Abstract
A structure-based approach is described for predicting the effects of amino acid substitutions on protein function. Structures were predicted using a homology modelling method. Folding and binding energy differences between wild-type and mutant structures were computed to quantitatively assess the effects of amino acid substitutions on protein stability and protein–protein interaction, respectively. We demonstrated that pathogenic mutations at the interaction interface could affect binding energy and destabilise protein complex, whereas mutations at the non-interface might reduce folding energy and destabilise monomer structure. The results suggest that the structure-based analysis can provide useful information for understanding the molecular mechanisms of diseases.
Keywords: Amino acid substitutions, homology modeling, folding energy, binding energy, protein stability, protein-protein interaction
1 Introduction
Revealing the effects of amino acid substitutions on protein structure and function is critical for understanding the complex mechanisms of human disease caused by single amino acid mutations. There are 67,000 – 200,000 non-synonymous Single Nucleotide Polymorphisms (nsSNPs) in the human population (Cargill et al., 1999), which give rise to a large number of amino acid substitutions in proteins. The residue changes at key sites within a protein may result in a series of conformation changes, including the breakage of salt bridges, alteration of interaction network, disruption of hydrogen bonds, which in turn may perturb the energy landscape. These changes can affect the kinetics of protein folding or cause protein aggregation and destabilisation (Dill et al., 1993). More than half of monogenic diseases are caused by single mutations, and a common mechanism by which amino acid substitutions cause human disease is protein stability change. Yue and Moult investigated the effect of amino acid substitutions on protein stability, and estimated that approximately 25% of nsSNPs in the human population might be deleterious to protein function (Yue and Moult, 2006). Of the known disease-causing missense mutations, the majority (83%) resulted in alternation of protein stability (Wang and Moult, 2001).
Amino acid substitutions can also affect protein-protein interactions. Approximately 88% of disease-associated nsSNPs are found to be located in the voids/pockets important for protein-protein interactions (Stitziel et al., 2003). The amino acid substitutions located at the binding interface or active site cleft could block the entrance to the active site, change the recognition, alter the specificity, or affect the binding affinity. For example, the substitution G2019S in leucine-rich repeat kinase 2 (LRRK2) was shown to be associated with familial and sporadic Parkinson’s disease (Aasly et al., 2005). Structure analysis indicates that this mutation is located at the interface of LRRK2’s N-terminal and C-terminal domains which is important for positioning of Mg2+ within the active site of the kinase (Albrecht, 2005, Mata et al., 2006). This finding is in agreement with the experimental result that G2019S enhances kinase activity in vitro (Kachergus et al., 2005). Recently, Teng et al. (Teng et al., 2009) examined the effects of nsSNPs at the interaction interfaces of 264 protein complexes using a homology modeling method and all atoms energy calculations. The results suggest that disease-causing mutations tend to destabilise protein-protein interactions. Therefore, understanding how amino acid substitutions affect protein stability and protein-protein interactions can provide new insights into the molecular mechanisms of human genetic diseases.
Protein structure modeling methods have been widely used for predicting the effects of disease-causing mutations on protein stability and protein-protein interaction. For instance, to predict the effects of the mutations related to the genetic disorder galactosemia, more than one hundred mutant structures of galactose-1-phosphate uridyltransferase were constructed using the homology modeling method, and the results suggested that most mutations might alter protein stability (Facchiano and Marabotti, 2009). By mapping disease-causing mutations onto known three-dimensional protein structures, Dimmic and coworkers (Dimmic et al., 2005) have shown that about 70% of the deleterious mutations are located in the structurally and/or functionally important sites. However, the effects of mutations were analyzed statically in these studies. The free energy perturbation (FEP) calculation has been used to quantitatively assess the effects of amino acid substitutions on protein stability. Dixit et al. (Dixit et al., 2009) used the AMBER force field and solvent-accessible surface area solvation methods to calculate the protein stability changes in terms of free energy differences caused by cancer-associated mutations in the RET and MET kinases, and showed that the amino acid substitutions could decrease the thermodynamical stability of the mutant structures. The FEP calculation was also used to assess the protein stability changes upon single amino acid substitutions in membrane proteins (Park and Lee, 2005). Nevertheless, these studies on FEP calculation did not take into account the effects of amino acid substitutions on protein-protein interactions.
The advent of high-throughput sequencing technology makes it possible to identify a large number of nsSNPs in the human genome. The dbSNP database, one of the primary data resources for genetic studies, contains the information of more than 23 million human SNPs (Smigielski et al., 2000). The records in the dbSNP database are linked to the Online Mendelian Inheritance in Man (OMIM) database, which contains disease gene information, including genetic polymorphisms, map locations, inheritance patterns and clinical descriptions (Wheeler et al., 2007). Computational analyses provide an efficient way for examining the effects of nsSNPs on protein stability and function, and for identifying potential disease-causing mutations. Ng and Henikoff (Ng and Henikoff, 2003) used a position-specific scoring matrix (PSSM) based method called Sorting Intolerant From Tolerant (SIFT) to predict whether an amino acid substitution affects protein function. We have recently developed the MuStab web server for predicting protein stability changes upon amino acid substitutions from sequence features (Teng et al., 2010). MuStab uses a support vector machine (SVM) model to discriminate between destabilizing and stabilizing amino acid substitutions in proteins. iPTREE-STAB (Huang et al., 2007) and I-Mutant 3.0 sequence version (Capriotti et al., 2008) are also available for sequence-based prediction of protein stability changes caused by point mutations. Structure-based methods, including PoPMuSiC-2.0 (Dehouck et al., 2009), Dmutant (Zhou and Zhou, 2002), Eris (Yin et al., 2007), I-Mutant 3.0 structure version (Capriotti et al., 2008) and FoldX (Schymkowitz et al., 2005), are available for examining the effects of mutations on protein stability and protein-protein interactions. In particular, the FoldX software tool can be used to provide quantitative estimations about the effects of amino acid substitutions on the stability of proteins or protein complexes using the empirical force field calculation (Schymkowitz et al., 2005). Among these protein stability predictors, I-Mutant3.0 structure version, Dmutant and FoldX gave the best predictive performances (Khan and Vihinen, 2010).
