Abstract
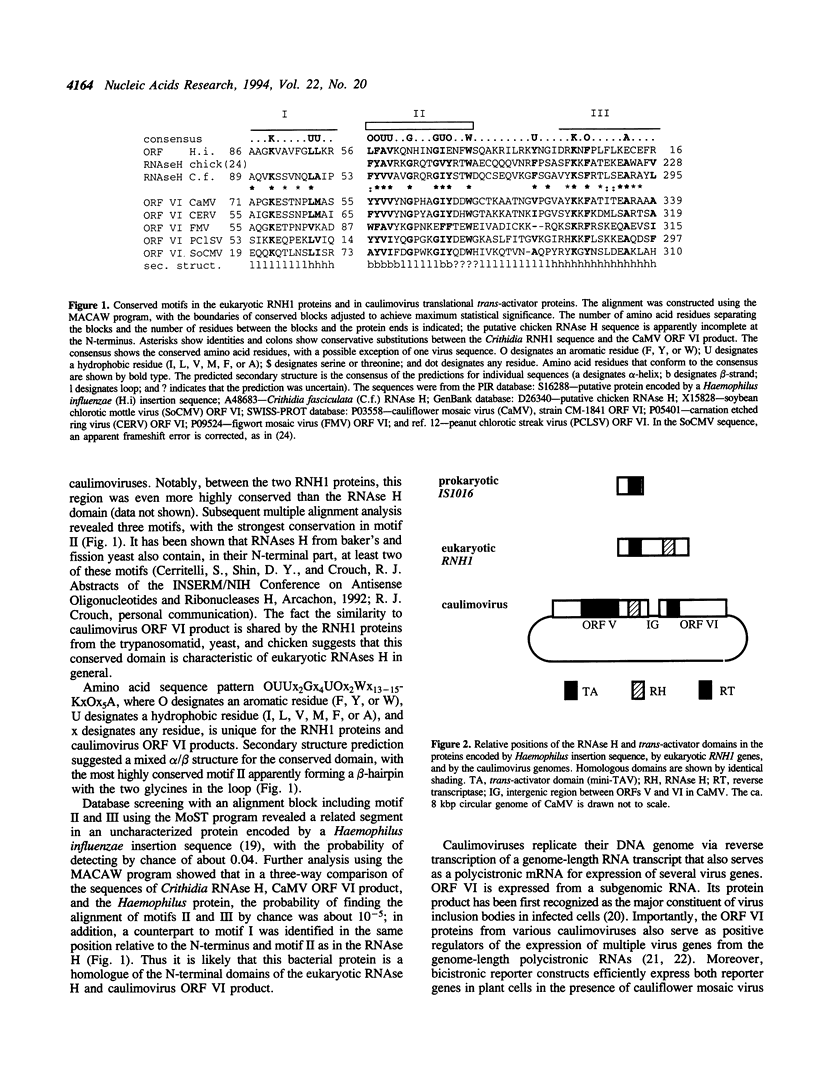
RNAse H (RNH1 protein) from the trypanosomatid Crithidia fasciculata has a functionally uncharacterized N-terminal domain dispensable for the RNAse H activity. Using computer methods for database search and multiple alignment, we show that the N-terminal domains of RNH1 and its homologue encoded by a cDNA from chicken lens are related to the conserved domain in caulimovirus ORF VI product that facilitates translation of polycistronic virus RNA in plant cells. We hypothesize that the N-terminal domain of eukaryotic RNAse H performs an as yet uncharacterized regulatory function, possibly in mRNA translation or turnover.
