of interconnectedness (Figure 2). A second group of 179 independent complexes (32%) defined by the map are not connected to other complexes. Among the baits that are expressed in S2R+ cells and part of the same MCL cluster, 36% (159/442) are found in direct reciprocal pull-downs. Some of the well-known complexes recovered in the DPiM are indicated in Figure 2.
Figure 2. Drosophila Protein interaction Map (DPiM).
Graphical representation of the DPiM comprising 10,969 high confidence co-complex membership interactions (at 0.05% FDR) involving 2,297 proteins. Protein interactions are shown as grey lines with thickness proportional to the HGSCore for the interaction in DPiM. The map defines 556 clusters, 377 of which are interconnected, representing nearly 80% of the proteins in the network. The remaining 179 clusters are not connected to members of other complexes. Depicted with different colors are 153 clusters enriched for GO terms, KEGG pathways or Pfam/Interpro domains. Proteins in other clusters that are not enriched are shown as grey circles. Selected complexes with known molecular function / biological role are indicated. See also Figure S4.
DPiM Quality Assessment
The quality of the DPiM was evaluated using four approaches. First, we examined whether the coAP-MS approach was capable of identifying known interactions. Second, we asked if the complexes tend to share Gene Ontology (GO) annotation. Third, we examined whether the genes encoding proteins of the same complex tend to be co-expressed. Finally, we tested the ability of DPiM interactions to be validated across species using human proteins as baits in HEK-293 cells.
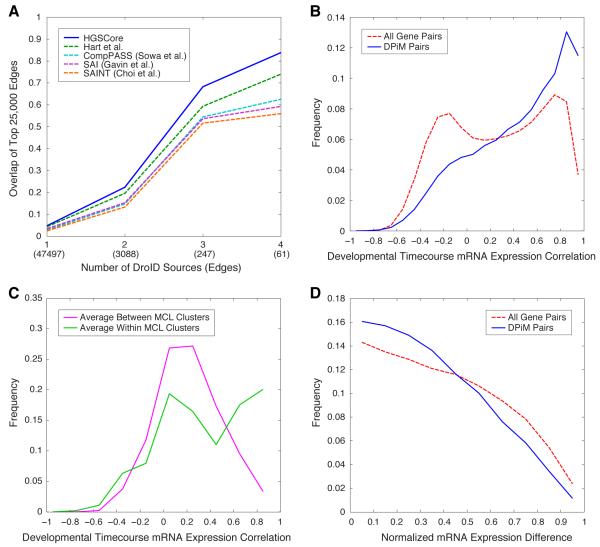
Defining a positive Drosophila reference set, in order to assess the sensitivity and specificity of different scoring methods is difficult, as existing data sets show little overlap (Yu et al., 2008) and there are no hand-curated databases similar to those available for the yeast and human proteomes. We hence used the extent of overlap from multiple diverse sources as an estimate of reliability of a given pairwise interaction. The DroID database (Murali et al., 2011) consolidates protein interaction data from seven discrete sources. Four bins of interactions were defined with increasing levels of confidence i.e., those supported by at least one, two, three or four independent DroID sources, and computed the overlaps with DPiM (Figure 3A). The coAP-MS data set was also analyzed using published scoring methods (Breitkreutz et al., 2010; Choi et al., 2011; Gavin et al., 2006; Hart et al., 2007; Sowa et al., 2009). Since these methods produce different numbers of interactions, we compared the top 25,000 interactions reported from each method with those listed in DroID. The HGSCore method recovered more interactions than other published scoring methods across all confidence levels, reflecting a 15% increase on average that is significant even when compared to the next best method (P-value 6.9×10−12) (Figure 3A). We find that the top 25,000 HGSCore interactions recover between 68% and 84% of the highest confidence interactions, i.e., physical interactions supported by either three or four independent DroID data sets (n=247 and 61 respectively). When considering only those interactions above the 0.05 FDR threshold of HGSCore, DPiM recovers between 56% and 71% of the highest confidence interactions. The overall increase in recall at increasing reference set confidence levels across multiple analysis methods suggests the underlying data in DPiM is of high quality, while the robust improvement HGSCore makes over established methods validates our approach. Nearly 86% of the interactions in DPiM are novel when considering all the interactions reported in DroID, which includes interolog data from three species (yeast, worm and human).
Figure 3. Evaluation of quality of DPiM protein interactions.
(A) Comparison of interactions in the DPiM data set and DroID. Four bins with increasing levels of confidence supported by at least one, two, three, or four DroID sources were defined. The overlap between the top 25,000 interactions defined by each of the co-occurrence analysis methods and DroID is shown. The number of interactions supported by given number of sources is indicated in parentheses along the X-axis. (B) Distribution of correlation coefficients between mRNAs corresponding to interacting proteins in DPiM compared to all gene pairs, based on the RNA-Seq data (Graveley et al., 2011). (C) Distribution of correlation coefficients of mRNAs corresponding to proteins within MCL clusters compared to those between MCL clusters, analysis similar to (B). (D) Normalized absolute mRNA expression differences between DPiM interactors and all gene pairs (Cherbas et al., 2011). See also Figure S2.
Proteins belonging to the same protein complex can be expected to be enriched for Gene Ontology (GO) annotations, share the same KEGG Pathways and contain similar protein domains. The DAVID Functional Annotation Tools (Huang da et al., 2009) were used to calculate enrichment for annotations, pathways and domains within each protein cluster generated by DPiM. About 28% of the MCL-derived protein clusters (153/556) are enriched for one or more of these features (multiple hypothesis testing-adjusted P<0.01) (Supplemental Figure S4). In total, almost half of the proteins in the DPiM network fall into a GO term enriched cluster (Supplemental Table S4). Due to the nature of MCL clustering, some components of larger complexes tend to separate into smaller independent clusters, making it statistically less likely to find significant enrichment due to the small sample size.
