Abstract
Molecular biology and genomics have made notable strides in the sharing of primary data and resources. In other domains of neuroscience research, however, there has been resistance to adopting formalized strategies for data exchange, archiving, and availability. In this article, we discuss how neuroscience domains might follow the lead of molecular biology on what has been successful and what has failed in active data sharing. This considers not only the technical challenges but also the sociological concerns in making it possible. Though, not a pain-free process, with increased data availability, scientists from multiple fields can enjoy greater opportunity for novel discoveries about the brain in health and disease.
“Pain shared is pain lessened; joy shared is joy increased. Thus we do refute entropy.”
–Spider Robinson, The Callahan Chronicles.
Whether or not we can refute the effect of entropy, it is widely assumed that scientific data are - like joy – an important thing to share. The ability to understand and even re-purpose data from others can only increase the likelihood of novel scientific discovery. However, the sharing of data does not come without some amount of pain. The question is whether different fields of biomedical research can learn from one another on how to best share and, thereby, lessen the pain of exchanging data.
Like many biomedical efforts, brain researchers routinely find themselves awash in a sea of complex data (Miles 2001). Neuroscientists, in particular, actively leverage functional genomics to study the brain, notably elements associated with spoken language (Oldham and Geschwind 2006), polygenic disease (Konradi 2005), and the role of entire evolutionary systems on the brain (Boguski and Jones 2004). More broadly, researchers use a range of sophisticated technologies attempting to better understand the fundamental elements comprising the multi-scale physiological basis of cognitive and behavioral processes of the brain. For instance, the relatively new capabilities of advanced neuroimaging have created the possibility of evaluating and predicting complex human behavior and disease in unprecedented ways (Raichle 2003). Functional brain mapping studies have explored the domain of cognitive function (D'Esposito 2000) and dysfunction in patient samples (Laurens, Kiehl et al. 2005). More recently, diffusion tensor imaging (DTI) has enabled the tracking in vivo of white matter fiber pathways (Basser and Jones 2002) (e.g. Fig. 1a). On a smaller scale, single unit recording studies examine the behavior of individual neurons from human (Hodaie, Cordella et al. 2006) and non-human (Walton, Bechara et al. 2007) samples. Across domains in neuroscience, a greater dependence is being placed on acquiring data in digital form, subjecting it to high-throughput analysis, and leveraging advanced graphical techniques as aids in visualization. All these steps require considerable processing power, storage, and rendering capability. Yet despite this amount of data and the different forms of representing the information they contain, little is made publicly available for other researchers to examine and explore (Kennedy 2003). Where neuroimaging data have been made available to the community (Van Horn, Grafton et al. 2004), intriguing results have been obtained when considered in new analytical light (Buckner, Snyder et al. 2000; Greicius, Krasnow et al. 2003).
