INTRODUCTION
Most organisms are subjected to daily fluctuations in light and temperature as a result of the full rotation of the Earth on its axis approximately every 24 h. Endogenous circadian biological clocks evolved that allow for the anticipation of these daily variations to control rhythmic gene expression, which in turn regulates metabolic and behavioral processes to provide a selective advantage (DeCoursey, 1961; DeCoursey et al., 2000; Johnson, 2005; Michael et al., 2003; Ouyang et al., 1998; Woelfle et al., 2004). For decades after their description, circadian clocks were believed to exist only in eukaryotes, as prokaryotes were thought to lack the complexity that would require a 24-h timing mechanism (Edmunds, 1983; Kippert, 1987). However, the existence of two incompatible biochemical processes – oxygenic photosynthesis and oxygen-sensitive nitrogen fixation – that occur in some unicellular cyanobacteria led to investigation of the mechanism used to separate these dissonant reactions (Mitsui et al., 1986). Ensuing research uncovered that these and other alternating rhythms in cyanobacteria display the three hallmark circadian characteristics: persistence under constant conditions, entrainment and phase resetting, and temperature compensation (Chen et al., 1991; Grobbelaar and Huang, 1992; Grobbelaar et al., 1986; Huang et al., 1990; Mitsui et al., 1986; Schneegurt et al., 1994; Sweeney and Borgese, 1989).
While several different cyanobacterial strains were identified as having bona fide circadian mechanisms, ultimately Synechococcus elongatus PCC 7942 was chosen as the model system due to its genetic malleability (Golden, 1988; Golden et al., 1987). This bacterium offered other genetic advantages in that it has a small (2.8 Mb) and now fully-sequenced genome (US Department of Energy Joint Genome Institute, www.jgi.doe.gov (Holtman et al., 2005)), is naturally transformable (Golden and Sherman, 1984), conjugates with Escherichia coli (Elhai and Wolk, 1988), and has a suite of vectors available for cloning S. elongatus genes (Golden and Sherman, 1983). In addition, an easily observable circadian “behavior” was genetically designed by fusing the promoter of the photosynthesis gene psbAI (PpsbAI) or any other S. elongatus PCC 7942 promoter to the luxAB bioluminescence genes from Vibrio harveyi (Kondo et al., 1993; Liu et al., 1995). The resulting bioluminescence obeys all three rules that deem a process under circadian control and can be monitored automatically in high-throughput assays; these reporter strains were paramount to the rapid discovery and characterization of the genes involved in the cyanobacterial circadian mechanism. As such, the S. elongatus PCC 7942 model now serves as the leader for understanding a biological clock system and its connection with metabolism, cell division and other fundamental cellular processes.
In the less than two decades since the development of a tractable prokaryotic clock model system (Kondo et al., 1993), we have made considerable headway in our understanding of the circadian mechanism in cyanobacteria. What has become increasingly apparent is that the prokaryotic circadian clock is quite complex, despite some historical prejudices regarding the lack of complexity in bacterial systems, and basic research into the Kai clock system has led to interesting insights into eukaryotic circadian mechanisms. Our purpose for this review is to summarize the most current information regarding input, oscillator, and output pathways in S. elongatus, highlighting the importance of genetic and mutational analyses that helped elucidate the players, biochemical modifications, and protein interactions that drive the circadian oscillator and the rhythms it controls in this single-celled, yet highly complex organism.
I. The Kai-Based Oscillator: Early Studies
A. Identification of the kai genes
Since the initial physiological characterization of circadian rhythms in cyanobacteria, genetic investigations into the underlying circadian clock have played a significant role in the discovery of genes that encode key clock proteins. Due to the photoautotrophic nature of S. elongatus, maintaining the cells in constant dark (DD) conditions to monitor their circadian activity is not possible. Instead, circadian rhythms of cyanobacterial cells are measured in constant light (LL); the time in LL in which the organism would ordinarily be in the light or the dark of an light/dark (LD) cycle is considered subjective day and subjective night, respectively. To determine the pervasiveness of the clock system on gene expression, a promoterless luxAB transposon was inserted throughout the genome randomly to report promoter activity via light production. This study showed that of the over eight hundred colonies that produced detectable bioluminescence, all displayed the same near 24-h periodicity of rhythmic promoter activity. Two distinct classes of rhythms were detected as displaying expression patterns that are 12 h out of phase from one another: class 1 peaked at subjective dusk, while class 2 peaked at subjective dawn (Liu et al., 1995). The use of the luxAB luciferase reporter system provided a practical method to screen tens of thousands of S. elongatus colonies to identify mutants defective in maintaining circadian time. Chemical mutagenesis generated a variety of circadian phenotypes that included arrhythmia and altered period lengths, which extended between the shortest at 16 h to the longest at 60 h (Kondo and Ishiura, 1994).