Genes expressing subunits of protein complexes often tend to be co-expressed (Jansen et al., 2002; Krogan et al., 2006). We therefore used the developmental time course transcription profiling data sets from the modEncode project (Graveley et al., 2011) to examine the mRNA expression profile correlation between genes encoding interacting proteins. The frequency distribution of the correlation coefficients calculated between genes connected by DPiM edges is clearly skewed toward co-regulated expression when compared with all-to-all gene correlations (Figure 3B). Similarly, transcripts corresponding to the same MCL clusters tend to be co-expressed more frequently than those belonging to different clusters (Figure 3C). Aside from correlated profiles, it has been suggested that both the expression profiles and the absolute level of expression of interacting partners may be maintained at similar levels in the cell as a consequence of co-regulation of complex subunit stoichiometry (Jansen et al., 2002). Following Jansen et al, we calculated the normalized differences between absolute mRNA expression levels from the modEncode RNA-Seq data (Cherbas et al., 2011) and confirmed this trend in flies (Figure 3D). Similar results involving both expression profiling and absolute levels were obtained from analogous analysis of gene expression data from 26 Drosophila tissues in FlyAtlas (Chintapalli et al., 2007) (Supplemental Figure S2).
Cross-Species Validation of DPiM Interactions
Using orthologous HA-tagged human proteins as coAP-MS baits in Human Embryonic Kidney (HEK) 293 cell line (Graham et al., 1977), we examined if DPiM defined interactions can be validated across species. A set of 118 human bait proteins was selected based on whether an ORF clone was available in the CCSB human ORFeome collection (Lamesch et al., 2007; Rual et al., 2004), and if the corresponding Drosophila ortholog involved high HGSCore interactions in DPiM.
After Gateway-cloning of the corresponding ORF inserts into the pHAGE-N-Flag-HA vector (Behrends et al., 2010), we successfully cloned and affinity-purified 80% (94/118) of the baits, but the data set was too small to be analyzed by the HGSCore method. In DPiM, a total of 2,641 interactions involve Drosophila orthologs of one of these 94 human proteins. Transcriptome data of HEK-293 cells (Shaw et al., 2002; Williams et al., 2004) suggested that several human orthologs of interactors predicted DPiM are not expressed in this cell type. Therefore, the analysis was restricted to 114 DPiM interactions that are found as “bait-prey” interactors in the raw Drosophila data set for which both human orthologs are expressed in 293 cells; the success rate was 51% (58/114) (Supplemental Table S5). This validation rate illustrates the high specificity of our coAP-MS approach and the value of DPiM as a reliable resource for biological hypothesis in human cells. A total of 268 human validated DPiM interactions were novel (Supplemental Table S5). Examples of these cross-species validated interactions are considered further below.
Proteasome and SNARE Complexes
To further assess the quality of the DPiM at protein complex level, we performed an in-depth analysis of two previously well-characterized complexes: the proteasome and the SNARE complex. The proteasome is a large multi-protein complex involved in protein degradation and has been extensively characterized in a variety of organisms but little-studied in Drosophila (Holzl et al., 2000). We used the KEGG database (Kanehisa et al., 2010), FlyBase (Tweedie et al., 2009) and original literature to generate a list of 51 putative Drosophila proteasome subunits (described in Supplemental Table S6).
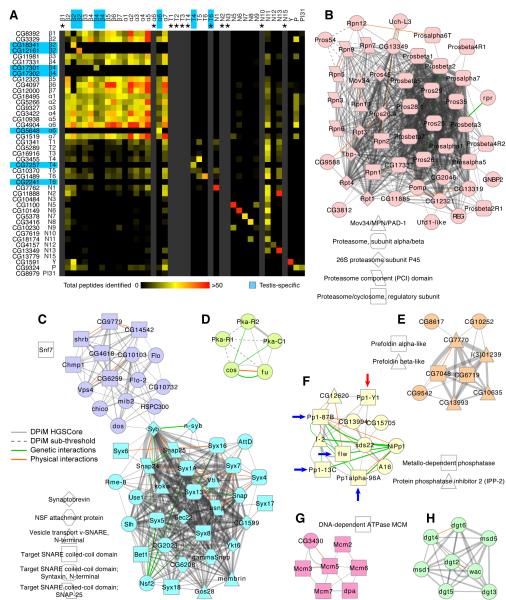
Affinity purification was performed for 32 individual proteasome subunits, and 42 of the 51 classified proteasome subunits were detected as co-purifying proteins in at least two bait purifications. On average, 70% of the co-purifying proteins are common between replicate proteasome bait purifications and 84% of the high confidence (DPiM) interactors were detected in both replicates (Supplemental Table S6). It is noteworthy that proteins predicted to be from the same proteasome substructure, i.e., core, base, or lid, consistently co-purified (Figure 4A). Consistent with yeast and human proteasome studies (Leggett et al., 2002; Wang et al., 2007), Rpn11 – a proteasomal lid subunit, pulled down the majority of the proteasome components. Consistent with its predicted role in maturation of the proteasome core (Fricke et al., 2007), the proteasome maturation protein (Pomp) co-purified with only a few core members (Figure 4A).
Figure 4. Biological implications of protein complexes in DPiM.