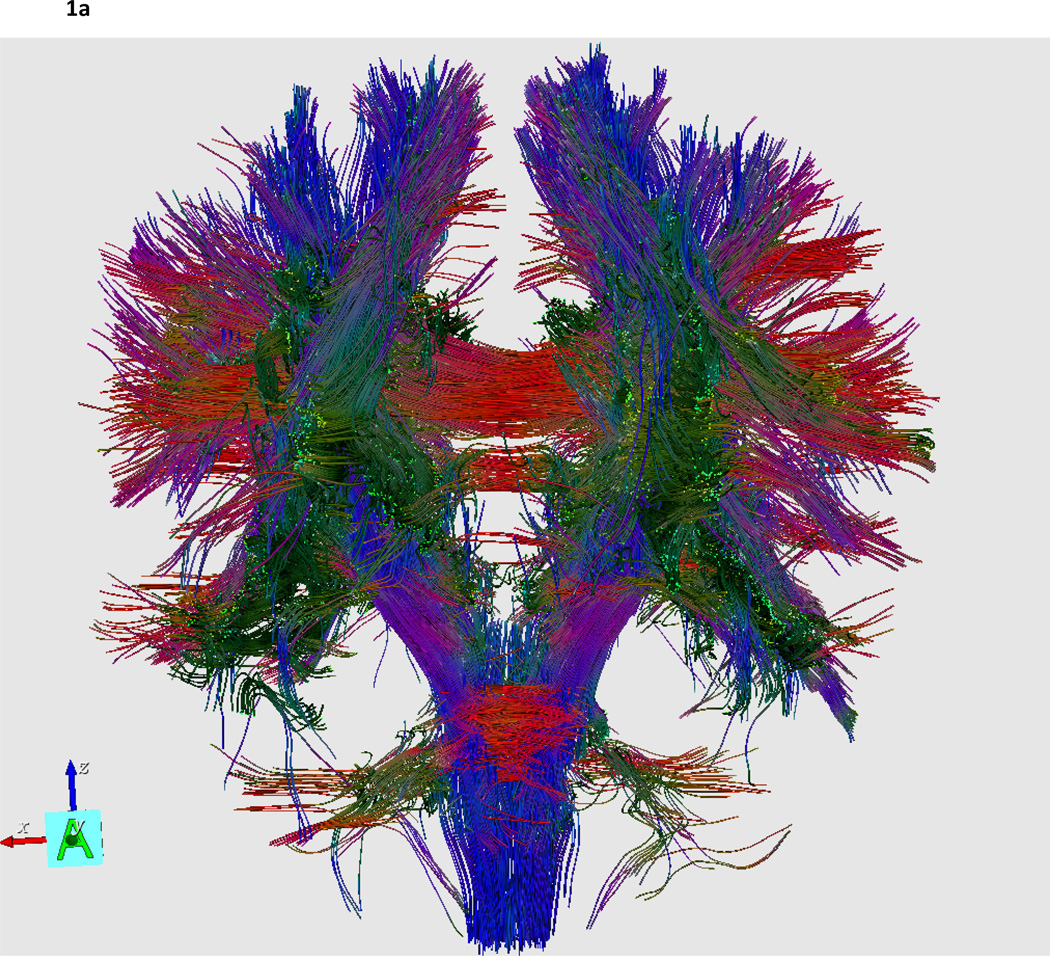
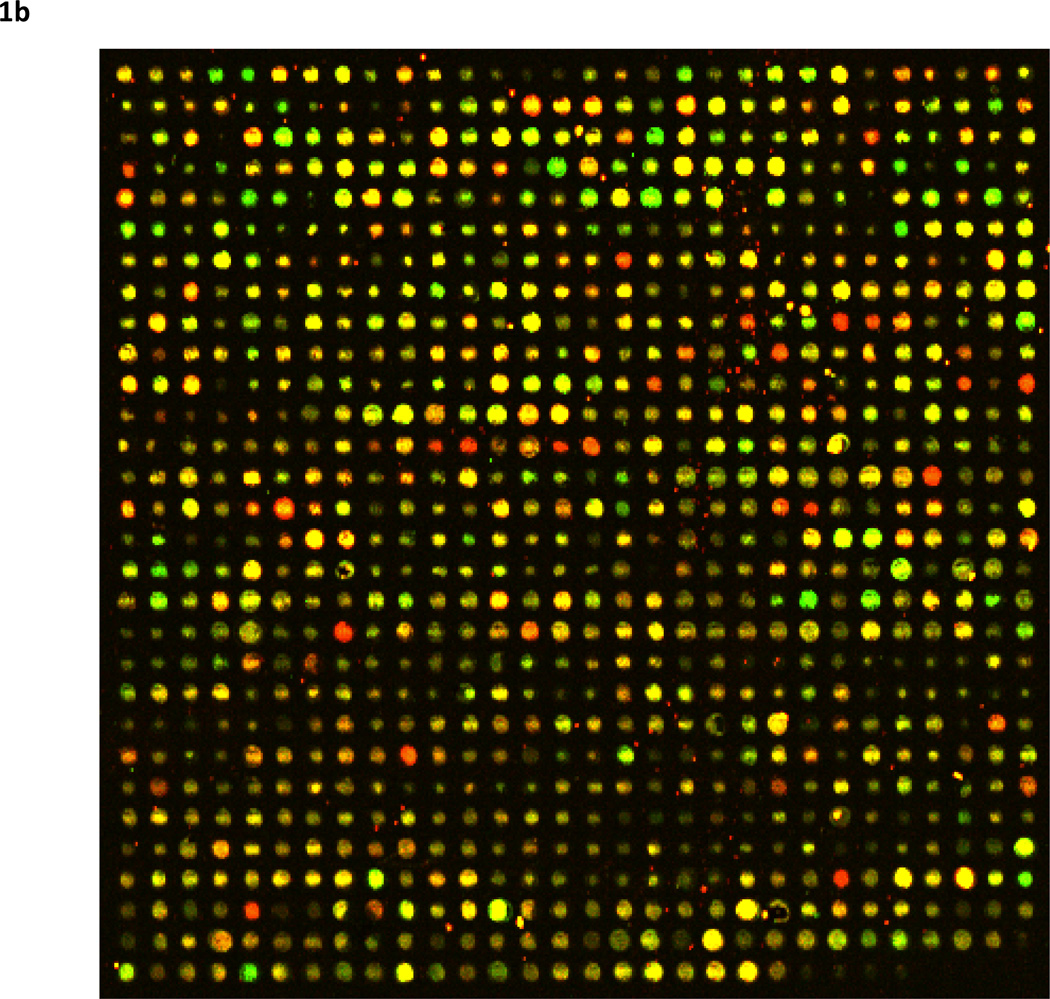
Figure 1. To share or not to share?
Neuroscience data is obtained in many forms - some are obtained using modern in vivo neuroimaging techniques to examine brain function or structure (1a), while some form are obtained from biological tissue samples to derive DNA sequences or gene expression profiles (1b). In the end, these data are represented as digital information, either as text, images, image volumes, time series, etc. Once they are digital, why not share them so that others might benefit from the information they contain? In this figure one might ask is one domain of more shareable than the other? Factors underlying the willingness of investigators to share their data include how difficult or costly the digital information was to obtain, a lack of efficient standards for data exchange, fear of being “scooped”, the overall amount of data to be shared, concerns over patient privacy, as well as the seniority of the investigator. Such factors can be difficult to quantify or overcome. But if they can be systematically surmounted with support from leading scientific organizations and government agencies then neuroscience will be enriched and new discoveries may be closer at hand. 1a) Diffusion tensor imaging (DTI; Philips 3.0 Tesla, 8-channel SENSE head coil, 32-gradient directions) white matter fiber tractography as determined via streamline projections along image voxels with maximal directional preference. Color denotes fiber orientation: Red=Left-to-Right, Green= anterior-posterior, and Blue=Inferior-to-Superior. 1b) A microarray segment from the frontal cortex chosen from the Stanford Microarray Database (SMD; http://genome-www5.stanford.edu): Experiment = 27745, SlideName = shcg212, Experiment = ”Brain(frontal)”, Category = Normal Tissue, Subcategory = Brain, Experimenter = JJUNKERM, ExptDate = 2002-04-16.
By contrast, in the field of molecular biology, microarray usage has expanded rapidly since the mid-1990s, where applications include basic research, target discovery/selectivity, biomarker determination, pharmacology, toxicology, the development of screening tests, and disease-subclass determination (Butte 2002). The development of high-throughput gene and microarray technology (Fig. 1b) and its applications in all facets of biology has been considered as one of the great success stories in the scientific community (Hood 2003). For example, public data repositories for microarray data (Parkinson, Kapushesky et al. 2007) and the availability of efficient data analysis tools (a PubMed search for “microarray data analysis tools” yields 327 articles) now mean that a generation of biological researchers are exploring these rich resources without physically collecting the data which they are studying.
