Abstract
Eukaryotic and Archaeal multisubunit RNA polymerases (Pols) are structurally related and require several similar components for transcription initiation. However, none of the Pol I factors were known to share homology with TFIIB or TFIIB-related proteins, key factors in the initiation mechanisms of the other Pols. Here we show that Rrn7, a subunit of the yeast Pol I Core Factor, and its human ortholog TAF1B are TFIIB-like factors. Although distantly related, Rrn7 shares many activities associated with TFIIB-like factors. Domain swaps between TFIIB-related factors show that Rrn7 is most closely related to the Pol III general factor Brf1. Our results point to the conservation of initiation mechanisms among multisubunit Pols and reveal a key function of yeast Core Factor/human SL1 in Pol I transcription.
Transcription initiation by most eukaryotic and archaeal multi subunit polymerases requires TFIIB or a TFIIB-related general transcription factor. TFIIB and TFIIB-like proteins (archaeal Tfb and the Pol III factor Brf) operate at multiple key steps during transcription initiation such as Pol binding, preinitiation complex formation, start site selection, promoter opening, and abortive initiation (1–4). Because of the fundamental and conserved role of TFIIB-like factors for other Pols, it was surprising that none of the Pol I-specific subunits or general initiation factors were known to share TFIIB homology.
Transcription of the S. cerevisiae rDNA locus by Pol I requires four general transcription factors: UAS-binding Upstream Activity Factor (UAF), TATA-binding protein (TBP), Core Factor (CF) and the regulatory factor Rrn3 (5). CF contains subunits Rrn6, 7, and 11 (6, 7), and interacts with UAF, TBP, Rrn3, and Pol I (5). CF is analogous to the human Pol I factor SL1, composed of four TBP-associated factors (TAFs), TAF1A, TAF1B, TAF1C, and TAF1D (8, 9), where TAF1A, B and C are orthologous to yeast CF subunits Rrn6, 7, and 11 (10). Both CF and SL1 recruit Pol I to its promoter but the function of the CF/SL1 subunits, apart from TBP and Pol binding, is unknown (5, 11, 12).
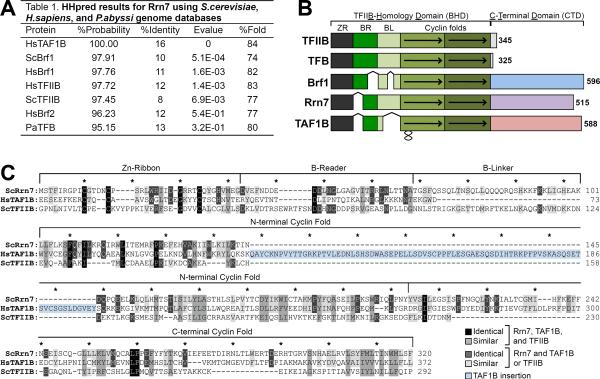
We guessed that a Pol I TFIIB-like factor would have diverged considerably from related factors (13) since Pol I subunits share relatively low protein sequence conservation with their Pol II and Pol III counterparts (14). Using the homology detection program HHpred, which uses pairwise hidden Markov model profile comparisons that are more sensitive than traditional web-based approaches (15), we detected high probability matches between the Rrn7 N-terminal 320 residues and the TFIIB family, indicating that Rrn7 is a TFIIB paralog. The probability scores ranged from 95%–98%, including a 100% match with the human SL1 subunit TAF1B (Fig 1A; Fig S1). No significant homology was detected between Rrn6 or Rrn11 with other Pol or basal factor subunits. Rrn7 and TAF1B share low sequence conservation between TFIIB family members and each other (8–16% identity), whereas the predicted secondary structures are similar (Fig 1A, Fig S2–S4). Rrn7 and TAF1B form a distinct Pol I specific clade, consistent with their sequence divergence from the other TFIIB-related proteins (Fig S5A). A plant-specific TFIIB-related protein (Brp1) was described to function in Pol I transcription since it crosslinks to the Pol I promoter in vivo and bound Pol I in vitro (16). However, Rrn7 and TAF1B match with 100% probability to Mee12, an uncharacterized TFIIB-related plant protein, while Brp1 is more TFIIB-related (Fig.S5B–D). This is consistent with a conserved Pol I function of Rrn7/Taf1B/Mee12 in all eukaryotes and plant-specific functions for the Brp proteins (16, 17).
Figure 1. Homology between S. cerevisiae Rrn7 and TFIIB family members.