Complementation of the mutants with an S. elongatus genomic DNA library showed that each of the circadian defects was rescued by a single locus that contained three adjacent genes (Ishiura et al., 1998). These genes were named kaiA, kaiB, and kaiC from the Japanese kanji for kaiten, which translates as a cycle of events suggestive of the turning of the heavens. The kai genes are essential for circadian rhythms to persist in S. elongatus. Deletion of each of the kai genes, singly, by operon, or all three together, renders the clock arrhythmic (Ditty et al., 2005; Ishiura et al., 1998). Importantly, kai genes are not essential for cell viability and kai mutants do not display a growth deficiency when grown in pure culture (Ishiura et al., 1998).
The kai genes are expressed from two promoters: one promoter drives expression of kaiA (PkaiA), and a promoter upstream of kaiB (PkaiBC) expresses a kaiBC dicistronic message (Ishiura et al., 1998). Expressing luxAB reporter genes from either of these promoters produces class 1 rhythms in bioluminescence that peak about 12 h after release into LL and every 24–25 h thereafter. The cyclic nature of kai transcription is mirrored in the accumulation of mRNA, with kaiA and kaiBC messages showing circadian fluctuations in absolute levels throughout the day (Ishiura et al., 1998). The kai genes and their protein products have been shown to possess autoregulatory properties similar those of eukaryotic circadian systems (Ishiura et al., 1998). The presence of KaiA positively regulates PkaiBC, as overexpression of KaiA increases the levels of bioluminescence from a PkaiBC::luxAB reporter. This activation is most likely indirect, as KaiA does not contain a predicted DNA-binding domain. When produced in excess, KaiC represses expression from the kaiBC promoter, suggesting a role in negative regulation of its own expression; however, some level of KaiC is necessary for full activity from the kaiBC promoter because levels of kaiBC expression are lower when kaiC is inactivated (Ishiura et al., 1998; Iwasaki et al., 2002; Nishiwaki et al., 2004). Together, the Kai proteins form a negative transcription-translation feedback loop, yet this feedback loop is not required for sustained circadian oscillation as is seen in other eukaryotic systems (Bell-Pedersen et al., 2005; Dunlap, 1999).
Interestingly, neither the kaiA nor kaiBC promoter region contains specific cis elements needed to maintain circadian rhythms in vivo. Placing kaiBC under control of the heterologous Ptrc promoter from E. coli restores wild-type rhythms of bioluminescence and kaiBC mRNA accumulation in a reporter strain that lacks endogenous kaiBC genes (Xu et al., 2003). Driving either kaiA or kaiBC expression from a class 2 promoter, such that peak levels of kai expression should be 12 h out of phase from that of their wild-type counterpart, can complement the respective kai null mutation (Ditty et al., 2005). These results provided preliminary evidence to hint that transcriptional-translational feedback is not the key parameter required to maintain rhythmicity in the prokaryotic model.
A short-term overexpression of KaiC can reset and shift the phase of peak expression in subsequent cycles (Ishiura et al., 1998; Xu et al., 2000). When the levels of kaiBC mRNA are on the rise during the subjective day, a pulse of elevated kaiC expression causes a phase advance. A short pulse of kaiC overexpression during the subjective night, when kaiBC levels are declining, causes a phase delay. The value of the phase shift is proportional to the phase difference between the time when the pulse is given and the time when kaiBC levels would normally be at their maximum (Ishiura et al., 1998). Murayama et al. (2008) quantitatively assessed KaiC-mediated transcriptional regulation; repression of PkaiBC exhibited dose-dependent kinetics, and kaiBC mRNA abundance increased proportionally with the KaiC phosphorylation ratio. KaiC phosphorylation state was found to be a major determinant in the activation of PkaiBC; however, no similar correlation between decreasing PkaiBC activity and decreasing KaiC phosphorylation ratio was found (Murayama et al., 2008).