To encourage greater data sharing and meta-data availability, the Society for Neuroscience (SfN) has addressed the issue of sharing data head-on by hosting the PubMed Plus New Directions in Publishing and Data Mining conference in St. Louis MO last June 18–19, 2007. The overarching goal of the SfN leadership was to explore methods for journals and on-line neuroscience data repositories to operate more collaboratively and to facilitate more effective data mining. Much of the discussion at this gathering of leading neuroscientists, informaticists, and science publishers was focused on finding more convenient and effective ways to share neuroscience data that cannot be adequately captured or represented in a traditional publication. Of particular interest was the use of ontologies (Whetzel, Parkinson et al. 2006; Thomas, Mi et al. 2007) and structured abstracts (Seringhaus and Gerstein 2007) to describe how experiments were designed, executed, and to highlight major conclusions. Since these new means for description are currently the subjects of a great deal of work and debate in biology, it seems likely that the neuroscience field can take advantage of the resulting tools and experiences. However, many attendees also expressed deep concern about whether the neuroscience community can make the sociological changes needed to either share data or to use tools and resources developed elsewhere. The meeting encouraged the development of a SfN Meta-Data Task Force to begin examining more closely how to encourage authors to provide more detailed information concerning study methods and materials. They also examined how best to link this information to formalized nomenclatures and online resources to further enrich these published studies. These developments suggest that, in fact, the neuroscience community as a whole is in an excellent position to 1) carefully examine the experiences of other disciplines so as to more quickly enjoy the benefits that can come from the sharing of primary and meta- data, and 2) to then act on what they learn to help neuroscientists in the field make the most from these rich sources of information. The experience of the genomics and microarray communities provide examples of where one might start.
To accommodate genomic and microarray data, biologists have worked closely with computer scientists to develop standards and tools to formalize and facilitate data sharing. The most successful example is the Gene Ontology (GO) project -- a collaborative effort to provide consistent descriptions of gene products in different databases (Ashburner, Ball et al. 2000; Harris, Clark et al. 2004). In addition, the MGED (Microarray Gene Expression Data) society has encouraged the adoption and use of concise data standards in order to promote data sharing and longevity, notably the Minimum Information About Microarray Experiments (MIAME) specification for the types of information that should be available (Brazma, Hingamp et al. 2001), the MAGE object model that specifies a format for exchanging data (Spellman, Miller et al. 2002) and the MGED Ontology that provides terms for annotating microarray experiments (Whetzel, Parkinson et al. 2006). While the microarray standards have provided a structure to formalize the description of microarray studies, they have not to date achieved the same widespread adoption as GO. Yet, interest is growing towards having a collection of highly focused and efficient standards for multiple forms of biological data that can link information across domains, thereby making it easier to see relationships that would have been difficult to examine otherwise. Efficient data standards can enable the formulation of testable predictions that lead to additional empirical investigation, testing, and assessment of disease.
While the genomics community has been proactive in building data sharing infrastructure (in terms of data standards and data repositories), neuroscientists have remained skeptical to the value of formalized standards and open data archives. This attitude persists despite a variety of brain-specific databases that have been created for this purpose (e.g. The SfN Neuroscience Database Gateway, http://www.sfn.org/index.cfm?pagename=NDG_main). Many of these resources contain data from published articles that can be re-used to evaluate new methods, confirm reported findings, or that can be combined in unique ways. For genomics, the situation is slightly mitigated by the ability to divide up the problem, with researchers agreeing to work on the pieces, share results, thereby working jointly to map the genes on a chromosome. However, buy-in to neuroscience database submission has been hard to achieve with many brain scientists expressing apprehension to data sharing efforts largely concerning being scooped on some un-recognized result present in their own data (Koslow 2000). In a field where the currency of merit is not how widely one’s data are being shared but how many papers can be written about each dataset, it should be no surprise that most neuroscientists have been resistant to pleas to share data.