(A) HHpred search results. % Fold compares the predicted secondary structure of the template and query. E-value is the average expected number of non-homologous proteins with a score higher than the database match. (B) Schematic of TFIIB family organization. BHD, TFIIB homology domain; CTD, C-terminal domain; ZR, zinc ribbon; BR, B-reader; BL, B-linker. Arrows indicate the cyclin fold repeats, and the black squiggle indicates a TAF1B insertion. The number to the right indicates protein length. (C) Alignment of Rrn7, TAF1B, and TFIIB. Residues colored according to conservation. Blue highlight: TAF1B insertion.
The TFIIB homology domains (BHDs) of Rrn7 and TAF1B contain the same four subdomains found in all TFIIB-like proteins: the zinc ribbon, B-reader, B-linker, and cyclin folds (Fig 1B). For TFIIB, the ribbon domain binds Pol II, the reader and linker domains are situated in the Pol II active site where they are involved in open complex formation and transcription start site selection, and the cyclin domains bind TBP, Pol II and promoter DNA (2, 3). The Brf proteins also contain a large C-terminal domain (CTD) that is essential for function, and mediates interaction between Pol III and the TFIIIB subunits Bdp1 and TBP (18, 19). Rrn7 and TAF1B contain a large CTD (Fig S6,7), but the Rrn7 CTD sequence is yeast-specific and the TAF1B CTD is conserved among metazoans. Both are unrelated to the Brf CTD.
The function of Rrn7 derivatives with deletions and single amino acid substitutions was tested in a yeast strain containing a chromosomal rrn7 deletion and where the 35S rRNA precursor is transcribed by Pol II from the GAL7 promoter (20). Strains lacking Pol I transcription activity are dependent on galactose for survival and cannot grow on glucose media. Disruption of every Rrn7 subdomain was lethal, indicating that all TFIIB- and Brf1-like domains are essential (Fig S8).
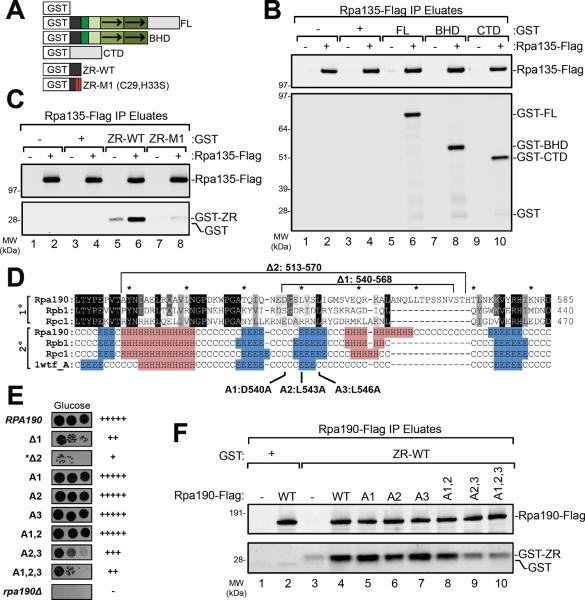
Since TFIIB-like factors directly interact with their respective Pols, we immobilized yeast Pol I via the second largest Pol I subunit (Rpa135) and measured the binding of GST-Rrn7 derivatives (Fig 2A,B). Full-length Rrn7 and the separate BHD and CTD domains specifically bind immobilized Pol I. This Pol-binding behavior is similar to Brf1, where both its BHD and CTD bind Pol III (21). The TFIIB ribbon domain binds with high affinity to the Rbp1 dock domain, where it positions the adjacent reader and linker regions in the enzyme active site (2, 3, 22, 23). As predicted for a TFIIB-like factor, the Rrn7 ribbon bound strongly to Pol I, whereas binding was severely reduced by a mutation in two Zn-coordinating residues (Fig 2A,C).
Figure 2. Rrn7 ribbon and Pol I dock domain mutations affect Rrn7-Pol I interaction.
(A) GST-Rrn7 fusion proteins. (B) Binding of Rrn7 Full length (FL), BHD, or CTD to immobilized Rpa135-Flag Pol I. (C) Binding of WT or Rrn7 ribbon mutant M1 to immobilized Pol I. (D) Sequence and secondary structure alignment of Rpa190 (Pol I), Rpb1 (Pol II), and Rpc1 (Pol III) dock domains. Residues are colored according to conservation and PSIpred predictions: helical (H, red), beta-sheet (E, blue), and coil (C, white). Known yeast Rpb1 secondary structure (1wtf_A) is shown. In the mutations Δ1 and Δ2, the deleted sequences were replaced by Gly-Ser-Gly. (E) Growth of yeast dock domain mutants. +++, WT growth; −, no growth. (F) Pol I with the indicated dock domain mutations was immobilized to a-Flag beads and tested for Rrn7 ribbon-binding. Panels B, C, F are Western blots probed with anti-Flag or -GST antibody.
