Abstract
Oxidation of DNA due to exposure to reactive oxygen species is a major source of DNA damage. One of the oxidation lesions formed, 5-hydroxy-2'-deoxycytidine, has been shown to miscode by some replicative DNA polymerases but not by error prone polymerases capable of translesion synthesis. The 5-hydroxy-2'-deoxycytidine lesion is repaired by DNA glycosylases that require the 5-hydroxycytidine base to be extrahelical so it can enter into the enzyme's active site where it is excised off the DNA backbone to afford an abasic site. The thermodynamic and NMR results presented herein, describe the effect of a 5-hydroxy-2'-deoxycytidine•2'-deoxyguanosine base pair on the stability of two different DNA duplexes. The results demonstrate that the lesion is highly destabilizing and that the energy barrier for the unstacking of 5-hydroxy-2'-deoxycytidine from the DNA duplex may be low. This could provide a thermodynamic mode of adduct identification by DNA glycosylases that require the lesion to be extrahelical.
The inadvertent oxidation of DNA is one of the most common types of DNA damage that occurs in cells, with thousands of oxidized DNA base lesions formed per cell per day.1–7 The most common oxidative modification is 8-oxoguanine (oxoG), which can interfere with DNA replication and cause an increase in mutations.8–17 Other sites of attack are at adenine and cytosine. In the case of the latter, 2'-deoxycytidine (dC) can be transformed into 5-hydroxy-2'-deoxycytidine (HO-dC) by 1e− oxidation and subsequent attack by water on the intermediate radical cation, or via dehydration of 5,6-dihydroxy-2'-deoxycytidine, another oxidation product that forms in cells.18–20 In turn, HO-dC can afford 5-hydroxy-2'-deoxyuridine (HO-dU) via deamination.
In vitro replication of DNA with a HO-dC residue in the template strand is prone to errors based upon the sequence context 21 so the modified base represents a potential promutagenic lesion. Other studies in E. coli and S. cerevisiae imply that a single-stranded template with a HO-dC lesion is read correctly, i.e., it codes for dG. 22,23 It has also been reported that the error prone polymerase Polι bypasses HO-dC without miscoding (24). In most studies, the deamination product, HO-dU, is dominant in terms of toxicity and mutagenicity derived from the initial formation of HO-dC. 21–23
Recently, the crystal structure of HO-dC in DNA has been reported in the context of the structural impact of the lesion on DNA polymerization by the bacteriophage RB69 polymerase.25 The study shows that HO-dC in the template strand can stabilize the incoming dGMP via canonical Watson-Crick base pairing, while incorporation of dAMP opposite the lesion leads to destabilizing unstacking. Similar to what has previously been reported, the misincorporation of damp opposite HO-dC occurs at 5-times the rate for dC.
Because the repair of HO-dC by different DNA repair glycosylase proteins requires the lesion to rotate out of the double helix and into the enzyme's active site,26–30 we initiated an investigation on the thermodynamic effects of HO-dC on DNA stability with a specific interest in whether it remains stably stacked in the double helix. We were also interested to see how the presence of a polar hydroxyl group in the major groove would affect the hydration of DNA and the association of cations. Both play an important role in the enthalpic stabilization of duplex DNA. The thermodynamic and NMR results show a remarkable destabilizing effect for the HO-dC•dG base pair and that it may readily adopt an extrahelical conformation, which may facilitate its initial recognition by DNA repair proteins. This endothermic change in free energy due to the HO-dC modification is accompanied by a large reduction in the release of cations and structural water from the DNA upon unfolding.
MATERIALS AND METHODS
Materials
The oligodeoxynucleotides were synthesized and HPLC purified by Invitrogen (Frederick, MD). The samples were desalted by gel-permeation chromatography, using a Sephadex G-25 column, lyophilized to dryness and characterized by MALDI-TOF-MS. The dry oligomers were annealed by dissolving the single-stranded oligodeoxynucleotides in appropriate buffer, heating the solution up to 90 °C for 10 min and allowing it to cool down slowly to room temperature.
The concentration of the oligomer solutions was determined at 260 nm and 80 °C using an extinction coefficient ~ 1.11 × 105 M−1 cm−1 (ODNs 1–4) and 1.08 × 105 M−1cm−1 (ODN5) at 260 nm and 25 °C assuming similar extinction coefficients for 5-OH-dC and dC. This value is obtained from the molar absorptivity at 25 °C, obtained from the tabulated values of the dimers and monomer bases,31 and extrapolated to high temperatures using the upper portions of the UV melting curves, following procedures described earlier.32 Stock solutions of different pH were prepared from 100 mM sodium phosphate buffer (100 mM mono- and dibasic forms) adjusted to the appropriate pH using either mono- or dibasic sodium phosphate solutions and diluted to 10 mM concentration when required. Na+ and osmolyte concentrations were adjusted using NaCl solution and ethylene glycol, respectively. All solutions were filtered through a 0.20 μm filter (Alltech Associates, Inc) and degassed before use.
Temperature-dependent UV-Spectroscopy
Absorption versus temperature profiles (UV melts) for each duplex were measured at either 260 or 275 nm using a Varian Cary 300 spectrophotometer (Palo Alto, CA) equipped with a Peltier temperature controller and interfaced with a computer for data acquisition and analysis. The temperature was scanned at heating rates of 1.00 °C/min. Melting curves as a function of strand concentration, 4–50 μM, were obtained to check for the molecularity of each molecule. Additional melting curves were obtained as a function of pH, salt and osmolyte concentration to determine the differential binding of counterions and water molecules that accompanies their helix→coil transitions.
UV melts were measured in the salt range of 10–200 mM NaCl at neutral pH, and at a constant total strand concentration of 5 μM, to determine the differential binding of counterions, ΔnNa+, which accompanied their helix–coil melting. This linking number was measured experimentally with the assumption that counterion binding to the helical and coil states of each oligonucleotide took place with a similar type of binding using the relationship;33
| Eq. 1 |
The numerical factor corresponded to the conversion of ionic activities into concentrations. The first term in parentheses, (ΔHcal/RTM2), was a constant determined directly from DSC experiments, where R is the gas constant. The second term in parenthesis was determined from UV experiments from the dependencies of TM on salt concentration.
For the determination of Δnw, UV melts were measured in the ethylene glycol concentration range of 0.5 – 3.0 m at pH 7.0 and 10 mM NaCl and at a constant total strand concentration of 5.0 μM. The osmolalities of the solutions were obtained with a Wescor Vapro vapor pressure osmometer, Model 5520 (Logan, UT). These osmolalities were then converted into water activities, aw, using the following equation:34
| Eq. 2 |
where Osm is the solution osmolality and Mw is the molality of pure H2O, equal to 55.5 mol/kg H2O. Differential binding of water, ΔnW, was calculated using the relationship:33
| Eq. 3 |
The ΔHcal/RTM2 term used in the determination of Δnw at higher salt concentration is the one obtained experimentally at the particular salt concentration.