The experimental approach for determining the effects of amino acid substitutions on protein stability is to obtain the mutant proteins and measure their thermal stability changes by melting experiments. However, the experimental approach is time-consuming and thus may not be applied to a large number of amino acid substitutions. In the present study, a structure-based approach was performed for predicting the effects of amino acid substitutions on protein stability and protein-protein interaction. The differences of folding energy and binding energy between the wild-type and mutant structures were calculated to predict the protein stability and protein-protein interaction changes caused by the mutations. The predictions were evaluated by using other bioinformatic methods. The results suggest that the structure-based approach can provide useful information for characterizing disease-causing mutations in human genetic studies.
2 Methods
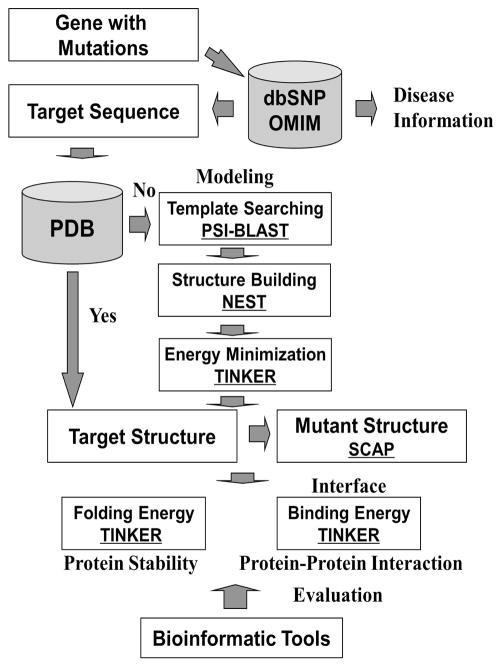
The schematic diagram of the structure-based approach is shown in Figure 1. The methodology was also investigated in two previous studies (Teng et al., 2008, Zhang et al., 2010). For a specific gene with mutations, the related sequence and disease information were extracted from the dbSNP and OMIM databases. If the structure of the target protein was available in the Protein Data Bank (PDB), no structure modeling was needed. Otherwise, target structures were constructed using the homology modeling method (Xiang, 2006). The suitable templates were identified in the PDB database using the PSI-BLAST program (Altschul et al., 1997), and then used to construct the target structures with the NEST program (Petrey et al., 2003). Energy minimization was performed to obtain the optimal structure with the TINKER program (Ponder, 1999), and the mutant structure was constructed using the SCAP program (Xiang and Honig, 2001). The folding energy of the wild-type or mutant structure was calculated using TINKER to estimate the effects of the mutations on protein stability. For amino acid substitutions located at the interface, the binding energy changes were also computed to predict the effects of the mutations on protein-protein interaction. At the end, the predictions were compared with several bioinformatics tools, including FoldX (Schymkowitz et al., 2005), PoPMuSiC-2.0 (Dehouck et al., 2009), Dmutant (Zhou and Zhou, 2002), Eris (Yin et al., 2007), MuStab (Teng et al., 2010), iPTREE-STAB (Huang et al., 2007) and I-Mutant 3.0 (both sequence and structure versions) (Capriotti et al., 2008).
Figure 1.
Schematic diagram of the approach for assessing the effects of amino acid substitutions on protein stability and protein-protein interaction. Underlined are the software tools used in this study.
2.1 Protein structure modeling
Homology modeling was applied to the proteins with no structures available in the PDB database. The structures were modeled as follows:
1) Template searching
The suitable templates were selected from the PDB database for the target protein. Position-Specific Iterated BLAST (PSI-BLAST) (Altschul et al., 1997) was used for the template searching. The structures with significant E-value (< 10−5) were selected as the suitable templates.
2) Structure building
The program NEST was used to the build structure models according to the sequence alignment between the target protein and its structural template (Petrey et al., 2003). NEST is an integrated model-building program, including the program LOOPY9 for loop prediction and SCAP10 for side-chain modeling.
3) Energy minimization
To generate the optimal structure, energy minimization was performed by using the TINKER package (Ponder, 1999) with the CHARMM27 force field parameters (Brooks et al., 1983). The MINIMIZE program in TINKER was used to minimize structures with the algorithm of Limited Memory BFGS Quasi-Newton Optimization (Ponder, 1999).
The mutant structures were derived in silico from the wild-type structure using the SCAP program (Xiang and Honig, 2001). The amino acid substitutions were introduced by side-chain replacements with the rest of the structure kept rigid. The MINIMIZE program in the TINKER package was used to minimize the mutant structures.