The Pol II (Rbp1) and Pol III (Rpc1) dock domains are moderately conserved with Pol I (Rpa190; 34–40% identity); however, their predicted secondary structure is nearly identical (Fig 3D). Replacement of Rpa190 dock residues with Glycine-Serine linkers (Δ1, Δ2; Fig 3D) gave moderate to severe growth defects whereas 2 out of 3 multiple alanine substitutions gave moderate growth defects (Fig 2E, compare A2,3 and A1,2,3 to A1,2). There was high correlation between alanine substitutions that reduced Rrn7 ribbon-Pol I binding and growth defects (A2,3 and A1,2,3; Fig 2F). Together, our findings suggest that the Rrn7 ribbon interacts with the Rpa190 dock domain, similar to the TFIIB ribbon-Pol II dock interaction.
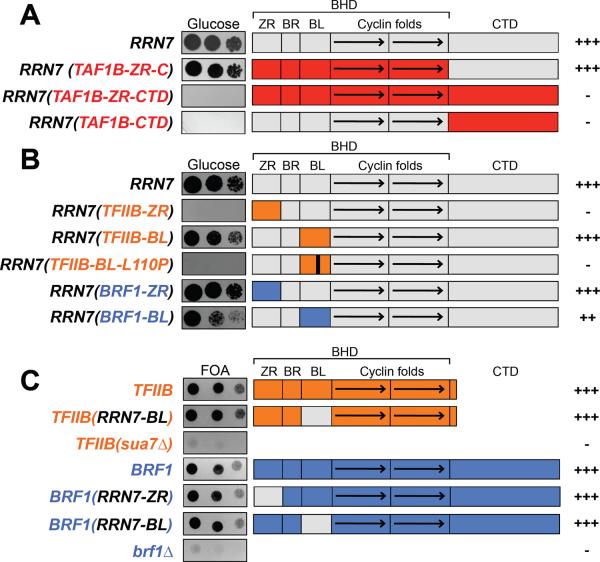
Figure 3. Interchangeable TFIIB family domains.
RRN7 yeast complementation assay with chimeric proteins containing (A) Rrn7 (grey) and TAF1B (red), (B) TFIIB (orange) or BRF1 (blue). (C) TFIIB and BRF1 yeast complementation assay with indicated Rrn7 domains (grey) in place of TFIIB or Brf1 domains. +++, WT growth; −, no growth.
TAF1B displays several differences from S. cerevisiae Rrn7, including a very short B-linker and a large insertion within the first cyclin repeat (Fig 1C, blue highlight). However, we note the low sequence similarity between Rrn7 and TAF1B in the linker and at the beginning of the first cyclin repeat, so the boundary between these domains in TAF1B is not certain. Substitution of the entire TAF1B BHD for the analogous domain in Rrn7 complemented yeast growth (Fig 3A), whereas substitution of the TAF1B CTD did not, explaining why full-length TAF1B will not substitute for Rrn7.
To test if any of the TFIIB or Brf1 domains could functionally substitute within Rrn7, we generated Rrn7 derivatives containing segments of TFIIB or Brf1. We found that the TFIIB and Brf1 B-linkers complemented Rrn7 function, (Fig 3B; Fig S9), and the Rrn7 B-linker could functionally substitute within TFIIB or Brf1 (Fig 3C). The TFIIB linker forms a helix proposed to lie at the junction of single and double stranded DNA in the open complex state (2, 3). Since B-linkers from the TFIIB-related factors substitute for one another, they may all fulfill this function, despite heterogeneity in length and sequence. Consistent with this proposal, we found that the TFIIB linker mutation L110P, which disrupts TFIIB function (2), also disrupted linker function in the Rrn7-TFIIB chimeria (Fig 3B).
The TFIIB and Brf1 cyclin and B-reader domains could not functionally complement the analogous Rrn7 domains (Fig S9). However, the Brf1 and Rrn7 ribbon domains were interchangeable, whereas the TFIIB ribbon was not. (Fig 3B,C). These results show that Rrn7 is more functionally related to Brf1, consistent with their similar domain architecture, interchangeable ribbon domains, and BHD and CTD-mediated Pol binding.