Differential Scanning Calorimetry
All calorimetric experiments were carried out using a VP-DSC differential scanning calorimeter (Microcal, Inc., Northampton, MA). The dry oligodeoxynucleotides were dissolved in 10 mM sodium phosphate buffer (pH 7.0) and adjusted to the desired ionic strength with NaCl for all unfolding experiments. In a typical DSC experiment ~ 0.75 ml of a dilute aqueous solution of oligonucleotide (125–200 μM) was loaded into a sample cell and a matched reference buffer solution loaded into a reference cell. Each solution was thermally scanned from 0–100 °C at a constant heating rate of 45 °C/hour over five forward scans. The DSC melting curves were normalized by the heating rate, and a buffer versus buffer scan was subtracted and normalized for the number of moles. The resulting curves were then analyzed with Origin version 7.0 (Microcal); their integration (∫ΔCp dT) yielded the molar unfolding enthalpy (ΔHcal), which was independent of the nature of the transition.35,36 The molar entropy (ΔScal) was obtained similarly, using ∫(ΔCp/T) dT. The Gibb's free energy change at any temperature T was then obtained with the Gibbs equation: ΔG°(T) = ΔHcal − TΔScal.
Circular Dichroism
Circular dichroism (CD) spectra were recorded on a Jasco (model J-815) CD spectrometer (Easton, MD, USA) equipped with a Peltier device and nitrogen purging capabilities. The spectrum of each duplex was obtained using a strain-free 1 cm quartz cell at low temperatures to ensure 100% duplex formation. Data was collected at 4 and 90 °C. Typically, 1 OD of a duplex sample was dissolved in 1 mL of a buffer containing 10 mM sodium phosphate (pH 7.0). After equilibration for 5 min at each sample temperature, the instrument collected spectral data in the 220 to 350 nm range every 1.0 nm.
NMR Studies
Samples for the observation of exchangeable protons were dissolved to a duplex concentration of 81 nM in 180 μL of 10mM NaH2PO4, 100 mM NaCl, 50 μM Na2EDTA buffer (pH 7.0) containing 9:1 H2O/D2O (v/v). One- and two-dimensional NMR experiments were performed on a Bruker Avance spectrometer operating at 900 MHz. Chemical shifts were referenced to the water resonance. NMR data were processed using TOPSPIN software (3.0, Bruker Inc., Karlsruhe, Germany). 1D NMR spectra for the exchangeable protons were recorded at 5, 15, 25, 35, 45, 55 and 60 °C. The 1H-1H NOESY3 spectra of unmodified and modified samples in H2O were collected at 5 °C, with 70 and 250 ms mixing times and relaxation delay of 2.0 s. These experiments were recorded using a field gradient Watergate pulse sequence4 for water suppression.
RESULTS
The HO-dC lesion, which is commercially available as the protected phosphoramidite, was incorporated into two different self-complementary oligodeoxynucleotide (ODN) sequences (Table 1). All ODNs were purified by HPLC and analyzed by MALDI-TOF to demonstrate purity and identity. ODN-1 and -2 are based on the well-studied Drew Dickerson dodecamer37 that has G/C rich termini and an A/T rich central core. ODN-3 and -4 lack the A/T central core, which minimizes the formation of hairpin structures that tend to form in solutions of ODN-1 under certain conditions.38
Table 1.
Standard thermodynamic profiles for 5-HO-dC (X) modified DNA at pH 7.0 in 10 mM sodium phosphate buffer.
| ODN | Sequence | NaCl (mM) | TM (°C) | ΔH (kcal/mol) | ΔG° (kcal/mol) | TΔS (kcal/mol) | ΔNNa+ (mol-1 DNA) | Δnw (mol-1DNA) |
|---|---|---|---|---|---|---|---|---|
| 1 | CGCGAATTCGCG GCGCTTAAGCGC |
10 | 33.3 | −116.0 | −6.9 | −109.0 | −2.3 ± 0.15 | −38.0 ± 2.0 |
| 2 | CGCGAATTXGCG GCGXTTAAGCGC |
10 | 31.5 | −74.3 | −2.8 | −71.5 | −1.0 ± 0.11 | −21.0 ± 3.0 |
| 3 | GAGAGCGCTCTC CTCTCGCGAGAG |
10 | 41.3 | −78.2 | −6.9 | −71.3 | −3.4 ± 0.2 | −41.0 ± 3.0 |
| 4 | GAGAGCGCTXTC CTXTCGCGAGAG |
10 | 15.0 | −41.7 | −0.7 | −41.0 | −2.3 ± 0.15 | −13.0 ± 1.0 |
| 5 | GACAGCGCTCTC CTCTCGCGACAG |
10 | 8.5 | −31.1 | +0.2 | −31.3 | n.d. | n.d. |
Oligomer concentration = 10 μM
CD
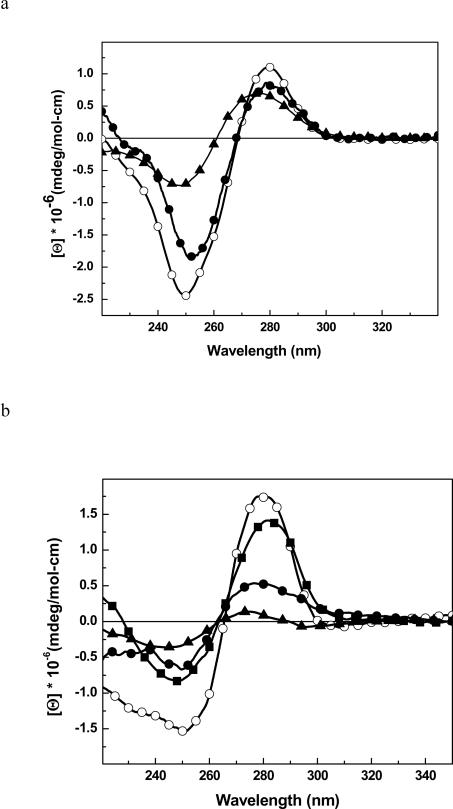
To confirm that the global conformation of the HO-dC modified DNAs, the CD spectrum of ODN-2 was obtained and compared to the control ODN-1 (Figure 1a). Both duplexes show an overall normal B-DNA conformation, but there is a modest reduction in the intensity of the negative band near 250 nm in ODN-2 that is indicative of reduced base stacking.39 A similar result is seen with ODN-4 vs. ODN-3 (Figure 1b). For comparison purposes, we synthesized an analogous self complementary DNA sequence (ODN-5) with a dC•dC mismatch in place of the HOdC•dG pairing and determined its CD spectrum at low temperature (Figure 1b). The intensities of the positive and negative bands fall midway between the unmodified DNA and the DNA with the HOdC•dG base pair.
Figure 1.