2.2 Folding energy calculation
The effects of amino acid substitutions on protein stability were assessed by the folding energy changes. The energy calculation was based on the monomer structure of the target protein, and was performed as described in the recent publication (Zhang et al., 2010). The folding energy is the energy difference between the folded and unfolded states:
| (1) |
where G(folded) or G(unfolded) is the total potential energy of the target protein in the folded or unfolded state, respectively.
The protein stability change (ΔΔGstability) is the folding energy difference between the wild-type (WT) structure and the structure with the amino acid substitution (AAS). It can be calculated using the following equation:
| (2) |
However, the energy difference between the wild-type and mutant proteins in the unfolded state, G(unfolded: WT) - G(unfolded: AAS), is difficult to calculate. In the present study, we assume that the difference of energy in the unfolded state can be estimated by using the substitution site and its neighboring residues. The total potential energy of the eleven-residue segment (S11) with the substitution site in the middle position was used to represent the folding energy of the full-length protein in the unfolded state. Equation (2) can thus be rewritten as:
| (3) |
All of the above total potential energy terms were calculated using the ANALYZE program in the TINKER package. A positive value of ΔΔGstability indicates that the amino acid substitution may make the protein more stable, whereas a negative value of ΔΔGstability suggests that the mutation can destabilise the protein.
2.3 Binding energy calculation
For an amino acid substitution located at the interaction interface, the binding energy difference of the protein complex between the wild-type and mutant structures was used to assess the effect of the mutation on protein-protein interaction. As described in the previous study (Teng et al., 2009), the binding energy was calculated using the rigid body approach, in which the structures of the monomers were kept as they were in the dimer complex. The binding energy, ΔΔG(binding), was the difference between the total potential energy of the dimer complex and the individual monomers:
| (4) |
where ΔG(folding: complex), ΔG(folding: A) and ΔG(folding: B) are the folding free energy values of the dimer complex, monomer A and monomer B, respectively. Since the internal mechanical energy values of the unbound and bound monomers are the same, the energy terms in the unfolded state can be canceled out in equation (4). Thus, the binding free energy can be calculated as below:
| (5) |
where G(folded: complex), G(folded) and G(folded) are the total potential energy values of the dimer complex, monomer A and monomer B in the folded state, respectively.
In this study, the total potential energy was computed using the ANALYZE program in the TINKER package. The effect of an amino acid substitution on protein-protein interaction was assessed by using the binding energy difference between the wild-type (WT) structure and the structure with the amino acid substitution (AAS):
| (6) |
A positive value of the binding energy change (ΔΔΔGbinding) indicates that the amino acid substitution may strengthen the binding affinity and make the protein dimer complex more stable. In contrast, a negative value of ΔΔΔGbinding suggests that the mutation can weaken the binding affinity and destabilise the dimer complex.
2.4 Prediction evaluation
Several bioinformatic tools were used to evaluate the predictive power of the structure-based approach used in this study, and the predictions were considered to be reliable if a consensus was reached by most of the predictors. Sequence-based prediction of the direction of protein stability change could give useful information. Three sequence-based tools were used to predict the directions of protein stability changes caused by amino acid substitutions from primary sequence data, including iPTREE-STAB (http://210.60.98.19/IPTREEr/iptree.htm), MuStab (http://bioinfo.ggc.org/mustab/) and I-Mutant3.0 (sequence version) (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi).
Structure-based prediction methods could provide quantitative assessment of the effects of amino acid substitutions on protein stability. Khan and Vihinen (Khan and Vihinen, 2010) compared the predictive performances of different protein stability predictors, and showed that three structure-based tools, including I-Mutant3.0 (structure version) (http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi), Dmutant (http://sparks.informatics.iupui.edu/hzhou/mutation.html) and FoldX (Schymkowitz et al., 2005) were the most reliable predictors. These three tools were used in this study. Other two structure-based predictors, including PoPMuSiC-2.0 (http://babylone.ulb.ac.be/popmusic/) and Eris (http://eris.dokhlab.org), were also used to calculate the folding energy for monomer structures, respectively. The difference of the folding energy between the wild-type and mutant structures was used to assess the protein stability change caused by an amino acid substitution, and compared with the ΔΔGstability value calculated using the approach applied in this paper. Furthermore, FoldX was also used to determine the interaction energy of complex protein. The effect of an amino acid substitution on protein-protein interaction was estimated by the interaction energy difference of the protein complex between the wild-type and mutant structures (ΔΔΔGFoldX), which was compared with ΔΔΔGbinding computed using the method utilized in this study.
In addition, ClustalX (Larkin et al., 2007) was used to perform the multiple sequence alignment for conservation analysis. Protein sequences from different species were downloaded from the NCBI Entrez database using the GENE search option with the gene name as the query.