In only the past few years have such data standards concepts began to appear for other neuroscience domains. For instance, several standards for the efficient description of neuroimaging studies (Keator, Gadde et al. 2006; Marcus, Olsen et al. 2007) have been put forward, accommodating analysis annotations, activation threshold parameters, clustering and voxel-level statistics. Neuroimaging file formats such as NIfTI (http://nifti.nimh.nih.gov/) have been devised to make tools and data more interoperable. Sophisticated informatics tools such as the LONI Pipeline (http://pipeline.loni.ucla.edu) help to capture the manner in which data have been processed that can be stored and exchanged between investigators. In particular, this documents the workflow provenance of data and records the computational processing tools applied to them. Though not traditionally thought of as integral to the final data analysis process, such factors associated with how the data were treated can have profound effects on reported findings (Lukic, Wernick et al. 2002).
The interest in the application of standards to describe experimental data can, however, be a source of some pain for biomedical researchers (Ball 2006). Ontologies (i.e. formal hierarchical frameworks used to describe experiments) and data standards require two minimal characteristics: they must be useful and they must be used. The development of ontologies has an appeal for the biological sciences, especially neuroscience, in being able to related disparate information across spatial, temporal, and paradigmatic scales while fully leveraging the emerging Semantic Web (http://sciencecommons.org/projects/data/, for example) in which the context of the data is its principle attribute. Unlike the genomics community’s GO, neuro-ontologies and standards describing experimental details and conclusions are not necessarily user-friendly and so have not been widely adopted. We do not doubt, however, that efficient meta-data frameworks for neuroscience are achievable with the careful consideration of the needs of the users they are meant to serve. With these standards in hand, researchers will be able to efficiently describe their research findings so that others may be better able to examine, combine, and re-use these data beyond their original scope.
The sociological process of acceptance of data sharing is of particular interest for other fields wishing to begin their own data sharing efforts in earnest. While it certainly appears to many as if the genetics community adopted data sharing willingly and easily overnight, it wasn’t always so. Intellectual property concerns (Chokshi, Parker et al. 2006) and research priority (Marshall 2002) have been widely debated, as well as what rules the research community should follow when sharing (Roberts 2002). There are examples where DNA sequence information from some studies had failed to be made available at all via GenBank (Noor, Zimmerman et al. 2006). Competitiveness between databases and the reliability of some data has led to a few squabbles (Soldatova and King 2005; Shields 2006). Willingness to share data has often been inversely related to how difficult that data was to acquire or analyze. The funding road has not always been smooth -- The Protein Data Bank (PDB), for instance, was founded in the 1970’s and struggled to stay afloat through many challenges only to now be considered the primary repository for protein structure data (Berman, Battistuz et al. 2002). When new interpretations of these genomic data began to appear in leading journals, the excitement was such that new domains of science were born that before did not exist -- notably the relatively new discipline of systems biology (Hood 2003). In time, many of the concerns over these aspects of data sharing have died down and the concept has gained acceptance as the benefits have become clearer. This movement toward digital biology and chemistry owes itself to achievements in organizing study data, in persevering through hard times, and in making the data widely available to all of those who can use it to conduct novel science.
For many brain researchers, determining the biological value of these ever-growing collections of data has, indeed, become one of their greatest challenges. Funding ways to accommodate this ever growing amount of brain data has required special consideration for infrastructure, database construction, privacy concerns, and user access (Ascoli, De Schutter et al. 2003). Despite support for data sharing and public archiving from the SfN, the International Neuroinformatics Coordinating Facility (INCF), and other major scientific bodies, the unclear directions for NIH funding, of particular concern to the biomedical research community (Mitka 2007), has been especially disruptive for those interested in informatics for the brain (Bloom 2006; De Schutter, Ascoli et al. 2006; Gazzaniga, Van Horn et al. 2006). In order to maximize efforts, data sharing projects in molecular biology and from neuroscience must learn from each other in terms of what has worked as well as what has failed and exchange ideas for gaining acceptance with a busy and skeptical community. This can occur more rapidly if journals call for deposition of primary data into recognized archives as a condition of publication; if scientific societies demand greater openness in scientific exchange; and, if data standards and useful software tools exist explicitly for these purposes. These are painful ideas for some. Yet, only through the sharing of this pain can uncertainty be reduced as we move toward a time when more data are obtained digitally and expectations increase over its online availability under new models for scientific publishing.