CD spectra of: (a) ODN-1 (black line, ④), ODN-2 (③) and ODN-1 (▲) heated to 90 °C; (b) ODN-3 (④), ODN-4 (③), ODN-5 (■) and ODN-3 (▲) heated to 90 °C.
Thermodynamic Characterization of DNA with HO-dC Lesions
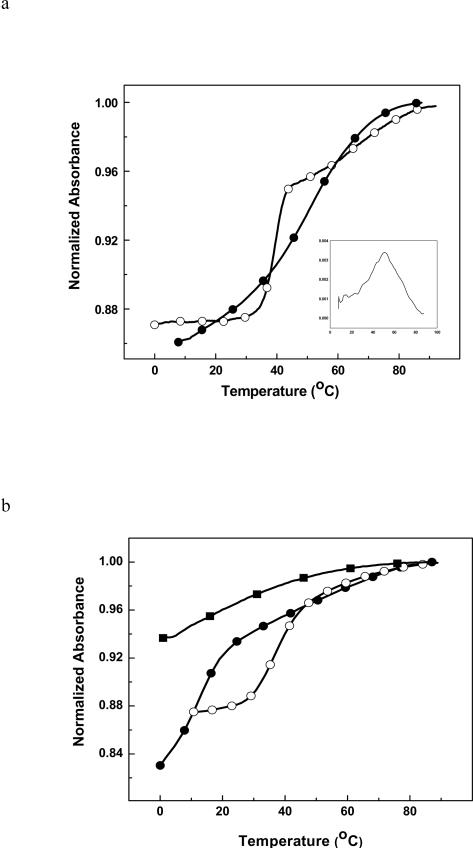
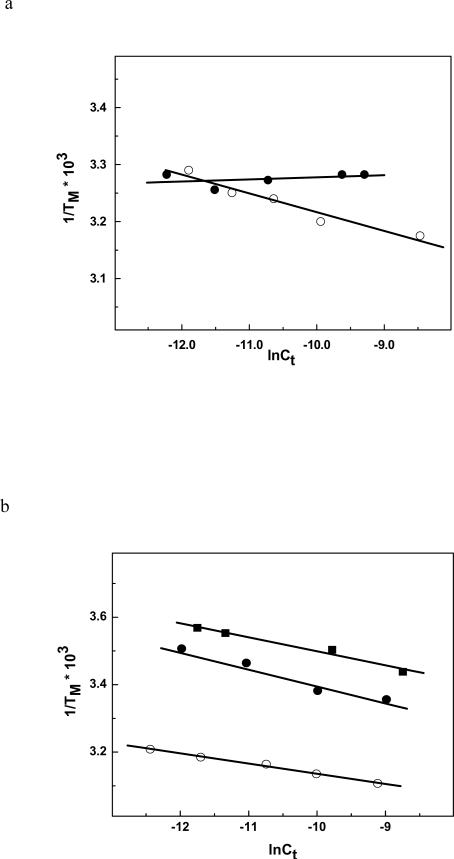
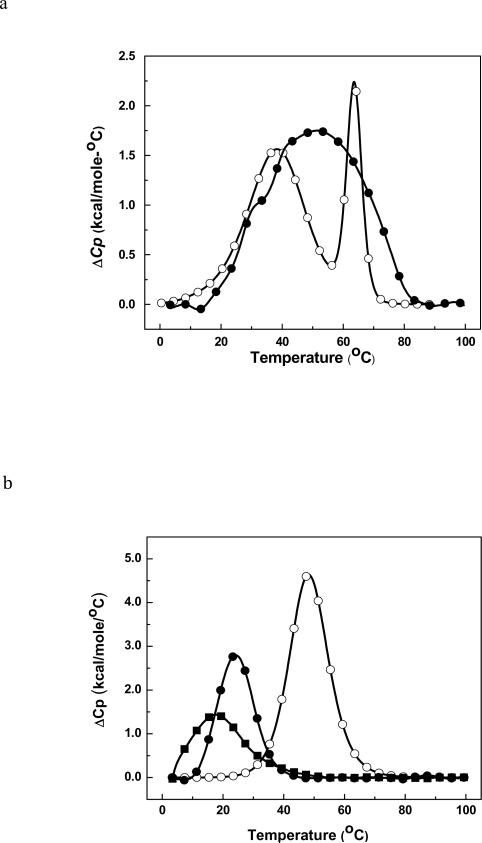
The thermal melts of the five duplexes were followed by monitoring the absorbance at 260 nm as a function of temperature. ODN-1 shows what appears to be a classical two state unfolding (Figure 2a) with TM of 33.3 °C (Table 1). The unfolding of ODN-2 occurs with a broad transition and there appears to be multiple transitions (Figure 2a). Because of this result, we looked at the effect of strand concentration to determine whether ODN-2 was forming a hairpin (Figure 3a). As expected, ODN-1 showed a TM dependence on strand concentration but the HO-dC modified ODN-2 did not. The linear response of ODN-2 suggests that it is preferentially forming a unimolecular hairpin. This would explain the broad melt in Figure 2a due to the presence of a mixture of duplex and hairpin structures even at the lower temperatures. ODN-3, ODN-4 and ODN-5 showed a TM dependence on strand concentration (Figure 3b). The DSC analysis of ODN-1 and -2 in low salt are also consistent with the latter having multiple folded structures with different melting transitions (Figure 4a). In contrast, ODN-1 shows a well-resolved melt indicating the presence of the duplex (the low temperature transition) and hairpin (the high temperature transition). Clearly, the presence of the HO-dC has a significant destabilizing effect on the central domain of the duplex form of ODN-2.
Figure 2.
(a) UV melting curves of ODN-1 (③) and ODN-2 (④); (b) ODN-3 (④), ODN-4 (③) and ODN-5 (■) in 10 mM sodium phosphate buffer (pH 7.0) at ~ 10 μM strand concentration.
Figure 3.
(a) TM dependence on strand concentration of ODN-1 (④) and ODN-2 (③); (b) ODN-3 (④), ODN-4 (③) and ODN-5 (■) in 10 mM sodium phosphate buffer (pH 7.0) in ~ 4-75μM strand concentration.
Figure 4.
Differential scanning calorimetry (DSC) curves in 10 mM sodium phosphate buffer (pH 7.0) at ~ 150-200 μM strand concentration for (a) ODN-1 (④) and ODN-2 (③); (b) at ~ 124–150 μM strand concentration for ODN-3 (④), ODN-4 (③) and ODN-5 (■).