3 Results and discussion
To evaluate the usefulness of the structure-based approach utilized in this paper, three case studies were carried out for four pathogenic mutations and one neutral nsSNP in three human genes (Table 1). One disease-causing mutation, A111V (dbSNP ID: rs28928889, OMIM ID: 141850.0029), and one neutral nsSNP, T119N (dbSNP ID: rs1058069), in the human HBA2 gene (haemoglobin subunit alpha) were used to show their different effects on protein stability and protein-protein interaction. Two pathogenic mutations, Q61K (dbSNP ID: rs28933406, OMIM ID: 190020.0002) and A146T (dbSNP ID: rs104894231, OMIM ID: 190020.0008), in the human HRAS gene (v-Ha-ras Harvey rat sarcoma viral oncogene homolog) were analyzed to assess the effects of mutations on different structural regions (interface or non-interface). The computational approach was also used to investigate the substitution, A693V, in the human ZBTB20 gene (zinc finger and BTB domain containing 20). As discussed in the following sections, the results suggest that the pathogenic mutations make the monomer structures less stable (ΔΔGstability < 0), and/or weaken the binding affinity to destabilise the dimer structures (ΔΔΔGbinding < 0). In contrast, the neutral nsSNP has only slight effects on protein stability and protein-protein interaction (ΔΔGstability and ΔΔΔGbinding close to 0).
Table 1.
The effects of five amino acid substitutions on protein stability. The unit of energy change is kcal/mol.
| Amino acid substitution | A111V (HBA2) | T119N (HBA2) | Q61K (HRAS) | A146T (HRAS) | A693V (ZBTB20) | |
|---|---|---|---|---|---|---|
| Structure-based Tools | ΔΔGstability | −0.75 | 0.06 | −4.42 | −1.39 | −2.69 |
| FoldX | −4.19 | 10.54 | −2.74 | −0.22 | −0.68 | |
| PoPMuSiC-2.0 | −0.49 | −0.50 | −0.24 | −0.38 | −0.05 | |
| Dmutant | −0.48 | 0.32 | −0.38 | −0.24 | −0.34 | |
| Eris | 4.28 | 2.29 | −2.63 | −1.24 | −0.72 | |
| I-Mutant 3.0 (structure version) | 0.13 | −0.30 | 0.29 | −0.79 | −0.03 | |
| Consensus | Decreased | Increased | Decreased | Decreased | Decreased | |
| Sequence-based Tools | I-Mutant 3.0 (sequence version) | Increased | Increased | Increased | Decreased | Increased |
| MuStab | Increased | Decreased | Increased | Decreased | Increased | |
| iPTREE-STAB | Increased | Decreased | Increased | Decreased | Decreased | |
| Consensus | Increased | Decreased | Increased | Decreased | Increased | |
It was shown that the predictions agree well with the results gave by the most of structure-based methods. However, the sequence-based tools often did not agree with the consensus predictions from the structure-based methods (Table 1). The structure-based predictors (I-Mutant3.0 structure version, Dmutant and FoldX) appeared to be more reliable for predicting protein stability changes caused by mutations (Khan and Vihinen, 2010). Thus, this study focused on the structure-based analyses.
3.1 Pathogenic mutation and neutral nsSNP in haemoglobin
Haemoglobin molecules in red blood cells transport oxygen from the lung to the peripheral tissues, and thus are important for maintaining cell viability. Human haemoglobin is made up of symmetric dimers of polypeptide chains, the α/β-globin dimers (Kan, 1991). Several point mutations in α-globin have been shown to cause α-thalassemia, which can result in Hydrops fetalis (Chui and Waye, 1998). In this study, the two amino acid substitutions of human haemoglobin subunit alpha (HBA2), A111V and T119N, were analyzed to show the different effects of disease-causing and neutral amino acid substitutions on protein stability and protein-protein interaction. The homodimer structure of HBA2 was built using the crystal structure of human deoxy haemoglobin (PDB: 1O1P) as the template.
The majority of disease-causing mutations cause protein destabilisation, whereas most neutral nsSNPs have limited effect on protein stability (Wang and Moult, 2001). In the present study, the predicted effects of A111V (disease-causing) and T119N (neutral) on protein stability agree well with the previous observations. As shown in Table 1, the folding energy change (ΔΔGstability) caused by A111V is −0.75 kcal/mol, suggesting that the mutation may destabilise haemoglobin monomer structure. The decreased protein stability is also predicted for the A111V mutation by three structure-based tools including FoldX, PoPMuSiC-2.0 and Dmutant (Table 1). In contrast, the neutral nsSNP (T119N) is predicted by our calculations and three structure-based tools (FoldX, Dmutant and Eris) to stabilize the protein monomer. PoPMuSiC-2.0 and I-Mutant3.0 (structure version) give the opposite predictions. The results suggest that T119N may not cause destabilisation of the monomer structure.
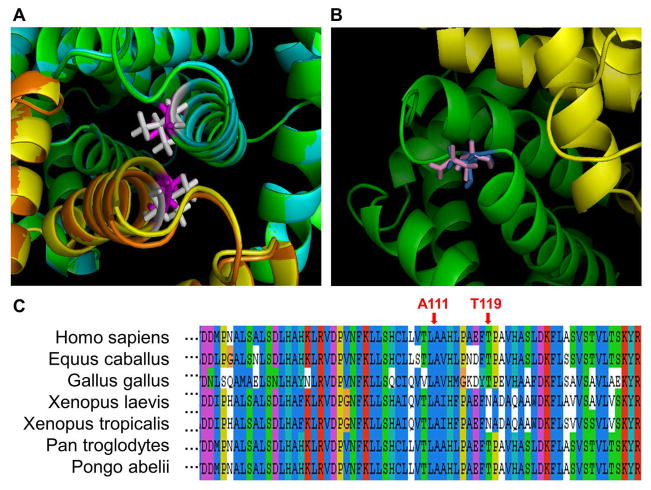
Amino acid substitutions at the interaction interface may result in binding affinity changes, and thus affect the structure of the protein complex. As shown in Figure 2a, the pathogenic mutation, A111V, is located in the α-helix of the HBA2 binding interface. Although most regions of the wild-type and mutant structures are similar, the structures are not overlapped in the α-helix interface region. This structural change may significantly affect the binding energy, and make the protein complex unstable. The observation has been confirmed by the binding energy calculation using both TINKER and FoldX (ΔΔΔGbinding = −11.56 kcal/mol and ΔΔΔGFoldX = −1.41 kcal/mol) (Table 2). In contrast, the neutral nsSNP (T119N) is located in the flexible loop region (Figure 2b). Since T119N is not located in the inner region of the interface, it may not significantly affect protein-protein interaction. The binding energy change caused by T119N is ΔΔΔGbinding = 0.90 kcal/mol (Table 2), which is smaller than the absolute value of binding energy change caused by A111V.