There is much to be taken from the intra-disciplinary experiences of the brain science community in organizing data, sharing, and developing tools for its efficient analysis. Examination of what has worked for one domain can be educational for another - leading to brand new disciplines within neuroscience. With prompting and support from leading societies, encouragement from government funders, and general interest in managing their extensive collections of information, brain scientists might readily overcome disorder and benefit from freely shared meta- and primary neuroscience data. Through cross-disciplinary discussion and interactivity, perhaps then the entropy associated with the collecting of large digital datasets on brain form and function can, indeed, be refuted – or most certainly reduced – easing the way toward new and joyous neuroscientific discoveries.
Acknowledgements
The authors would like to thank Professor Floyd Bloom of the Scripps Research Institute and Dr. Balaji Srinivasan of the Statistics Department at Stanford University for useful discussion and comments on this article.
References
- Ascoli GA, De Schutter E, et al. An information science infrastructure for neuroscience. Neuroinformatics. 2003;1(1):1–2. doi: 10.1385/NI:1:1:001. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CA. Are we stuck in the standards? Nat Biotechnol. 2006;24(11):1374–1376. doi: 10.1038/nbt1106-1374. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Jones DK. Diffusion-tensor MRI: theory, experimental design and data analysis - a technical review. NMR Biomed. 2002;15(7–8):456–467. doi: 10.1002/nbm.783. [DOI] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 6 No 1):899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- Bloom F. Prying open the black box. Science. 2006;314(5796):17. doi: 10.1126/science.1135216. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Jones AR. Neurogenomics: at the intersection of neurobiology and genome sciences. Nat Neurosci. 2004;7(5):429–433. doi: 10.1038/nn1232. [DOI] [PubMed] [Google Scholar]
- Brazma A, Hingamp P, et al. Minimum information about a microarray experiment (MIAME)- toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, et al. Functional brain imaging of young, nondemented, and demented older adults. J Cogn Neurosci. 2000;12 Suppl 2:24–34. doi: 10.1162/089892900564046. [DOI] [PubMed] [Google Scholar]
- Butte A. The use and analysis of microarray data. Nat Rev Drug Discov. 2002;1(12):951–960. doi: 10.1038/nrd961. [DOI] [PubMed] [Google Scholar]
- Chokshi DA, Parker M, et al. Data sharing and intellectual property in a genomic epidemiology network: policies for large-scale research collaboration. Bull World Health Organ. 2006;84(5):382–387. doi: 10.2471/blt.06.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Esposito M. Functional neuroimaging of cognition. Semin Neurol. 2000;20(4):487–498. doi: 10.1055/s-2000-13182. [DOI] [PubMed] [Google Scholar]
- De Schutter E, Ascoli GA, et al. On the future of the human brain project. Neuroinformatics. 2006;4(2):129–130. doi: 10.1385/NI:4:2:129. [DOI] [PubMed] [Google Scholar]
- Gazzaniga MS, Van Horn JD, et al. Continuing Progress in Neuroinformatics. Science. 2006;311(5758):176. doi: 10.1126/science.311.5758.176a. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, et al. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc Natl Acad Sci U S A. 2003;100(1):253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M, Clark J, et al. Nucleic Acids Research. 2004;32(Database Issue):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodaie M, Cordella R, et al. Bursting activity of neurons in the human anterior thalamic nucleus. Brain Res. 2006;1115(1):1–8. doi: 10.1016/j.brainres.2006.07.085. [DOI] [PubMed] [Google Scholar]