To avoid the complication associated with intramolecular hairpin formation, we turned to the analysis of ODN-3 and -4. ODN-3 does not form a hairpin due to the G/C rich central core. The UV melts for this pair of duplexes are shown in Figure 4a and reveal a dramatic reduction in the TM due to HO-dC (ΔΔTM is ~24 °C at high concentration). Even at 0 °C, the UV melt of ODN-4 does not afford a linear baseline. It is also apparent that the hyperchromicity, which reflects changes in base stacking, is significantly reduced in ODN-4 vs. ODN-3. The DSC of ODN-3 and -4 (Figure 4b) corroborate the difference in stability observed in the UV melting experiment and provide quantification of the difference in ΔΔG°, which is more than 6 kcal/mol (Table 1). The DSC plots show that both ODN-3 and -4 unfold in single sharp transitions. The loss of stabilization is mainly due to the 36 kcal/mol reduction in the stabilizing enthalpy term that is not fully compensated by the increase in entropy (Table 1).
The UV melt and DSC of the DNA with the dC•dC mismatch (ODN-5) provides a comparison to ODN. The UV melt of ODN-5 shows a broad transition (TM ~ 8 °C) with little hyperchromicity (Figure 2b). The DSC thermogram of ODN-5 has a similar pattern with a broad low temperature curve with a ΔΔG° of ~ +7 kcal•mol−1 relative to ODN-3 (Table 1) due to an endothermic ΔΔH of 47 kcal•mol−1.
NMR Studies
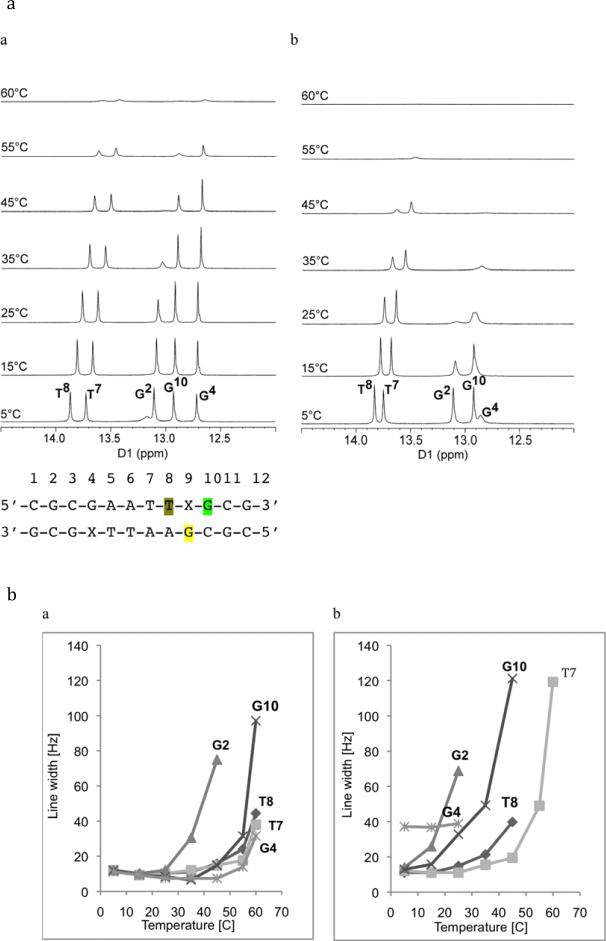
To determine how the HO-dC residue affected base pairing within the DNA sequence, the imino proton resonances of ODN-1 and -2 were assigned. Figure 5 shows the NOE connectivity of the purine N1 and pyrimidine N3 imino protons. The imino protons were assigned based on their sequential connectivities in NOESY spectra, and these assignments were supported by their NOE cross-peaks to Watson-Crick base-paired amino protons.40 The sequential connectivities were obtained from base pairs G2·C11 → G10·C3 → G4·X9/C9 → T8·A5 → T7·A6. For both duplexes the imino-proton resonances of the terminal base pairs C1·G12 are lost through fast exchange with water. The imino resonance from G4, which is base paired with X9 was less intense and broader compared to G4 for unmodified duplex. Moreover G4 imino peak was shifted downfield (by 0.15 ppm), which reflected the effect of base pairing with the opposing X9. Figure 6 shows the region of the NOESY spectrum showing the NOEs between the imino and amino protons. The G4H1 imino proton appeared as a broad peak at 12.6 ppm (Figure 5b); it exhibited weak cross-peaks with X9-N4-H1, X9-N4-H1 and A5-H2. In addition, X9 amino protons with T8-H3 cross-peaks were also observable.
Figure 5.
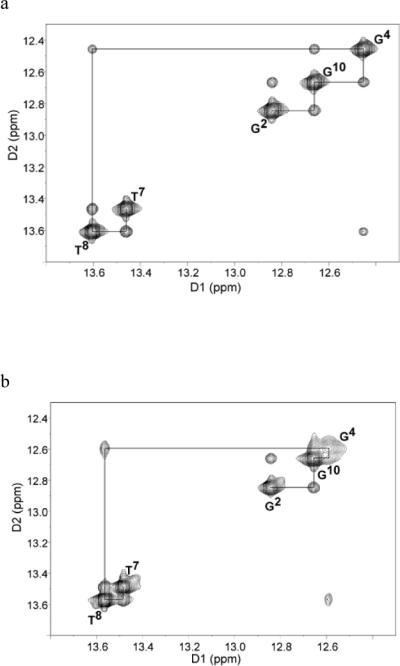
1H-1H NMR NOESY spectrum showing resonances for the thymine and guanine imino protons and sequential NOE connectivity for the imino protons of the base pairs G2·C11 to A6·T7 for (a) unmodified ODN-1 and (b) 5-HO-dC modified ODN-2.
Figure 6.
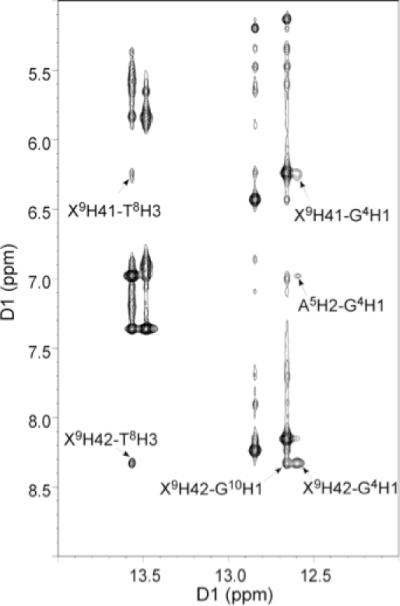
Expansion of the 1H-1H NOESY spectrum for 5-OH-dC modified ODN-2, showing the conservation of Watson-Crick base pairing and base stacking.