Figure 2. Illustration of two amino acid substitutions (A111V and T119N) in human HBA2.
a) Structural representation of the A111V mutation. The wild-type chain A is shown in green color, mutant chain A in cyan, wild-type chain B in yellow, and mutant chain B in orange. The amino acid residue Ala 110 (wild-type) is shown in magenta, and Val 110 (mutant) in white.
b) Structural representation of T119N. Chains A and B are shown in green and yellow, respectively. The residue Asn 119 (wild-type) is shown in pink, and Thr 119 (mutant) in blue.
c) Multiple sequence alignment of HBA2 with the amino acid substitution sites indicated.
Table 2.
The effects of five amino acid substitutions on protein-protein interaction. The unit of energy change is kcal/mol.
| Amino acid substitution | A111V (HBA2) | T119N (HBA2) | Q61K (HRAS) | A146T (HRAS) | A693V (ZBTB20) |
|---|---|---|---|---|---|
| ΔΔΔGbinding | −11.56 | 0.90 | −7.29 | −0.21 | −0.31 |
| ΔΔΔGFoldX | −1.41 | 1.39 | −2.40 | −0.11 | 0.00 |
In addition, the multiple sequence alignment shown in Figure 2c suggests that the residue, Ala 111, is well conserved, but Thr 119 is not conserved in Xenopus laevis and Xenopus tropicalis. The result agrees with the previous observation that pathogenic mutations tend to be located at evolutionarily conserved positions (Miller and Kumar, 2001).
3.2 Pathogenic mutations at the interface or non-interface of HRAS
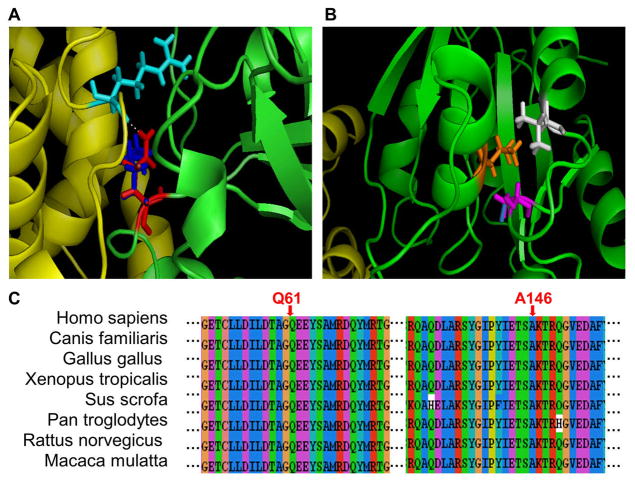
Follicular carcinoma is the second most common thyroid cancer, which accounts for about 15% of all thyroid malignancies. The v-Ha-ras Harvey rat sarcoma viral oncogene homolog (HRAS) encodes a follicular cancer-related protein located at the inner surface of cell membrane. The protein plays an important role in the transduction of signals arising from tyrosine kinase and G protein-coupled receptors. One pathogenic mutation (Q61K) in HRAS was found to cause constitutive activation of the downstream signaling pathways (Nikiforova et al., 2003). Another disease-causing mutation (A146T) was identified in patients with Costello syndrome, and was shown to affect the GTP/GDP binding of HRAS (Zampino et al., 2007). In this study, the heterodimer structure of HRAS has been built using the crystal structure of the transforming protein RhoA (PDB: 1OW3) as the template. The amino acid substitution Q61K is located at the interaction interface (Figure 3a), and A146T lies in a non-interface region of HRAS (Figure 3b). These two mutations in different structural regions have been analyzed to assess their effects on protein stability and protein-protein interaction.
Figure 3. Illustration of two disease-causing mutations (Q61K and A146T) in human HRAS.
a) Structural representation of the Q61K mutation. Chains A and B are shown in green and yellow, respectively. The residue Gln 61 (wild-type) is shown in red, Lys 61 (mutant) in blue, and Arg 47 of chain B in cyan. The hydrogen bond is represented as a white dash line.
b) Structural representation of the A146T mutation. Chains A and B are shown in green and yellow, respectively. Ala 146 (wild-type) is shown in magenta, and Thr 146 (mutant) in blue. Two neighboring residues, Leu 15 in orange and Val 148 in white, are also shown.
c) Multiple sequence alignment of HRAS with the amino acid substitution sites indicated.