Full text
PDF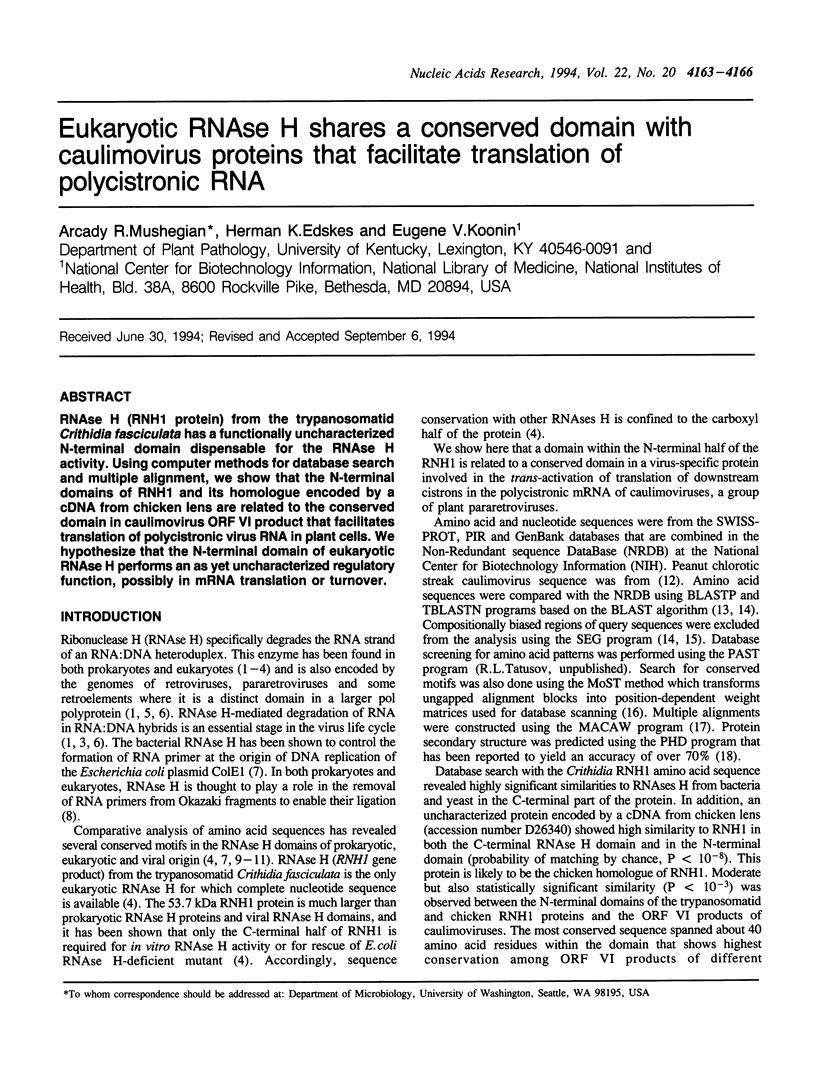


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Boguski M. S., Gish W., Wootton J. C. Issues in searching molecular sequence databases. Nat Genet. 1994 Feb;6(2):119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bonneville J. M., Sanfaçon H., Fütterer J., Hohn T. Posttranscriptional trans-activation in cauliflower mosaic virus. Cell. 1989 Dec 22;59(6):1135–1143. doi: 10.1016/0092-8674(89)90769-1. [DOI] [PubMed] [Google Scholar]
- Campbell A. G., Ray D. S. Functional complementation of an Escherichia coli ribonuclease H mutation by a cloned genomic fragment from the trypanosomatid Crithidia fasciculata. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9350–9354. doi: 10.1073/pnas.90.20.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazenave C., Frank P., Büsen W. Characterization of ribonuclease H activities present in two cell-free protein synthesizing systems, the wheat germ extract and the rabbit reticulocyte lysate. Biochimie. 1993;75(1-2):113–122. doi: 10.1016/0300-9084(93)90032-n. [DOI] [PubMed] [Google Scholar]
- Crouch R. J. Ribonuclease H: from discovery to 3D structure. New Biol. 1990 Sep;2(9):771–777. [PubMed] [Google Scholar]
- De Tapia M., Himmelbach A., Hohn T. Molecular dissection of the cauliflower mosaic virus translation transactivator. EMBO J. 1993 Aug;12(8):3305–3314. doi: 10.1002/j.1460-2075.1993.tb06000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Fütterer J., Hohn T. Translation of a polycistronic mRNA in the presence of the cauliflower mosaic virus transactivator protein. EMBO J. 1991 Dec;10(12):3887–3896. doi: 10.1002/j.1460-2075.1991.tb04958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fütterer J., Potrykus I., Valles Brau M. P., Dasgupta I., Hull R., Hohn T. Splicing in a plant pararetrovirus. Virology. 1994 Feb;198(2):663–670. doi: 10.1006/viro.1994.1078. [DOI] [PubMed] [Google Scholar]
- Goulian M., Richards S. H., Heard C. J., Bigsby B. M. Discontinuous DNA synthesis by purified mammalian proteins. J Biol Chem. 1990 Oct 25;265(30):18461–18471. [PubMed] [Google Scholar]
- Itoh T., Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980 May;77(5):2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M. S., McClure M. A., Feng D. F., Gray J., Doolittle R. F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll J. S., Loynds B. M., Moxon E. R. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol Microbiol. 1991 Jun;5(6):1549–1560. doi: 10.1111/j.1365-2958.1991.tb00802.x. [DOI] [PubMed] [Google Scholar]
- McClure M. A. Evolution of retroposons by acquisition or deletion of retrovirus-like genes. Mol Biol Evol. 1991 Nov;8(6):835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993 Jul 20;232(2):584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Svitkin Y. V., Meerovitch K., Lee H. S., Dholakia J. N., Kenan D. J., Agol V. I., Sonenberg N. Internal translation initiation on poliovirus RNA: further characterization of La function in poliovirus translation in vitro. J Virol. 1994 Mar;68(3):1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcriptases. Retrons in bacteria. Nature. 1989 May 25;339(6222):254–255. doi: 10.1038/339254a0. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Reverse transcription in the eukaryotic genome: retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol Biol Evol. 1985 Nov;2(6):455–468. doi: 10.1093/oxfordjournals.molbev.a040365. [DOI] [PubMed] [Google Scholar]
- Walder R. Y., Walder J. A. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990 Oct;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]