A series of 1D NMR spectra for the exchangeable protons for ODN-1 and -2 were recorded at 5, 15, 25, 35, 45, 55 and 60 °C (Figure 7a). The temperature dependences of the line widths for base pairs of unmodified and modified duplexes are compared in Figure 7b. The N1-imino proton of the X9·G4 modified base pair in ODN-2 was already broad at 5 °C and disappeared at higher temperatures. In the unmodified ODN-1, the same imino proton resonance remained sharp even at as high temperature as 45–50 °C. The NMR data also show that the N1-imino proton of the G2·C11 base pair in modified ODN-2 was sharp only at 5 °C; when the temperature was increased the peak started to broaden and finally disappeared at 25 °C. In unmodified ODN-1, the G2·C11 imino proton was sharp up to room temperature, above which it started to broaden. The G10·C3 base pair, which is adjacent to the X9·G4 pair was almost non-observable at 35 °C in ODN-2, while it is still sharp even at 45 °C for ODN-1. The peak corresponding to T8, which is immediately adjacent to the X9·G4 pair in ODN-2, started to broaden at approximately 35 °C, while the same imino proton in ODN-1 was still visible at 55 °C. The imino resonance of T7·A6 base pair remained sharp for ODN-2 and disappeared above 45 °C; however, for ODN-1 the same imino proton remained sharp and intense at this temperature.
Figure 7.
(a) 1H-NMR of imino proton resonances as a function of temperature for (a) unmodified ODN-1 and (b) 5-HO-dC modified ODN-2. (b) Temperature dependence of line widths of the imino proton resonances of (a) unmodified ODN-1 and (b) 5-HO-dC modified ODN-2.
The temperature-dependent NMR data show increased exchange rates between the G4·X9, G2·C11 and G10·C3 base pairs and solvent, which suggest that base pairing is destabilized compared to the unmodified duplex ODN-1. In addition, the expanded NOESY spectrum showed broad cross-peak between modified X9 and the complementary G4, indicating weaker Watson-Crick base pairing in comparison to unmodified duplex. Presence of the cross-peaks between amino X9 and imino G10 and T8 suggests conserved base stacking of OH-dC with neighboring bases. However this interaction appears as a set of broad cross-peaks that implies weaker interaction than for other peaks, but also reflects structural changes, which are still under investigation.
Effect of HO-dC on DNA Hydration and Cation Binding
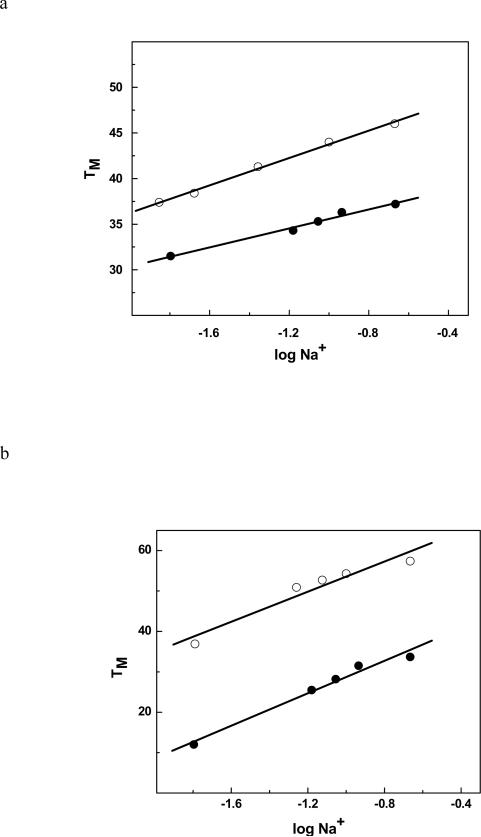
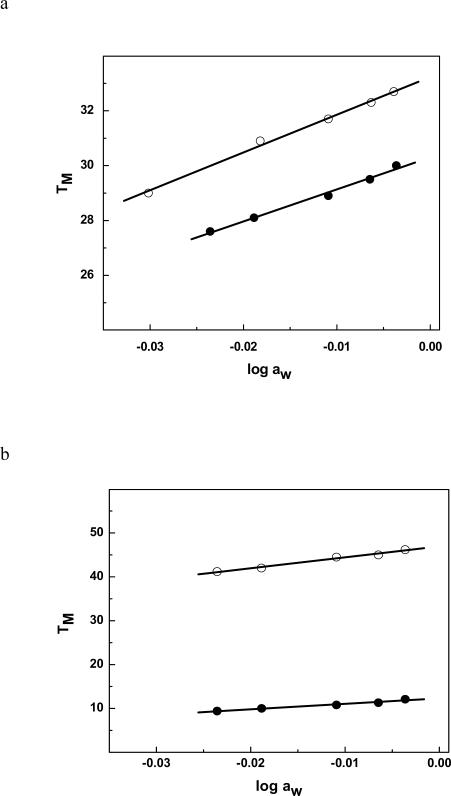
To understand the origin of the destabilization observed in the DSC and NMR experiments, we probed the effect of the HO-dC lesion on the binding of water and cations to ODN-2 vs. -1 and ODN-4 vs. -3 (Figures 8 and 9, respectively). The ΔΔnW for ODN-2 relative to ODN-1 is 17 waters/mol DNA, while the change is even greater for ODN-4 relative to ODN-3 (ΔΔnW = 28 waters/mol) (Table 1). There was also a significant reduction in the release of cations from the modified duplexes ΔΔnNa+ of 1.3 and 1.1 Na+/mol DNA for ODN-1 vs. -2 and ODN-3 vs. -4, respectively. These changes are consistent with a reduction in base pair stacking; double-stranded DNA is more hydrated and has more cations associated with it than single-strand DNA despite the higher number of polar heteroatoms that are accessible in single-strand DNA.41,42
Figure 8.
(a) TM dependence on salt concentration for ODN-1 (④) and ODN-2 (③) in 10 mM sodium phosphate buffer (pH. 7.0); (b) ODN-3 (④), ODN-4 (③) at ~ 8 μM strand concentration.
Figure 9.
(a) TM dependence on osmolyte concentration (function of ethylene glycol) for for ODN-1 (④) and ODN-2 (③); (b) ODN-3 (④), ODN-4 (③) in 10 mM sodium phosphate buffer (pH. 7.0) at ~ 8 μM strand concentration.