Both amino acid residues in the HRAS protein, Gln 61 and Ala 146, are conserved in other species (Figure 3c), suggesting that they may be functionally important sites. As shown in Table 1, the folding energy changes (ΔΔGstability) caused by Q61K and A146T are −4.42 kcal/mol and −1.39 kcal/mol (Table 1), respectively, suggesting that both mutations may destabilise the HRAS monomer structure. Consistent with the above results, the predictions made by structure-based tools show decreased protein stability for both mutations (excluding I-Mutant3.0 structure version for Q61K). Furthermore, all the sequence-based methods also predict that A146T could make HRAS protein unstable.
The Q61K mutation is located at the interaction interface (Figure 3a), and the binding energy change caused by Q61K is ΔΔΔGbinding = −7.29 kcal/mol, or ΔΔΔGFoldX = −2.40 kcal/mol (Table 1), suggesting that the mutation may significantly affect protein-protein interaction. The distance between Gln 61 and its interaction partner, Arg 47 from the other chain, is only 1.88 A, which is within the distance of hydrogen bond formation. When the polar residue Gln is replaced by positively charged residue Lys, the hydrogen bonds may be affected, and thus make strongly unfavorable interactions with Arg 47. In contrast, the A146T mutation located in a non-interface region (Figure 3b) does not appear to have a significant effect on protein-protein interaction. As shown in Table 1, the binding energy change caused by A146T is ΔΔΔGbinding = −0.21 kcal/mol, or ΔΔΔGFoldX = −0.11 kcal/mol. Nevertheless, Ala 146 and its neighboring residues (Leu 15 and Val 148) may form the hydrophobic pocket, which is involved in the binding of the purine ring of GTP/GDP. The substitution of Ala 146 by the polar residue Thr may alter the hydrophobic environment in the pocket, and thus affect the binding of GTP or GDP.
3.3 Application: the A693V substitution in ZBTB20
The structure-based approach was also used to investigate the amino acid substitution, A693V, in the human ZBTB20 gene (zinc finger and BTB domain containing 20). ZBTB20 plays important roles in neurogenesis (Mitchelmore et al., 2002), postnatal survival and glucose homeostasis (Sutherland et al., 2009). The A693V substitution is implicated to impair the function of ZBTB20 in the brain. Thus, predicting the effects of A693V on protein stability and function may help determine the pathogenic potential of the amino acid substitution.
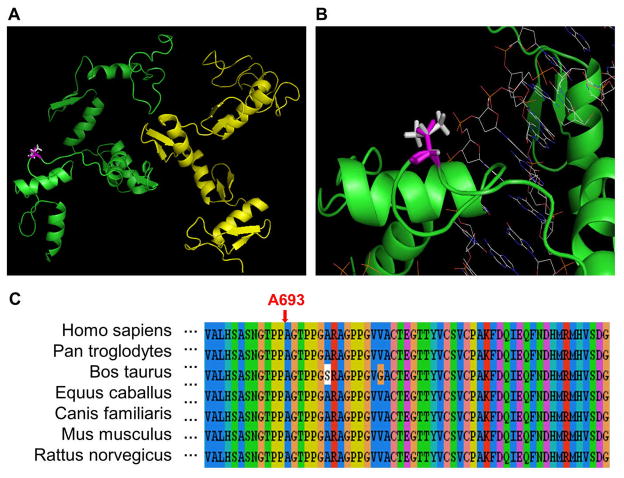
The structure of the C-terminal region (560-739) of ZBTB20, including five zinc finger domains, was constructed using the homology modeling method with the six-finger zinc finger peptide (PDB: 2I13) as the template. As shown in Figure 4a, although ZBTB20 may form a homodimer structure, the A693V mutation is not located at the interaction interface. The binding energy change caused by A693V is ΔΔΔGbinding = −0.31 kcal/mol, or ΔΔΔGFoldX = 0 kcal/mol (Table 2), suggesting that the amino acid substitution has little effect on dimer formation. The folding energy change was also calculated for A693V using TINKER (ΔΔGstability = −2.69 kcal/mol, Table 1). In addition, all of the structure-based methods predicted that A693V will decrease protein stability. Thus, the consensus prediction is that A693V will slightly destabilise the monomer structure of ZBTB20.
Figure 4. Illustration of the A693V mutation in human ZBTB20.
a) Structural representation of the A693V mutation. Chains A and B are shown in green and yellow, respectively. Ala 693 (wild-type) is shown in magenta, and Val 693 (mutant) in white.
b) Representation of the modeled structure of ZBTB20 in complex with DNA. Shown are chain A in green, Ala 693 in magenta, Val 693 in white, and the DNA molecule as wireframe.
c) Multiple sequence alignment of ZBTB20 with the amino acid substitution site indicated.
Since the ZBTB20 protein was previously shown to bind DNA (Mitchelmore et al., 2002), the structure of ZBTB20 in complex with DNA has been modeled using the six-finger zinc finger peptide (PDB: 2I13) as the template. As shown in Figure 4b, the amino acid residue, Ala 693, is located close to the phosphate group of DNA backbone. Therefore, another possibility is that the A693V substitution may be involved in protein-DNA interaction. The multiple sequence alignment shown in Figure 4c also suggests that Ala 693 is highly conserved in other species, and thus may be important for the normal function of ZBTB20.