(A) Two-dimensional heat map showing the number of peptides identified for each proteasome subunit. Each column corresponds to proteins co-purified in a particular proteasome bait experiment. Grey columns (marked by asterisks) were added if a bait was unavailable. Both axes are arranged according to proteasome subunit classification, i.e., core (beta and alpha) or regulatory (base and lid). Seven testis-specific subunits are highlighted in blue. “P” refers to the Pomp protein. (B) The proteasome cluster in DPiM with subunits shaped according to Pfam/Interpro domains; circles represent nodes without domain enrichment. The thickness of the each grey line is proportional to the HGSCore of interaction. Additional physical (red lines) and genetic (green lines) evidence from literature are shown, with line thickness proportional to number of sources. (C) Clusters #7 and #162, the Snap/SNARE complex, connected by Syb to several members of Cluster #22, the Flotillin complex. (D) Cluster #117 includes proteins belonging to the Hedgehog signaling pathway. Protein Pka-R1 has interactions with HGSCores below threshold (dotted lines). (E) Cluster #42, the Prefoldin complex, all six predicted members are connected, along with three additional proteins, none of which are well studied. (F) Cluster #26, Protein Phosphatase type 1 complex has multiple genetic and physical interactions described in the literature. The known subunits PP1α87B, PP1α13C, PP1α96A and PP1β9C (blue arrows) and testis-specific subunit Pp1-Y1 (red arrow) are shown (G) Cluster #60, MCM (helicase) complex, has all six known members along with CG3430 (connected to Mcm3 and Mcm5). (H) Cluster #47, the Augmin complex, involved in mitotic spindle organization, is a standalone complex in the DPiM network. See also Figure S3 and Table S6.
Six of the 51 annotated proteasome subunits were detected only when they were used as baits. Interestingly, these were all recently described as testis-specific proteasome proteins (Belote and Zhong, 2009) and indeed, expression profiling analysis confirmed that they are not expressed in the Drosophila embryo-derived S2R+ cells (Cherbas et al., 2011). Nevertheless, when used as baits, the testis-specific proteins interacted with other proteasome components with profiles similar to those of their respective ubiquitous paralogs (Figure 4A). The fact that paralogous proteins produce similar interaction profiles illustrates the reproducibility of our coAP-MS approach and also suggests that DPiM provides valuable information that can reach beyond the S2R+ proteome.
Importantly, this study also uncovered a set of seven additional subunits not originally predicted to be part of the proteasome complex: CG12321, CG11885, CG2046, CG13319, GNBP2, CG3812, and RPR (Figure 4B). Sequence similarity analysis revealed that CG12321 and CG11885 are the Drosophila homologs of proteasome assembly chaperone 2 and 3 respectively (KEGG). Nothing is known about the functions of CG2046 or CG13319, and the sequences or domain structures of GNBP2, CG3812 and RPR do not suggest a plausible relationship to the proteasome. Direct experimentation will be essential to explore their functionality and potential role in the proteasome complex.
We next examined the SNARE [SNAP (Soluble NSF Attachment Protein) Receptor] complex. SNARE proteins are a large protein superfamily implicated in mediating membrane fusion events during protein trafficking (Sudhof and Rothman, 2009). In Drosophila, 23 SNARE proteins have been described (KEGG pathway: dme04130) and all of them are well connected in DPiM. All SNARE proteins with the exception of Syntaxin 6 fall into two clusters (Clusters #7 and #162; Figure 4C). Among nine proteins in Cluster #7 (Supplemental Table S4) that are not classified in KEGG as SNARE proteins, seven (Syb, Snap, Slh, gammaSnap, Syx13, CG6208 and Nsf2) have “SNAP receptor activity” or “SNAP activity” GO annotations and thus represent potential genuine interactors of the SNARE proteins. The remaining two proteins in Cluster #7 (AttD and Rme-8) do not have prior annotation related to SNAP receptor activity. We also found that Syb is linked to several proteins in the map, which suggests that it is a shared component of multiple complexes. Connections of particular interest are the ones that link Syb with members of Cluster #22 (the Flotillin complex), which is involved in protein transport and control of subcellular localization (Figure 4C). In total, 57 interactions (31 novel) from the SNAP/SNARE complex and 10 interactions (9 novel) from the Flotillin complex were independently validated in Human 293F cells (Supplemental Table S5).
The analyses of the proteasome and SNARE complexes confirm previously reported interactions, further validating the quality of the DPiM. Consequently, this also strengthens the potential of DPiM to formulate functional hypotheses at the levels of both pair-wise interactions and protein complex definition.
Functional Implications of DPiM
Slightly over half of the Drosophila protein-coding genes have associated experimental annotation (based on FlyBase release 5.23). Another 12% are annotated purely in silico [by Inferred Electronic Annotation (IEA)] and the remainder (1/3rd of protein-coding genes) has no functional annotation. DPiM provides the first empirical evidence and functional validation for 376 uncharacterized gene products and another 210 that were until now only annotated with IEA evidence. A total of 153 MCL clusters in the map show significant enrichment for GO terms, KEGG pathways and Pfam/InterPro domains (multiple hypothesis - adjusted P<0.01) indicating that members share common biological or functional attributes. These 153 annotation-enriched clusters include 167 proteins that lacked any annotation, for which DPiM provides functional associations and biological context (Supplemental Table S4). Inspection of individual protein complexes provides insights into specific as well as general functional aspects of the map. To illustrate this, six protein clusters with members sharing GO terms and pleiotropic cellular functions are described below (Figure 4).
The Hedgehog pathway is presumed to be “off” in the S2R+ cell line (Cherbas et al., 2011) but was represented by a few known pathway members (Pka-C1, Pka-R2, Cos and Fu) as an autonomous cluster (Figure 4D). Interestingly three of the four members of this cluster are protein kinases. Pka-R1 has only sub-threshold HGSCore interactions with members of this cluster (Figure 4D). Pka-C1, known to interact with the transcription factor Costa, was not detected in our analysis of S2R+ cells.