DISCUSSION
Why is the HO-dC•dG base pair in DNA so unstable? To check the stability of the HO-dC modified oligomers, the MALDI-TOF of ODN-4 was rerun after the thermodynamic studies were completed and it showed the same spectrum as it did when we initially purified it so chemical degradation of the lesion cannot account for the NMR and thermodynamic observations. The equilibrium between the amino-imino tautomeric forms for the deoxynucleoside has been studied by NMR43 and UV resonance Raman spectroscopy.44 In the former, there was no change in the preference for the “normal” amino tautomer within the detection limits of the method when the pH was adjusted so the nucleoside was in the neutral form. In the Raman study, a 100-fold increase in the imino tautomer was reported, but it still comprised less than 0.1 % of the predominant amino tautomer. The increase in the rare imino tautomer could be in part responsible for the infrequent G→A transitional mutations that arise from this lesion during DNA replication, but it cannot explain the dramatic effect in the thermodynamic or NMR measurements. Moreover, the structure of HO-dC in the primer strand paired with a 3'-dGMP within the active site of a DNA polymerase adopts a classical Watson-Crick alignment with no evidence for a wobble pair arrangement in the crystal structure.25
In the non-ionized (neutral) form, the 5-hydroxy group of HO-dC can approach to within 3.7 Å from the 5'-non-bridging phosphate oxygen that points into the major groove in a canonical B-DNA conformation. This type of H-bond interaction could generate a locally distorted base pairing structure. The 5-hydroxyl group on HO-dC can ionize near neutral pH: the pKa for this process was calculated to be 7.3 for the nucleoside and 8.5 for the nucleotide.45 The ionization of the 5-hydroxy group could locally destabilize the DNA due to a repulsive electrostatic interaction between the ionized −O-dC (i.e., anionic) and the polyanionic phosphodiester backbone. To explore this possibility, we ran UV melts of ODN-3 and -4 at pH's that bracketed the reported pKa of HO-dC. We observed that the ΔTM between ODN-3 and -4 at the different pH values remained fairly constant, although both unmodified and modified duplexes were marginally less stable at the more acidic conditions (Table 2). There was also little change in the hyperchromicity observed in the melts suggesting minimum pH dependent change in base stacking. There are several potential explanations for this result. The pKa of HO-dC in a double helix may be significantly altered due to reduced water activity in the major groove.46 This has been observed for the pKa of dC where protonation of the N3-position is required for the formation of stable C+-G•C triplets.47, 48 The pKa of dC is ~4.3,49 while in an intermolecular triplex it can be as high as 6, and as high as 7 in an intramolecular triplex.50 If this is the case for the 5-hydroxyl group, the pH range used in our stability studies may not have captured the ionized form.
Table 2.
Effect of pH on the TM and hyperchromicity of unmodified DNA and 5-HO-dC (X) substituted DNA at 100 mM NaCl in 10 mM sodium phosphate buffer.
| pH | GAGAGCGCTCT TM(°C) | Hyperchromicity (%) | GAGAGCGCTXTC TM (°C) | Hyperchromicity (%) |
|---|---|---|---|---|
| 5.5 | 52.7 | 13.4 | 22.1 | 11.9 |
| 6.4 | 55.1 | 12.5 | 27.4 | 9.2 |
| 7.0 | 55.3 | 12.9 | 28.2 | 10.2 |
| 8.5 | 54.7 | 14.1 | 25.8 | 13.3 |
An alternative explanation for the effect of HO-dC is the effect of the 5-hydroxy group on the direct and magnitude of the base's dipole moment. Multi-configuration self-consistent field (MCSCF) calculations indicate that there is a dramatic drop in the dipole moment for the amino-keto tautomer of HO-dC: 4.6151 or 4.8 – 5.952 D vs. 6.08 –7.61 D for dC,53 which will electrostatically destabilize the pairing with the strong anti-parallel dipole moment of dG.
The magnitude of the destabilization suggested that HO-dC•dG may not be base pairing in solution via a 3 H-bond Watson-Crick motif despite the canonical crystal structure of HO-dC with dGMP at the active site of bacteriophage DNA polymerase.25 A duplex was prepared with a dC•dC mismatch to see how it compared to ODN-4. The dC•dC mismatch, which is generally the most destabilizing base pair arrangement,54 does not form a hairpin (Figure 4b) and is even more destabilizing than the HO-dC•dG (Table 1). The CD of the mismatched ODN-5 is actually more similar to that of ODN-3 than to ODN-4. However, the ΔΔG° and ΔTM between ODN-4 and ODN-5 from the thermodynamic studies indicate that the pairing between HO-dC and dG behaves similar to a dC•dC mismatch that can only form 1 H-bond assuming the predominance of the neutral amino tautomer.
While the destabilization induced by the HO-dC•dG pair is pronounced, we have reported similar thermodynamic parameters for other oxidized bases and for alkylated lesions. For example, an 8-oxo-dG•dC base pair causes a significant destabilization (ΔΔG > 3 kcal•mol−1) that is driven by a >35 kcal•mol−1 reduction in the ΔH term.55 As seen with the HO-dC•dG modified DNA, the effect is observable by temperature-dependent monitoring of the imino proton resonances and there is a concomitant reduction in the release of water and cations upon the unfolding of the DNA with the oxidized lesion. Despite these thermodynamic differences, the crystal56 and high resolution NMR57 structures of DNA with 8-oxo-dG•dC are indistinguishable from wild type DNA. As is often the case, the similarity in structures for unmodified and 8-oxoguanine modified DNA by these two structural techniques does imply that the molecules will have similar thermal or thermodynamic characteristics. They clearly do not.
The pattern is the same for DNA with 3-methyl-3-deaza-dA•dT,58 3-deazadA•dT,58 7-deaza-dG•dC59 and 7-deaza-dA•dT60 base pairs. In all cases we observed reduced stability due to an unfavorable enthalpic change that is not fully compensated for by the increase in the entropy term. Moreover, these modified duplexes are characterized by reduced release of hydrophobic water and cations upon unfolding.
We propose that the local destabilization of DNA affords a thermodynamic signature that can be exploited by base excision repair glycosylases in the initial screening of the genome for lesions, a suggestion originally suggested by Plum and Breslauer.61 While the lesions may not extensively populate an extrahelical conformation, the energetic penalty required to extrude them from the base stack and deform the DNA backbone62 will be significantly reduced. It is known that glycosylase binding is accelerated when the lesion is in a mismatch.63,64 Thus, as the glycosylases “scan” the DNA the low energy barrier to form a DNA conformation that initially stabilizes the interaction of the protein with the DNA would constitute a thermodynamic based mechanism of lesion detection. If the lesion is a substrate for the glycosylase, it will be excised off the backbone. If not, the complex will collapse and the glycosylase can continue to scan the DNA for lesions. A related proposal is the explanation for the specificity of the alkyladenine DNA glycosylase for substrate lesions derived from a detailed study of the kinetics of base flipping and excision of 1,N6-ethenoadenine by alkyladenine DNA glycosylase.65 Related to this suggestion of specificity being based on the ease of base extrusion is the report that the bacterial AlkD glycosylase catalyzes the hydrolysis of N3-methyladenine off the DNA backbone by stabilizing it as an extrahelical lesion.66 In this case, no enzymatic step is required since the rate of hydrolysis of 3-methyladenine from the deoxyribose in single-stranded DNA is quite rapid even at neutral pH.67
Acknowledgments
Funding This work was supported in part by the National Cancer Institute/National Institutes of Health (CA29088).