4 Conclusion
In this paper, a structure-based approach is described for assessing the effects of amino acid substitutions on protein stability and protein-protein interaction. Homology modeling and free energy calculation methods were used to compute the differences of folding energy and binding energy between the wild-type and mutant structures. Three case studies showed that the disease-causing mutations at the interaction interface might reduce the binding energy, and thus weaken the affinity in the protein complex. The pathogenic mutations in the non-interface region could reduce the folding energy and thus destabilise the monomer structure. Therefore, the structure-based approach can be used to quantitatively assess the effects of amino acid substitutions on protein stability and protein-protein interaction. The approach may be useful for understanding the molecular mechanisms by which gene mutations cause human diseases.
Acknowledgments
This work is supported by the CSREES/USDA, under project number SC-1700355. EA acknowledges the support from NIH/NLM, grant number R03 LM009748.
Biographies
Shaolei Teng is currently a PhD student in the Department of Genetics and Biochemistry at Clemson University. He received his MS degree in Molecular Biology from Gyeongsang National University in South Korea, and his BS degree in Biotechnology from Soochow University in China. His current research interests include machine learning and structure modeling. He has published 7 papers in peer-reviewed journals and conference proceedings.
Anand K. Srivastava is a Senior Research Scientist and Associate Director of the Center for Molecular Studies, J.C. Self Research Institute of Human Genetics, Greenwood Genetic Center and Professor (adjunct) in the Dept. of Genetics and Biochemistry, Clemson University. He received his MS degree in Biochemistry from the University of Allahabad, and PhD in Biochemistry from Banaras Hindu University, India. His research interest is to understand the etiology of inherited disorders of human brain development, cognitive function, and epithelial organogenesis. He has published more than 65 peer-reviewed papers including reviews and book chapters.
Charles E. Schwartz is Director of Research at the Greenwood Genetic Center and Head of the J.C. Self Research Institute. He received his Ph.D. in Biochemistry from Vanderbilt University and his M.S. degree in Biochemistry from Oklahoma State University. For over 25 years his research has focused on the identification of genes that cause intellectual disability. He has published over 250 peer reviewed articles and has been a reviewer for many journals.
Emil Alexov got his MS in Physics in 1984 and in 1990 received PhD in Plasma Physics from Sofia University, Bulgaria. Since 1990 he was an Assistant Professor at the Department of General Physics at Sofia University. In 1995 he came to United States as a Research Associate at the Department of Physics, City College of New York and later in 2000 joined Columbia University, New York, as Howard Hughes Bioinformatics Specialist. In 2005 he became Associate Professor at the Department of Physics, Clemson University. His research is focused on further development of DelPhi and modeling disease-causing missense mutations.
Liangjiang Wang is an Assistant Professor in the Department of Genetics and Biochemistry at Clemson University, and an adjunct faculty member at Greenwood Genetic Center. He received his PhD in Molecular Biology from the University of Georgia, and his MS degree in Computer Science from Mississippi State University. His research focuses on biological knowledge discovery and genomic data integration. He has published more than 40 peer-reviewed papers in journals and conference proceedings. He has been in the editorial board for three international journals, and a reviewer for more than 20 journals.
Contributor Information
Shaolei Teng, Email: steng@clemson.edu, Department of Genetics and Biochemistry, Clemson University, Clemson, SC 29634, USA.
Liangjiang Wang, Department of Genetics and Biochemistry, Clemson University, Clemson, SC 29634, USA.
Anand K. Srivastava, Email: anand@ggc.org, J.C. Self Research Institute of Human Genetics, Greenwood Genetic Center, Greenwood, SC 29646, USA
Charles E. Schwartz, Email: ceschwartz@ggc.org, J.C. Self Research Institute of Human Genetics, Greenwood Genetic Center, Greenwood, SC 29646, USA
Emil Alexov, Email: ealexov@clemson.edu, Department of Physics and Astronomy, Clemson University, Clemson, SC 29634, USA.
References
- Aasly JO, Toft M, Fernandez-Mata I, Kachergus J, Hulihan M, White LR, Farrer M. ‘Clinical features of LRRK2-associated Parkinson’s disease in central Norway. Ann Neurol. 2005;57(5):762–5. doi: 10.1002/ana.20456. [DOI] [PubMed] [Google Scholar]
- Albrecht M. LRRK2 mutations and Parkinsonism. Lancet. 2005;365(9466):1230. doi: 10.1016/S0140-6736(05)74810-8. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization and dynamic calculations. J Comp Chem. 1983;4(187–217) [Google Scholar]
- Capriotti E, Fariselli P, Rossi I, Casadio R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics. 2008;9(Suppl 2, No. S6) doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22(3):231–8. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Chui DH, Waye JS. Hydrops fetalis caused by alpha-thalassemia: an emerging health care problem. Blood. 1998;91(7):2213–22. [PubMed] [Google Scholar]
- Dehouck Y, Grosfils A, Folch B, Gilis D, Bogaerts P, Rooman M. Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics. 2009;25(19):2537–43. doi: 10.1093/bioinformatics/btp445. [DOI] [PubMed] [Google Scholar]