Eukaryotic prefoldin is a multi-subunit complex composed of two alpha and four beta subunits that are required for stabilization of nascent proteins as they are translated and delivered to chaperonins for protein folding (Ohtaki et al., 2010). The complex is not well characterized in flies and the subunits have been inferred from in silico approaches. This complex in DPiM (Figure 4E) contains all six components (CG7770, CG6719, l(3)01239, CG7048, CG13993, CG10635) as well as three additional putative complex members (CG9542, CG8617, and CG10252) (Figure 4E); essentially nothing is known about these proteins except for their sequences.
The complex related to Protein Phosphatase type 1 (PP1), one of the major classes of eukaryotic serine/threonine protein phosphatases (Dombradi et al., 1990) includes all four known catalytic subunits, PP1c’s, as well as the testis-specific subunit Pp1-Y1 (arrows in Figure 4F). In DPiM, this complex includes the two inhibitory subunits (I-2 and CG12620), two regulatory subunits (sds22 and A16). The two additional components CG15705 and CG13994 in this cluster were also found by Y2H analysis (Giot et al., 2003). Based mainly on Y2H interactions, it has been suggested that the Drosophila PP1c-interactome may include 40 putative PP1c-binding proteins (Bennett et al., 2006). Our coAP-MS analysis suggests that the PP1c complex in this cell type may be composed of fewer (twelve) proteins (Figure 4F).
The MCM (minichromosome maintenance 2-7) complex implicated in replication associated helicase activity is suggested to be composed of six proteins in Drosophila (Forsburg, 2004). DPiM defines a complex that contains all six as well as a seventh putative member, the uncharacterized protein CG3430 (Figure 4G).
The Augmin complex (Figure 4H), which is essential for spindle formation, has been defined through a series of biochemical studies, which in addition to the dgt protein core (dgt2-6), identified wac, msd1 and msd5 as members of the complex (Goshima and Kimura, 2010). The DPiM identified the Augmin complex in its entirety as a standalone cluster (Figure 4H). Additional examples of known protein complexes with diverse biological and molecular functions are shown in Supplemental Figure S3. The map also identified several IEA annotated proteins, which, while sharing GO terms, were not known to be members of a complex. For example: Cluster #166 (Supplemental Table S4) is made up of three members (CG12171, CG31549, CG31548) with a high average HGSCore (388). All three share a Glucose/ribitol dehydrogenase domain, a NAD(P)-binding domain, and Short-chain dehydrogenase/reductase (SDR) conserved sites. DPiM results suggest that these previously uncharacterized proteins form a functional complex. In contrast, DPiM also predicts the existence of complexes with members sharing experimentally derived annotation but no common GO terms (for example: Cluster #27, Supplemental Table S4).
Inter-Complex Interactions and Functional Relationships
The predictive value of DPiM for individual protein complexes is exemplified by the aforementioned analysis, but probing the interconnectedness of complexes within the map is far more challenging. On a global level, the interconnectedness of DPiM complexes is visualized in Supplemental Figure S4. In numerous cases, we observed that functionally related complexes are well connected in the map. For a better understanding of protein function, it is important to examine possible functional relationships that involve not only immediate complex neighbors, but also complexes that are associated with each other indirectly via intermediate protein assemblies.
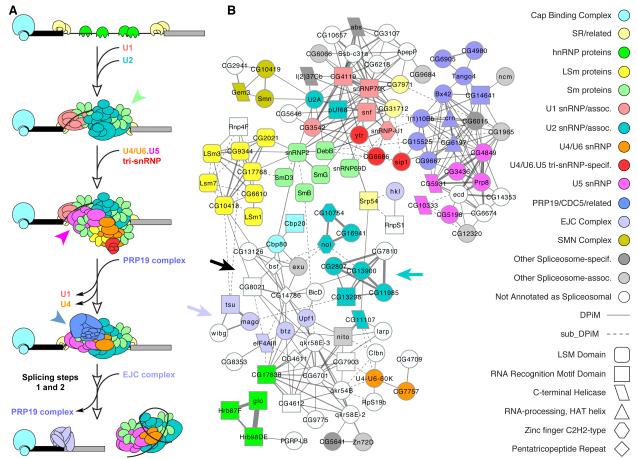
Given the level of functional characterization and modularity of the spliceosome, we chose it to examine whether functionally significant first- and second-degree neighboring interactions and clusters could be identified in DPiM. The conformation and composition of the spliceosome is highly dynamic and is responsible for the accuracy as well as the flexibility of the splicing machinery. It is composed of several well-defined snRNPs that associate sequentially with pre-mRNA to guide intron splicing (Figure 5A). Each snRNP consists of one or two snRNAs, a common set of seven Sm (or LSm) proteins, and a variable number of unit-specific proteins (Will and Luhrmann, 2010).
Figure 5. Modularity of the Spliceosome subnetwork.
(A) Schematic representation of step-wise interaction of snRNPs with pre-mRNA and other proteins in the process of splicing introns, as described in the literature. (B) The spliceosome subnetwork in DPiM consists of a dozen clusters that are well connected. The ~80 nodes in this subnetwork constitute a very substantial portion of the spliceosome pathway as defined in KEGG (pathway: dme03040) and (Herold et al., 2009). The major spliceosome sub-complexes are colored according to functional annotation (same as in A for comparison) and proteins are shaped according to Pfam domain enrichment. Protein interactions are shown as grey lines with thicknesses proportional to HGSCore and those with scores below the statistical cut-off are shown as dotted lines. Other proteins that are not classified as spliceosome components in KEGG or elsewhere but connected to these complexes in the DPiM network are uncolored. A majority of such non-spliceosomal proteins have “mRNA binding” annotation. The modularity of this multi-subunit molecular machinery is preserved in DPiM in the form of subnetworks that cluster together. Colored arrows and arrowheads denote complexes referred to in the text.