References
- (1).Wagner JR, Hu CC, Ames BN. Endogenous oxidative damage of deoxycytidine in DNA. Proc. Natl. Acad. Sci. U S A. 1992;89:3380–3384. doi: 10.1073/pnas.89.8.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- (3).O'Brien PJ. Radical formation during the peroxidase catalyzed metabolism of carcinogens and xenobiotics: the reactivity of these radicals with GSH, DNA, and unsaturated lipid. Free Radic. Biol. Med. 1988;4:169–183. doi: 10.1016/0891-5849(88)90025-1. [DOI] [PubMed] [Google Scholar]
- (4).von Sonntag C. The chemistry of free-radical-mediated DNA damage. Basic Life Sci. 1991;58:287–317. doi: 10.1007/978-1-4684-7627-9_10. [DOI] [PubMed] [Google Scholar]
- (5).Dizdaroglu M. Chemical determination of free radical-induced damage to DNA. Free Radic. Biol. Med. 1991;10:225–242. doi: 10.1016/0891-5849(91)90080-m. [DOI] [PubMed] [Google Scholar]
- (6).Halliwell B, Gutteridge JM. Biologically relevant metal ion-dependent hydroxyl radical generation. An update. FEBS Lett. 1992;307:108–112. doi: 10.1016/0014-5793(92)80911-y. [DOI] [PubMed] [Google Scholar]
- (7).Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutat. Res. 1992;275:331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- (8).Floyd RA. The role of 8-hydroxyguanine in carcinogenesis. Carcinogenesis. 1990;11:1447–1450. doi: 10.1093/carcin/11.9.1447. [DOI] [PubMed] [Google Scholar]
- (9).Dizdaroglu M. Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on gamma-irradiation in aqueous solution. Biochemistry. 1985;30:4476–4481. doi: 10.1021/bi00337a032. [DOI] [PubMed] [Google Scholar]
- (10).Kasai H, Nishimura S. Hydroxylation of guanine in nucleosides and DNA at the C-8 position by heated glucose and oxygen radical-forming agents. Environ. Health Perspect. 1986;67:111–116. doi: 10.1289/ehp.8667111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Kasai H, Crain PF, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Carcinogenesis. 1986;7:1849–1851. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- (12).Abu-Shakra A, Zeiger E. Formation of 8-hydroxy-2'-deoxyguanosine following treatment of 2'-deoxyguanosine or DNA by hydrogen peroxide or glutathione. Mutat. Res. 1997;390:45–50. doi: 10.1016/s0165-1218(96)00164-4. [DOI] [PubMed] [Google Scholar]
- (13).Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- (14).Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 15.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi JY, Kim HS, Kang HK, Lee DW, Choi EM, Chung MH. Thermolabile 8-hydroxyguanine DNA glycosylase with low activity in senescence accelerated mice due to a single-base mutation. Free Radical Biol. Med. 1999;27:848–854. doi: 10.1016/s0891-5849(99)00141-0. [DOI] [PubMed] [Google Scholar]
- 17.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 18.Wagner JR, Cadet J. Oxidation reactions of cytosine DNA components by hydroxyl radical and one-electron oxidants in aerated aqueous solutions. Acc. Chem. Res. 2010;43:564–571. doi: 10.1021/ar9002637. [DOI] [PubMed] [Google Scholar]
- 19.Luo Y, Henle ES, Linn S. Oxidative damage to DNA constituents by iron-mediated fenton reactions. The deoxycytidine family. J. Biol. Chem. 1996;271:21167–21176. [PubMed] [Google Scholar]
- 20.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic. Biol. Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 21.Purmal AA, Kow YW, Wallace SS. Major oxidative products of cytosine, 5-hydroxycytosine and 5-hydroxyuracil, exhibit sequence context-dependent mispairing in vitro. Nucleic Acids Res. 1994;22:72–78. doi: 10.1093/nar/22.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C --> T transitions in vivo. Proc. Natl. Acad. Sci. U.S.A. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Negishi K, Sekine D, Morimitsu T, Suzuki T, Okugawa Y, Kawakami A, Otsuka C, Oyama H, Loakes D. Oligonucleotide transformation for the study of mutagenic specificities of DNA lesions in yeast. Nucleic Acids Symp. Ser. 2007;51:211–212. doi: 10.1093/nass/nrm106. [DOI] [PubMed] [Google Scholar]
- 24.Vaisman A, Woodgate R. Unique misinsertion specificity of pol iota may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zahn KE, Averill A, Wallace SS, Doublié S. The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site. Biochemistry. 2011;50:10350–10358. doi: 10.1021/bi201219s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eide L, Luna L, Gustad EC, Henderson PT, Essigmann JM, Demple B, Seeberg E. Human endonuclease III acts preferentially on DNA damage opposite guanine residues in DNA. Biochemistry. 2001;40:6653–6659. doi: 10.1021/bi0028901. [DOI] [PubMed] [Google Scholar]
- 27.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes. 5-Hydroxy-2'-deoxycytidine and 5-hydroxy-2'-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2'-deoxyuridine is a substrate for uracil DNA N-glycosylase. J. Biol. Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 28.Tremblay S, Wagner JR. Dehydration, deamination and enzymatic repair of cytosine glycols from oxidized poly(dG-dC) and poly(dI-dC) Nucleic Acids Res. 2008;36:284–293. doi: 10.1093/nar/gkm1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fromme JC, Verdine GL. Structure of a trapped endonuclease IIIDNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantor CR, Warshow MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 32.Marky LA, Blumenfeld KS, Kozlowski S, Breslauer KJ. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCGAATTCGCG): evidence for hairpin formation. Biopolymers. 1983;22:1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- 33.Kaushik M, Suehl N, Marky LA. Calorimetric unfolding of the bimolecular and i-motif complexes of the human telomere complementary strand, d(C3TA2)4. Biophys Chem. 2007;126:154–164. doi: 10.1016/j.bpc.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 34.Courtenay ES, Capp MW, Anderson CF, Record MT., Jr. Vapor pressure osmometry studies of osmolyte – protein interactions: implications for the action of osmoprotectants in vivo and for the interpretation of “osmotic stress” experiments in vitro. Biochemistry. 2000;39:4455–4471. doi: 10.1021/bi992887l. [DOI] [PubMed] [Google Scholar]
- 35.Marky LA, Breslauer KJ. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 36.Rentzeperis D, Marky LA, Dwyer TJ, Geierstanger BH, Pelton JG, Wemmer DE. Interaction of minor groove ligands to an AAATT/AATTT site: correlation of thermodynamic characterization and solution structure. Biochemistry. 1995;34:2937–2945. doi: 10.1021/bi00009a025. [DOI] [PubMed] [Google Scholar]