- Dill KA, Fiebig KM, Chan HS. Cooperativity in protein-folding kinetics. Proc Natl Acad Sci U S A. 1993;90(5):1942–6. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmic MW, Sunyaev S, Bustamante CD. Inferring SNP function using evolutionary, structural, and computational methods. Pac Symp Biocomput. 2005;(382–4) [PubMed] [Google Scholar]
- Dixit A, Torkamani A, Schork NJ, Verkhivker G. Computational modeling of structurally conserved cancer mutations in the RET and MET kinases: the impact on protein structure, dynamics, and stability. Biophys J. 2009;96(3):858–74. doi: 10.1016/j.bpj.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchiano A, Marabotti A. Analysis of galactosemia-linked mutations of GALT enzyme using a computational biology approach. Protein Eng Des Sel. 2009;23(2):103–13. doi: 10.1093/protein/gzp076. [DOI] [PubMed] [Google Scholar]
- Huang LT, Gromiha MM, Ho SY. iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics. 2007;23(10):1292–3. doi: 10.1093/bioinformatics/btm100. [DOI] [PubMed] [Google Scholar]
- Kachergus J, Mata IF, Hulihan M, Taylor JP, Lincoln S, Aasly J, Gibson JM, Ross OA, Lynch T, Wiley J, Payami H, Nutt J, Maraganore DM, Czyzewski K, Styczynska M, Wszolek ZK, Farrer MJ, Toft M. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet. 2005;76(4):672–80. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan YW. Molecular biology of haemoglobin: its application to sickle cell anemia and thalassemia. Schweiz Med Wochenschr Suppl. 1991;43(51–4) [PubMed] [Google Scholar]
- Khan S, Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31(6):675–84. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29(5):286–93. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 2001;10(21):2319–28. doi: 10.1093/hmg/10.21.2319. [DOI] [PubMed] [Google Scholar]
- Mitchelmore C, Kjaerulff KM, Pedersen HC, Nielsen JV, Rasmussen TE, Fisker MF, Finsen B, Pedersen KM, Jensen NA. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem. 2002;277(9):7598–609. doi: 10.1074/jbc.M110023200. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn GW, 2nd, Tallini G, Kroll TG, Nikiforov YE. RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab. 2003;88(5):2318–26. doi: 10.1210/jc.2002-021907. [DOI] [PubMed] [Google Scholar]
- Park H, Lee S. Prediction of the mutation-induced change in thermodynamic stabilities of membrane proteins from free energy simulations. Biophys Chem. 2005;114(2–3):191–7. doi: 10.1016/j.bpc.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Petrey D, Xiang Z, Tang C, Xie L, Gimpelev M, Mitros T, Soto C, Goldsmith-Fischman S, Kernytsky A, Schlessinger A, Koh I, Alexov E, Honig B. Uning Multiple Structure Alignments, Fast Model Builing, and Energetic Analysis in Fold Recognition and Homology Modeling. Proteins. 2003;53(430–435) doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- Ponder JW. TINKER-software tools for molecular design. 3.7. St. Luis: Washington University; 1999. [Google Scholar]
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28(1):352–5. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitziel NO, Tseng YY, Pervouchine D, Goddeau D, Kasif S, Liang J. Structural location of disease-associated single-nucleotide polymorphisms. J Mol Biol. 2003;327(5):1021–30. doi: 10.1016/s0022-2836(03)00240-7. [DOI] [PubMed] [Google Scholar]
- Sutherland AP, Zhang H, Zhang Y, Michaud M, Xie Z, Patti ME, Grusby MJ, Zhang WJ. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol Cell Biol. 2009;29(10):2804–15. doi: 10.1128/MCB.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Madej T, Panchenko A, Alexov E. Modeling effects of human single nucleotide polymorphisms on protein-protein interactions. Biophys J. 2009;96(6):2178–88. doi: 10.1016/j.bpj.2008.12.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Michonova-Alexova E, Alexov E. Approaches and resources for prediction of the effects of non-synonymous single nucleotide polymorphism on protein function and interactions. Curr Pharm Biotechnol. 2008;9(2):123–33. doi: 10.2174/138920108783955164. [DOI] [PubMed] [Google Scholar]
- Teng S, Srivastava AK, Wang L. Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genomics. 2010 doi: 10.1186/1471-2164-11-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17(4):263–70. doi: 10.1002/humu.22. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Geer LY, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Ostell J, Miller V, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35(Database issue):D5–12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z. Advances in homology protein structure modeling. Curr Protein Pept Sci. 2006;7(3):217–27. doi: 10.2174/138920306777452312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Honig B. Extending the Accuracy Limits of Prediction for Side-chain Conformations. J Mol Biol. 2001;311(421–430) doi: 10.1006/jmbi.2001.4865. [DOI] [PubMed] [Google Scholar]
- Yin S, Ding F, Dokholyan NV. Eris: an automated estimator of protein stability. Nat Methods. 2007;4(6):466–7. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- Yue P, Moult J. Identification and analysis of deleterious human SNPs. J Mol Biol. 2006;356(5):1263–74. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- Zampino G, Pantaleoni F, Carta C, Cobellis G, Vasta I, Neri C, Pogna EA, De Feo E, Delogu A, Sarkozy A, Atzeri F, Selicorni A, Rauen KA, Cytrynbaum CS, Weksberg R, Dallapiccola B, Ballabio A, Gelb BD, Neri G, Tartaglia M. Diversity, parental germline origin, and phenotypic spectrum of de novo HRAS missense changes in Costello syndrome. Hum Mutat. 2007;28(3):265–72. doi: 10.1002/humu.20431. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Teng S, Wang L, Schwartz CE, Alexov E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum Mutat. 2010;31(9):1043–9. doi: 10.1002/humu.21310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11(11):2714–26. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]