The spliceosome subnetwork in DPiM (Figure 5B), is composed of a dozen clusters containing most of the known spliceosome-related proteins. This clustering of spliceosome components in an unbiased systematic analysis of whole cell lysates illustrates the power of our approach. Importantly, these spliceosome clusters are interconnected in the network, consistent with the notion that they share functionality, while remaining spatially and temporally modular. The complex defined by the six Sm proteins (green arrowhead Figure 5A) is connected to other first-degree and second-degree neighboring clusters composed of specific U1, U2, U4, U5 and U6 related factors. Most Prp19/CDC5L complex members (magenta arrowhead, Figure 5A) are well connected to all U5 specific factors (blue arrowhead, Figure 5A and Figure 5B). Similarly, the U2 snRNP-specific factors (CG2807, CG7810, CG13900, CG13298, CG11985, cyan arrow Figure 5B) and members of Exon Junction Complex (EJC, blue gray arrow, Figure 5B) are connected to Sm/LSm proteins via CG14786 (Figure 5B) and other members of Cluster #62 (black arrow, Figure 5B). Although none of the Cluster #62 members are classified spliceosome components, two are predicted as members of EJC (Upf1 and btz) and two others (CG8021 and bsf) have GO term annotation related to mRNA binding (not enriched at P<0.01). Thus a second-degree neighboring cluster defines functionally related protein assemblies in DPiM.
Protein Complex Evolution
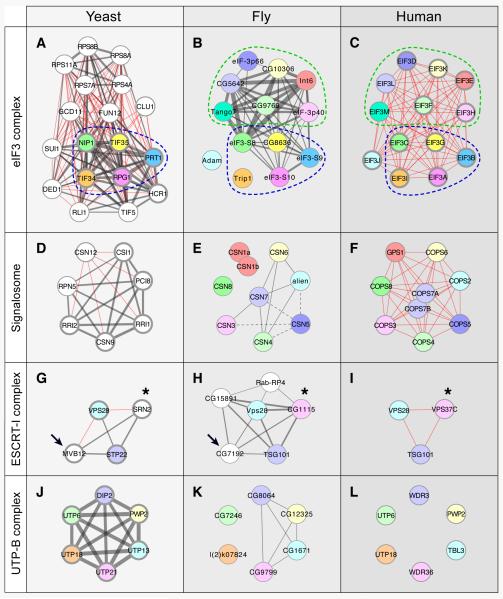
Examining the extent of conservation of individual protein subunits as well as the overall complex composition across organisms can shed valuable insight into their cellular roles. The most extensive manually curated annotations of protein complexes exist for yeast (MIPS, CYC2008) and human (REACTOME, CORUM). We aligned complexes defined by DPiM clusters with those described in yeast and human. Several complexes, for example: MCM (Figure 4G, Cluster #60), CCT (chaperonin containing TCP1, Supplemental Figure S3, Cluster #32) and prefoldin (Figure 4E, Cluster #42)] showed almost complete conservation of composition between clearly orthologous subunits. Below, we focus on examples where orthology relationships are less obvious (Figure 6).
Figure 6. Examples of protein complex evolution.
Comparison of four complexes defined in fly by DPiM (center panels) with yeast (left panels) and human complexes (right panels). Grey lines show physical interactions that have weighted scores and red lines show interactions implied by the curated data sets. For comparison, Inparanoid orthologs in all three species are depicted with identical colors. Proteins that do not have homologs in other species are shown in white. Complex members for which evidence exists in both high-throughput and curated datasets (yeast) or both REACTOME and CORUM databases (human) are distinguished by thicker nodes (A-C) The eIF3 complex (Cluster #24). The fly and human complexes share seven interconnected proteins (within green dotted region), which are not present in yeast. Five proteins are conserved in all three species (within blue dotted region). (D) The signalosome complex in yeast is composed of proteins sharing little sequence similarity with metazoan counterparts. The eukaryotic signalosome is composed of eight subunits (CSN1-8) as seen in the human complex (F) but CSN 1 (a and b) and CSN8 are not part of the fly signalosome in S2R+ cells. (E). ESCRT-I function is conserved from yeast to humans, but only VPS28 and STP22 in yeast and their respective fly and human orthologs are readily apparent (G-I). Additional analysis suggests a distant relationship between MVB12 in yeast and Drosophila complex member CG7192, a protein of unknown function (arrows). The yeast SRN2 also shares the Mod_r domain with CG1115 and VPS37C (asterisks). (J) The yeast UTP B complex involved in RNA processing has six well-connected members. (K) In DPiM only four members are connected but CG7246 and l(2)kO7824 are not included in the DPiM Cluster #160. (L) There is no evidence suggesting physical interaction among the complex members in human.