- 37.Wing R, Drew H, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 38.Marky LA, Blumenfeld KS, Kozlowski S, Breslauer KJ. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide d(CGCGAATTCGCG): evidence for hairpin formation. Biopolymers. 1983;22:1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- 39.Bloomfield VA, Crothers DM, Tinoco I Jr., editors. Nucleic Acids: Structures, Properties, and Functions. University Science Books; Sausalito, CA: 1999. Electronic and Vibrational Spectroscopy; pp. 185–196. [Google Scholar]
- 40.Boelens R, Scheek RM, Dijkstra K, Kapten R. Sequential assignment of imono- and amino-proton resonances in 1H NMR spectra of oligonucleotides by two-dimensional NMR spectroscopy. Application to a lac operator fragment. J. Magn. Reson. 1985;62:378–386. [Google Scholar]
- 41.Lee CH, Mizusawa H, T Kakefuda T. Unwinding of double-stranded DNA helix by dehydration. Proc. Natl. Acad. Sci. U. S. A. 1981;78:2838–2842. doi: 10.1073/pnas.78.5.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bastos M, Castro V, Mrevlishvili G, Teixeira J. Hydration of ds-DNA and ss-DNA by neutron quasielastic scattering. Biophys. J. 2004;86:3822–3827. doi: 10.1529/biophysj.104.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaFrancois CJ, Fujimoto J, Sowers LC. Synthesis and characterization of isotopically enriched pyrimidine deoxynucleoside oxidation damage products. Chem. Res. Toxicol. 1998;11:75–83. doi: 10.1021/tx970186o. [DOI] [PubMed] [Google Scholar]
- 44.Suen W, Spiro TG, Sowers LC, Fresco JR. Identification by UV resonance Raman spectroscopy of an imino tautomer of 5-hydroxy-2'-deoxycytidine, a powerful base analog transition mutagen with a much higher unfavored tautomer frequency than that of the natural residue 2'-deoxycytidine. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4500–4505. doi: 10.1073/pnas.96.8.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.La Francois CJ, Jang YH, Cagin T, Goddard WA, 3rd, Sowers LC. Conformation and proton configuration of pyrimidine deoxynucleoside oxidation damage products in water. Chem. Res. Toxicol. 2000;13:462–470. doi: 10.1021/tx990209u. [DOI] [PubMed] [Google Scholar]
- 46.Young MA, Jayaram B, Beveridge DL. Local dielectric environment of B-DNA in solution:▯ Results from a 14 ns molecular dynamics trajectory. J. Phys. Chem. B. 1998;102:7666–7669. [Google Scholar]
- 47.Thuong NT, Hêlène C. Sequence-specific recognition and modification of double-helical DNA by oligonucleotides. Angew. Chem. Internal. Ed. Engl. 1993;32:666–690. [Google Scholar]
- 48.Singleton SF, Dervan PB. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992;31:10995–11003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- 49.H. R. The Modified Nucleosides in Nucleic Acids. Columbia University Press; New York: 1971. pp. 192–194. [Google Scholar]
- 50.D., Schröder W, Weisz K. Influence of sequence-dependent cytosine protonation and methylation on DNA triplex stability. Biochemistry. 2000;39:5886–5892. doi: 10.1021/bi992630n. [DOI] [PubMed] [Google Scholar]
- 51.Krauss M, Osman R. Electronic spectra of the H and OH adducts of cytosine. J. Phys. Chem. A. 1997;101:4117–4120. [Google Scholar]
- 52.Cysewski P. Structure and properties of hydroxyl radical modified nucleic acid components: tautomerism and miscoding propoerties of 5-hydroxycytosine. J. Mol. Struct. (Theochem) 1999;466:49–58. [Google Scholar]
- 53.Czerminski R, Lesyng B, Pohorille A. Tautomerism of pyrimidine bases-uracil, cytosine, isocytosine: Theoretical study with complete optimization of geometry. Int. J. Quant. Chem. 1979;16:605–613. [Google Scholar]
- 54.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr. Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 55.Singh SK, Szulik MW, Ganguly M, Khutsishvili I, Stone MP, Marky LA, Gold B. Characterization of DNA with an 8-oxoguanine modification. Nucleic Acids Res. 2011;39:6789–6801. doi: 10.1093/nar/gkr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD. X-ray structure of a DNA 5'-d(CGC-oxoG-AATTCGCG) decamer containing 7,8-dihydro-8-oxoguanine. Proc. Natl. Acad. Sci. USA. 1995;92:719–723. doi: 10.1073/pnas.92.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E. NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res. 1991;19:1407–1412. doi: 10.1093/nar/19.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ganguly M, Wang R-W, Marky LA, Gold B. Thermodynamic characterization of DNA with 3-deazaadenine and 3-methyl-3-deazaadenine substitutions. J. Phys. Chem. B. 2010;114:7656–7661. doi: 10.1021/jp101004k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ganguly G, Wang F, Kaushik M, Stone MP, Marky LA, Gold B. A study of 7-deaza-2'-deoxyguanosine•2'-deoxycytidine base pairing in DNA. Nucleic Acids Res. 2007;35:6181–6195. doi: 10.1093/nar/gkm670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kowal E, Ganguly M, Pallan P, Marky LA, Gold B, Egli M, Stone MP. Altering the electrostatic potential in the major groove: thermodynamic and structural characterization of 7-deaza-2'-deoxyadenosine•dT base pairing in DNA. J. Phys. Chem. B. 2011;115:13925–13934. doi: 10.1021/jp207104w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plum GE, Breslauer KJ. DNA lesions. A thermodynamic perspective. Ann. N. Y. Acad. Sci. 1994;726:45–55. doi: 10.1111/j.1749-6632.1994.tb52796.x. [DOI] [PubMed] [Google Scholar]
- 62.Mol CD, Parikh SS, Putnam CD, Lo TP, Tainer JA. DNA repair mechanisms for the recognition and removal of damaged DNA bases. Annu. Rev. Biophys. Biomol. Struct. 1999;28:101–128. doi: 10.1146/annurev.biophys.28.1.101. [DOI] [PubMed] [Google Scholar]
- 63.Biswas T, Clos LJ, 2nd, SantaLucia J, Jr., Mitra S, Roy R. Binding of specific DNA base-pair mismatches by N-methylpurine-DNA glycosylase and its implication in initial damage recognition. J. Mol. Biol. 2002;320:503–513. doi: 10.1016/S0022-2836(02)00519-3. [DOI] [PubMed] [Google Scholar]
- 64.O'Brien PJ, Ellenberger T. Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase. J. Biol. Chem. 2004;279:9750–9757. doi: 10.1074/jbc.M312232200. [DOI] [PubMed] [Google Scholar]
- 65.Wolfe AE, Patrick J, O'Brien PJ. Kinetic mechanism for the flipping and excision of 1,N6-ethenoadenine by human alkyladenine DNA glycosylase. Biochemistry. 2009;48:11357–11369. doi: 10.1021/bi9015082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rubinson EH, Gowda ASP, Spratt TE, Gold B, Eichman BF. An unprecedented nucleic acid capture mechanism for excision of DNA damage. Nature. 2010;468:406–411. doi: 10.1038/nature09428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fujii T, Saito T, Nakasaka T. Purines. XXXIV. 3-Methyladenosine and 3-methyl-2'-deoxyadenosine: their synthesis, glycosidic hydrolysis, and ring fission. Chem. Pharmaceut. Bull. 1989;37:2601–2609. [Google Scholar]