- Hood L. Systems biology: integrating technology, biology, and computation. Mech Ageing Dev. 2003;124(1):9–16. doi: 10.1016/s0047-6374(02)00164-1. [DOI] [PubMed] [Google Scholar]
- Keator DB, Gadde S, et al. A general XML schema and SPM toolbox for storage of neuro- imaging results and anatomical labels. Neuroinformatics. 2006;4(2):199–212. doi: 10.1385/ni:4:2:199. [DOI] [PubMed] [Google Scholar]
- Kennedy DN. Share and share alike. Neuroinformatics. 2003;1(3):211–213. doi: 10.1385/NI:1:3:211. [DOI] [PubMed] [Google Scholar]
- Konradi C. Gene expression microarray studies in polygenic psychiatric disorders: applications and data analysis. Brain Res Brain Res Rev. 2005;50(1):142–155. doi: 10.1016/j.brainresrev.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Koslow SH. Should the neuroscience community make a paradigm shift to sharing primary data? Nature Neuroscience. 2000;3(4):863–865. doi: 10.1038/78760. [DOI] [PubMed] [Google Scholar]
- Laurens KR, Kiehl KA, et al. Attention orienting dysfunction during salient novel stimulus processing in schizophrenia. Schizophr Res. 2005;75(2–3):159–171. doi: 10.1016/j.schres.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Lukic AS, Wernick MN, et al. An evaluation of methods for detecting brain activations from functional neuroimages. Artif Intell Med. 2002;25(1):69–88. doi: 10.1016/s0933-3657(02)00009-x. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, et al. The extensible neuroimaging archive toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Marshall E. Data sharing. DNA sequencer protests being scooped with his own data. Science. 2002;295(5558):1206–1207. doi: 10.1126/science.295.5558.1206. [DOI] [PubMed] [Google Scholar]
- Miles MF. Microarrays: lost in a storm of data? Nat Rev Neurosci. 2001;2(6):441–443. doi: 10.1038/35077582. [DOI] [PubMed] [Google Scholar]
- Mitka M. Scientists warn NIH funding squeeze hampering biomedical research. Jama. 2007;297(17):1867–1868. doi: 10.1001/jama.297.17.1867. [DOI] [PubMed] [Google Scholar]
- Noor MA, Zimmerman KJ, et al. Data sharing: how much doesn't get submitted to GenBank? PLoS Biol. 2006;4(7):e228. doi: 10.1371/journal.pbio.0040228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Geschwind DH. Deconstructing language by comparative gene expression: from neurobiology to microarray. Genes Brain Behav. 2006;5 Suppl 1:54–63. doi: 10.1111/j.1601-183X.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- Parkinson H, Kapushesky M, et al. Nucleic Acids Research. 2007;35(Database Issue):D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. Functional brain imaging and human brain function. J Neurosci. 2003;23(10):3959–3962. doi: 10.1523/JNEUROSCI.23-10-03959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L. Genome research. A tussle over the rules for DNA data sharing. Science. 2002;298(5597):1312–1313. doi: 10.1126/science.298.5597.1312. [DOI] [PubMed] [Google Scholar]
- Shields R. MIAME we have a problem. Trends in Genetics. 2006;22:65–66. doi: 10.1016/j.tig.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Soldatova LN, King RD. Are the current ontologies in biology good ontologies? Nature Biotechnology. 2005;23(9):1095–1098. doi: 10.1038/nbt0905-1095. [DOI] [PubMed] [Google Scholar]
- Spellman P, Miller M, et al. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biology. 2002;3(9):46. doi: 10.1186/gb-2002-3-9-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Grafton ST, et al. Sharing neuroimaging studies of human cognition. Nat Neurosci. 2004;7(5):473–481. doi: 10.1038/nn1231. [DOI] [PubMed] [Google Scholar]
- Walton MM, Bechara B, et al. The role of the primate superior colliculus in the control of head movements. J Neurophysiol. 2007 doi: 10.1152/jn.00258.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetzel P, Parkinson HE, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22(7):866–873. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]