The eIF3 complex defines the largest eukaryotic initiation factor, which directs the multitude of steps essential for initiating translation. Comparison of the complexes from yeast and human to that of Drosophila (Cluster #24, DPiM) reveals significant differences. The metazoan Drosophila and human complexes share seven interconnected proteins (Figure 6, A-C, within green-dotted region), which are not present in unicellular yeast, suggesting structural and functional remodeling specific to multicellular organisms. A group of four interconnected proteins is conserved in all three species (Figure 6, A-C, within blue-dotted region). Neither the raw data nor the HGSCore analysis supports Trp1 or Adam being part of the eIF3 complex, though their homologs are predicted to be members in other species. These findings allow us to raise the testable hypothesis that the role of yeast and human orthologs of Adam and Trip1 are not essential to the function of eIF3. We also compared Pfam domain compositions across the three species, revealing a gain of six domains in the metazoans in comparison to yeast and the loss of an unclassified domain in yeast with respect to metazoans (Supplemental Table S7A). It is worth noting that none of the eIF3 complex members were used as bait; its recovery illustrates the power of our scoring approach.
The signalosome is a functionally conserved complex that catalyzes the deneddylation of proteins and promotes degradation through the cullin family of ubiquitin E3 ligases (Kato and Yoneda-Kato, 2009). Yeast proteins share surprisingly little sequence similarity with metazoan counterparts, despite the fact that the yeast complex has been shown to be functionally homologous to metazoan signalosomes (Wee et al., 2002) (Figure 6D). The eukaryotic signalosomes are composed of eight subunits (CSN1-8) as seen in the human complex (Figure 6F). The Drosophila signalosome has also been suggested to comprise eight subunits (Freilich et al., 1999) but our coAP-MS data raise the possibility that CSN1a, CSN1b and CSN8 are not part of the complex, at least in S2R+ cells (Figure 6E). Domain analysis shows a linear growth in the number of PCI domains from yeast to humans, which cannot be attributed to the growth in the number of protein subunits (Supplemental Table S7B).
The three member ESCRT-I (endosomal sorting complex required for transport) complex is well known in flies and humans (Michelet et al., 2010) (Figure 6, G-I). In DPiM, the ESCRT-I complex clustered with three other proteins that have no human homologs according to InParanoid (Figure 6H). The yeast complex shows some interesting characteristics. First, Vps28 is linked to STP22, a conserved interaction also evident in Drosophila and humans. On the other hand, MVB12, a multivesicular body associated protein in yeast (arrow, Figure 6G) does not have a clear fly ortholog nor does it share a Pfam domain with any of the fly complex components. However, the Drosophila complex member CG7192, a protein of unknown function (arrow, Figure 6H) shares weak sequence similarity with the Caenorhabditis elegans protein C06A6.3, which has recently been shown to be functionally homologous to the yeast MVB12 (Audhya et al., 2007). Moreover, the yeast SRN2, while not identified as an ortholog of any metazoan gene, shares the Mod_r Pfam domain with fly CG1115 as well as human VPS37C (marked by asterisks, Figure 6, G-I), suggesting a weak evolutionary relationship.
Cluster #160 in DPiM links four proteins associated with UTP-B complex, a subcomplex of the SSU processome, a large ribonucleoprotein essential for RNA processing (Figure 6K). In yeast, two additional proteins (UTP6 and UTP18) are clearly part of this complex, but the corresponding proteins in Drosophila (CG7246 and l(2)kO7824) are not included in Cluster #160 (Figure 6, J-K). Both these proteins have been used as baits in the coAP-MS analysis and they did not co-purify other UTP complex members. Though the homologous proteins exist in humans, neither the interactions nor the complex have been extensively studied. The contrast of evolutionary information between yeast and fly provides an entry point for further investigation to see which of the interactions have been lost or retained in humans.
DISCUSSION
Understanding how functional units in the cell integrate their actions to control development and homeostasis defines a quintessential biological problem. Essential insights into this come from the definition of proteome architecture such as the map we present here, enabled by the knowledge of genome sequences and the development of sensitive mass spectrometry-based approaches. Though there are several studies focused on specific sub-proteomic spaces, no large-scale unbiased proteome map exists for higher eukaryotes [see review (Gavin et al., 2011)]. Our study defines a global metazoan protein complex network based on expression of a large library of affinity tagged baits. The map includes a majority of proteins expressed in S2R+ cells and is based on the HGSCore which includes a semi-quantitative measure of protein abundance (TSCs) thus improving the sensitivity in comparison to other existing scoring methods. However, we note that several known interactions are detected in our analysis but fall below the statistical threshold (Supplemental Table S3).
Several independent criteria indicate that the quality of the map is high, and clearly the algorithms we use successfully clustered proteins that have been grouped previously as multimeric complexes. The broad recovery of known interactions and the remarkable enrichment of GO terms in individual clusters suggests that novel interactions predicted by DPiM define important biological hypotheses as well as a powerful annotation tool. The analysis of the human protein orthologs we tested indicates that DPiM reflects general features of metazoan proteomes and thus will be directly useful in probing protein interactions across species. We expect that the experimental and analytical resources we established will be useful as the proteome analysis is expanded to include additional Drosophila proteins and cells lines or tissues and provide a paradigm for proteomic studies in other organisms.
DPiM, like its yeast counterparts (Gavin et al., 2006; Krogan et al., 2006), defines protein complex membership and suggests inter-complex relationships linking together functional units. Both issues are essential for understanding the network of functional relationships that govern the physiology of the cell. First, it Experimentally probing such relationships is not trivial but the availability of sophisticated genetic tools in Drosophila offers a unique opportunity to explore interactions using in vivo assays. Indeed, 118 of the DPiM direct interactions have been validated independently through genetic interactions involving mutant combinations (see FlyBase). Integration of protein and genetic interaction networks will afford us important insights that may provide a molecular basis for relationships only defined by genetics and hence generate mechanistic hypotheses.