Rrn7 shares many basic properties with TFIIB and Brf1 despite significant sequence variation. The finding that the Rrn7 and Brf1 ribbon domains are interchangeable suggests that they promote the same functions, such as providing an anchor, so that the reader and linker segments are positioned in the active site to facilitate DNA opening. In contrast, the TBP binding activity of CF seems split between Rrn7 and Rrn6 (6, 12). It remains unclear whether the Rrn7 cyclin repeats bind TBP and whether they interact with Pol I, or if these functions have been distributed among other CF subunits.
All Pols must overcome similar obstacles to initiate transcription including specific promoter binding, template opening, transcription start site recognition, and the transition to processive elongation. The best understood TFIIB family member is TFIIB, where each subdomain performs a specific and essential role in PIC formation, promoter opening and/or start site selection. Bacterial Pol, which lacks a TFIIB-related factor, uses σ factor for many of the same functions in promoter binding, open complex formation and initiation. Although there is no sequence or significant structural similarity, TFIIB and σ70 show striking topological similarities in the way they bind to their respective Pols in near identical locations (2, 3). Combined with previous findings, our results point to the conservation of initiation mechanisms among all multisubunit Pols.
Supplementary Material
Acknowledgments
We thank Beth Moorefield for yeast strains and plasmids, Joost Zomerdijk for comments and sharing results, and members of the Hahn laboratory, Beth Moorefield, and Ted Young for their comments and encouragement. Supported by NIH GM053451 to SH and T32 CA09657 to BAK.
References and Notes
- 1.Hahn S. Nat Struct Mol Biol. 2004 May;11:394. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kostrewa D, et al. Nature. 2009 Nov 19;462:323. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Science. 2010 Jan 8;327:206. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schramm L, Hernandez N. Genes Dev. 2002 Oct 15;16:2593. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 5.Aprikian P, Moorefield B, Reeder RH. Mol Cell Biol. 2001 Aug;21:4847. doi: 10.1128/MCB.21.15.4847-4855.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lalo D, Steffan JS, Dodd JA, Nomura M. J Biol Chem. 1996 Aug 30;271:21062. doi: 10.1074/jbc.271.35.21062. [DOI] [PubMed] [Google Scholar]
- 7.Lin CW, et al. Mol Cell Biol. 1996 Nov;16:6436. doi: 10.1128/mcb.16.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorski JJ, et al. EMBO J. 2007 Mar 21;26:1560. doi: 10.1038/sj.emboj.7601601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zomerdijk JC, Beckmann H, Comai L, Tjian R. Science. 1994 Dec 23;266:2015. doi: 10.1126/science.7801130. [DOI] [PubMed] [Google Scholar]
- 10.Boukhgalter B, et al. Gene. 2002 May 29;291:187. doi: 10.1016/s0378-1119(02)00597-8. [DOI] [PubMed] [Google Scholar]
- 11.Friedrich JK, Panov KI, Cabart P, Russell J, Zomerdijk JC. J Biol Chem. 2005 Aug 19;280:29551. doi: 10.1074/jbc.M501595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steffan JS, Keys DA, Dodd JA, Nomura M. Genes Dev. 1996 Oct 15;10:2551. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- 13.Hahn S. Nature. 2009 Nov 19;462:292. doi: 10.1038/462292a. [DOI] [PubMed] [Google Scholar]
- 14.Carter R, Drouin G. Mol Biol Evol. 2009 Nov;26:2515. doi: 10.1093/molbev/msp164. [DOI] [PubMed] [Google Scholar]
- 15.Soding J, Biegert A, Lupas AN. Nucleic Acids Res. 2005 Jul 1;33:W244. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imamura S, Hanaoka M, Tanaka K. EMBO J. 2008 Sep 3;27:2317. doi: 10.1038/emboj.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavel E, et al. PLoS One. 2011;6:e17216. doi: 10.1371/journal.pone.0017216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassavetis GA, Driscoll R, Geiduschek EP. J Biol Chem. 2006 May 19;281:14321. doi: 10.1074/jbc.M601702200. [DOI] [PubMed] [Google Scholar]
- 19.Liao Y, Willis IM, Moir RD. J Biol Chem. 2003 Nov 7;278:44467. doi: 10.1074/jbc.M308354200. [DOI] [PubMed] [Google Scholar]
- 20.Nogi Y, Yano R, Nomura M. Proc Natl Acad Sci U S A. 1991 May 1;88:3962. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khoo B, Brophy B, Jackson SP. Genes Dev. 1994 Dec 1;8:2879. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 22.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Science. 2004 Feb 13;303:983. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 23.Chen HT, Hahn S. Mol Cell. 2003 Aug;12:437. doi: 10.1016/s1097-2765(03)00306-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.