The experimental approach we used has certain a priori limitations. We rely on the transient expression of epitope-tagged bait proteins, which are not expressed normal levels, and tagging of the proteins may interfere with their functions. Nevertheless, the quality testing of the map indicates that despite these potential limitations our experimental approach is generally reliable. We also note that several recent studies of sub-proteomic spaces using a similar experimental approach have produced valid results (Behrends et al., 2010; Sowa et al., 2009). Any cell type used will inevitably involve only a fraction of the predicted proteome and expanding the analysis to different cell lines and tissues in the future will improve the overall proteomic coverage and define possible tissue-specific aspects of the map. We presume that some of the baits that failed to produce high quality coAP-MS results may be due to interference of a C-terminal tag with protein function. For the future, we note that the C-terminally tagged baits have also been tagged at the N-terminus [(Yu et al., 2011) http://www.fruitfly.org/EST/proteomics.shtml], possibly circumventing such inactivation.
The evolutionary comparisons illustrated in Figure 6 provide valuable means to explore gene function and to recognize functionally important protein interactions implied by the map. Examining the evolution of protein complex architecture across species can help establish or confirm distant orthologous relationships and improve annotation of orphan genes. The extent of protein conservation is linked to their ability to interact with other proteins, the nature of interactions and how essential a protein function is for the cell (Mintseris and Weng, 2005; Wuchty, 2004). Our data support models of protein network evolution that are driven by the acquisition or loss of protein complex members rather than re-wiring of existing components (van Dam and Snel, 2008; Yamada and Bork, 2009). A more detailed structural analysis will be necessary to examine the subunit interactions in those complexes where the level of conservation is low.
DPiM establishes a singular resource and a baseline to explore dynamic properties of the protein interaction network in a metazoan proteome. It also enables the analysis of specific subproteomic spaces at greater depth. It is now possible to examine if and how the protein complex relationships derived from S2R+ cells change in different developmental or genetic backgrounds. To promote such studies, we are producing transgenic fly lines carrying the same FLAG-HA tagged version of the proteins under the control of a UAS promoter. The expression of tagged proteins can be spatio-temporally regulated by the use of different Gal4 drivers. Exploring the dynamic nature of the protein complex network defined here, enhanced through the use of quantitative mass spectrometry, will be of fundamental value and will likely provide system-wide insights into the molecular defects underlying pathogenic conditions. We expect that analogies of protein interaction relationships between Drosophila and humans will be helpful in the analysis of disease-related pathways and indeed the identification and evaluation of disease-related targets.
EXPERIMENTAL PROCEDURES
Cloning, Expression and Purification
Open reading frames were transferred from the BDGP Drosophila melanogaster expression-ready clone set to the pMK33-C-FLAG-HA acceptor vector (Yu et al., 2011). Each clone was transiently transfected into a 54 ml culture of Drosophila S2R+ cells. Protein expression was induced with 0.35mM CuSO4 and whole-cell lysates prepared in Lysis Buffer (25 mM NaF, 1 mM Na3VO4, 50 mM Tris pH 7.5, 1.5 mM MgCl2, 125 mM NaCl, 0.2% IGEPAL, 5% glycerol and Complete™). Each clarified lysate was bound overnight to 75 μl of cross-linked immunoaffinity resin (Sigma). Unbound proteins were washed off with Lysis Buffer followed by PBS and then bound protein complexes were competitively eluted using synthetic HA peptide YPYDVPDYA (250 μg/ml) in PBS.
Mass Spectrometry and Data Analysis
The co-purified proteins were precipitated using trichloroacetic acid, washed with acetone, dried, digested overnight with trypsin and analyzed by LC-MS/MS. The spectral data was searched with SEQUEST (Eng et al., 2008) against a database of D. melanogaster proteins derived from FlyBase version 5.23. The LC-MS/MS identifications were filtered to, on average, a 1.2% protein FDR and 0.3% peptide FDR. The compiled data set was filtered to a combined 0.8% FDR and further post-processing was used to correct for column carry-over issues.
Bioinformatic Analysis
Both bait-prey and prey-prey protein interactions from coAP-MS data were analyzed and scored using HGSCore – a hypergeometric distribution error model, incorporating total spectral counts (TSC) to improve the accuracy of co-occurrence prediction. A randomized data set of similar size was created to estimate false discovery rate. Protein interactions were clustered using MCL (Enright et al., 2002). Other algorithms were implemented as described in original literature. Additional details are provided in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from the National Institutes of Health (NIH 5RO1HG003616) to SAT and a fellowship from the Deutsche José Carreras Leukämie-Stiftung e. V. to JFR. Generation of the clone set was supported by a grant from the National Human Genome Research Institute (NHGRI P41HG3487 to SEC). Special thanks to Anne-Claude Gavin, Bertrand Kuster and Charlie Cohen whose help was critical in the initiation of the project, as well as Gerry Rubin for his help throughout. We thank Norbert Perrimon for S2R+ cells, Lucy and Peter Cherbas for their generous help in cell culture, William Gelbart and the FlyBase team for making DPiM data available on FlyBase, and Alexey Veraksa, Ashim Mukherjee, Kadalmani Krishnan, Mathew Sowa, Dan Finley, Robin Reed, Angeliki Louvi and members of the Artavanis-Tsakonas, Celniker, Gygi and VijayRaghavan labs for helpful discussion and comments. We thank members of CCSB, Vidal and Harper labs, David Hill and Eric Bennett in particular, for help with the human ORFeome collection. We also thank Manolis Kellis and Rogerio Candeias for their help.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
Supplemental Data including four figures, seven tables and Supplemental Experimental Procedures can be found with this article online.
REFERENCES
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Audhya A, McLeod IX, Yates JR, Oegema K. MVB-12, a fourth subunit of metazoan ESCRT-I, functions in receptor downregulation. PLoS ONE. 2007;2:e956. doi: 10.1371/journal.pone.0000956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belote JM, Zhong L. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity. 2009;103:23–31. doi: 10.1038/hdy.2009.23. [DOI] [PubMed] [Google Scholar]
- Bennett D, Lyulcheva E, Alphey L, Hawcroft G. Towards a comprehensive analysis of the protein phosphatase 1 interactome in Drosophila. J Mol Biol. 2006;364:196–212. doi: 10.1016/j.jmb.2006.08.094. [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombradi V, Axton JM, Brewis ND, da Cruz e Silva EF, Alphey L, Cohen PT. Drosophila contains three genes that encode distinct isoforms of protein phosphatase 1. Eur J Biochem. 1990;194:739–745. doi: 10.1111/j.1432-1033.1990.tb19464.x. [DOI] [PubMed] [Google Scholar]
- Eng JK, Fischer B, Grossmann J, Maccoss MJ. A fast SEQUEST cross correlation algorithm. J Proteome Res. 2008;7:4598–4602. doi: 10.1021/pr800420s. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, Chamovitz DA. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–1190. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- Fricke B, Heink S, Steffen J, Kloetzel PM, Kruger E. The proteasome maturation protein POMP facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO Rep. 2007;8:1170–1175. doi: 10.1038/sj.embor.7401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gavin AC, Maeda K, Kuhner S. Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr Opin Biotechnol. 2011;22:42–49. doi: 10.1016/j.copbio.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Goshima G, Kimura A. New look inside the spindle: microtubule-dependent microtubule generation within the spindle. Curr Opin Cell Biol. 2010;22:44–49. doi: 10.1016/j.ceb.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C, Milenkovic T, Przulj N, Kaiser P, Huang L. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc Natl Acad Sci U S A. 2008;105:13333–13338. doi: 10.1073/pnas.0801870105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GT, Lee I, Marcotte ER. A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality. BMC Bioinformatics. 2007;8:236. doi: 10.1186/1471-2105-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Luhrmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol. 2009;29:281–301. doi: 10.1128/MCB.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Holzl H, Kapelari B, Kellermann J, Seemuller E, Sumegi M, Udvardy A, Medalia O, Sperling J, Muller SA, Engel A, et al. The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol. 2000;150:119–130. doi: 10.1083/jcb.150.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Janga SC, Babu M, Diaz-Mejia JJ, Butland G, Yang W, Pogoutse O, Guo X, Phanse S, Wong P, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biol. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato JY, Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells. 2009;14:1209–1225. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Michelet X, Djeddi A, Legouis R. Developmental and cellular functions of the ESCRT machinery in pluricellular organisms. Biol Cell. 2010;102:191–202. doi: 10.1042/BC20090145. [DOI] [PubMed] [Google Scholar]
- Mintseris J, Weng Z. Structure, function, and evolution of transient and obligate protein-protein interactions. Proc Natl Acad Sci U S A. 2005;102:10930–10935. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali T, Pacifico S, Yu J, Guest S, Roberts GG, 3rd, Finley RL., Jr. DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res. 2011;39:D736–743. doi: 10.1093/nar/gkq1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtaki A, Noguchi K, Yohda M. Structure and function of archaeal prefoldin, a co-chaperone of group II chaperonin. Front Biosci. 2010;15:708–717. doi: 10.2741/3641. [DOI] [PubMed] [Google Scholar]
- Rees JS, Lowe N, Armean IM, Roote J, Johnson G, Drummond E, Spriggs H, Ryder E, Russell S, Johnston DS, et al. In Vivo Analysis of Proteomes and Interactomes Using Parallel Affinity Capture (iPAC) Coupled to Mass Spectrometry. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002386. M110 002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, Vidalain PO, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, Zhang H, Zhong J, Finley RL., Jr. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Sudhof TC, Rothman JE. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Dam TJ, Snel B. Protein complex evolution does not involve extensive network rewiring. PLoS Comput Biol. 2008;4:e1000132. doi: 10.1371/journal.pcbi.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- Wee S, Hetfeld B, Dubiel W, Wolf DA. Conservation of the COP9/signalosome in budding yeast. BMC Genet. 2002;3:15. doi: 10.1186/1471-2156-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome Structure and Function. Cold Spring Harb Perspect Biol. 2010 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RD, Hing SN, Greer BT, Whiteford CC, Wei JS, Natrajan R, Kelsey A, Rogers S, Campbell C, Pritchard-Jones K, et al. Prognostic classification of relapsing favorable histology Wilms tumor using cDNA microarray expression profiling and support vector machines. Genes Chromosomes Cancer. 2004;41:65–79. doi: 10.1002/gcc.20060. [DOI] [PubMed] [Google Scholar]
- Wuchty S. Evolution and topology in the yeast protein interaction network. Genome Res. 2004;14:1310–1314. doi: 10.1101/gr.2300204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Bork P. Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol. 2009;10:791–803. doi: 10.1038/nrm2787. [DOI] [PubMed] [Google Scholar]
- Yu C, Wan KH, Hammonds AS, Stapleton M, Carlson JW, Celniker SE. Development of expression-ready constructs for generation of proteomic libraries. Methods Mol Biol. 2011;723:257–272. doi: 10.1007/978-1-61779-043-0_17. [DOI] [PubMed] [Google Scholar]
- Yu J, Pacifico S, Liu G, Finley RL., Jr. DroID: the Drosophila Interactions Database, a comprehensive resource for annotated gene and protein interactions. BMC Genomics. 2008;9:461. doi: 10.1186/1471-2164-9-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.