B. Basic Kai protein properties
Original primary motif searches within the Kai proteins revealed little about how these proteins worked together to form a circadian oscillator, as they have no homology to known eukaryotic circadian clock proteins; this turned out to be a fortuitous characteristic in that there were few pre-conceived notions about their biochemical functions. It was foreshadowed that phosphorylation of KaiC would be integral to its activity as bioinformatic analysis of the KaiC primary amino acid sequence revealed ATP-binding domains that predicted testable phosphorylation events (Ishiura et al., 1998). KaiC is composed of two tandemly duplicated domains (CI and CII) with each domain containing a Walker A P-loop ATP-binding motif, an imperfect Walker B motif, and catalytic carboxylate glutamate residues that are also present in other proteins known to bind ATP (Figure 1). The CI domain also contains two DXXG motifs (where X represents any amino acid residue) that are conserved among members of the GTPase family (Ishiura et al., 1998). Site-directed mutagenesis resulting in a K52H mutant protein of the P-loop 1 ATP-binding motif of CI disrupted the binding of ATP in vitro, and led to an inability to complement a kaiC null mutation in vivo. A corresponding K294H mutant in the P loop 2 of CII had no effect on the ability of KaiC to bind ATP and was able to restore rhythmicity to the kaiC null strain, albeit with a 70 h period (Nishiwaki et al., 2000). Neither mutation affected GTP binding. Mutations in the DXXG motifs also resulted in mutant rhythm phenotypes. The G71A mutant in CI had a lowered amplitude and distorted waveform in PkaiBC expression, and the G114A mutant had a 27 h period and a bimodal waveform. A glutamine residue immediately following the CII DXXG motif, when changed to Q115R, abolished the circadian rhythm (Nishiwaki et al., 2000).
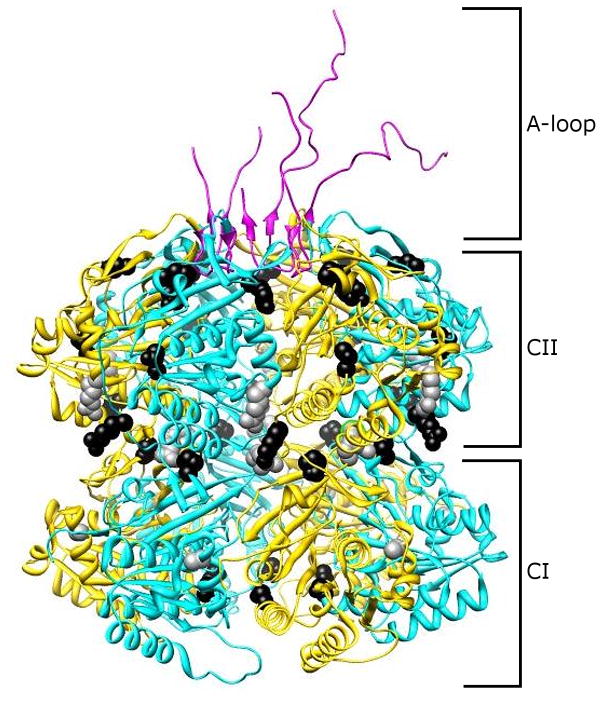
Figure 1.
Period-altering amino acid substitutions mapped to structure of a KaiC hexamer. KaiC monomers consist of two tandemly duplicated domains, CI and CII. These monomers oligomerize into hexameric structures with a thin “waist” region connecting the CI and CII regions. At the C-terminus of each KaiC monomer exists an “A-loop,” which determines the steady-state level of KaiC phosphorylation. Amino acid substitutions in the KaiC protein (Ishiura et al., 1998) that result in short-period rhythms (A87V, R215C, and R321Q) are denoted by gray circles, while those substitutions that give rise to period lengths greater than that of wild-type cells (S157C, P236S, R253H, M273I, T409A, and Y442H) are depicted as black circles. Image provided by Yong-Ick Kim, UC-San Diego.
Functions for KaiA and KaiB became apparent when they were incubated with KaiC in the presence of ATP. Although neither KaiA nor KaiB can undergo an autophosphorylation reaction, both alter the rate of KaiC autokinase activity (Iwasaki et al., 2002; Kitayama et al., 2003; Williams et al., 2002). The addition of KaiA to KaiC in vitro increases the rate at which KaiC autophosphorylates by 2.5-fold while the addition of only KaiB to KaiC does not have any effect. KaiB does, however, abrogate the positive action of KaiA on KaiC when combined together, such that autophosphorylation by KaiC is reduced to about half of the KaiA/KaiC combination. The phosphorylation state of KaiC as detected by immunoblot from cell extracts, cycles throughout the day, which led to the hypothesis that KaiC phosphorylation would be critical for circadian timing (Iwasaki et al., 2002). Mutations in the kai genes or other clock components that affect the ability of KaiC to phosphorylate in a rhythmic fashion alter the functionality of the clock (Iwasaki et al., 2002; Mehra et al., 2006; Xu et al., 2003).
C. Kai protein structure, homotypic and dynamic heterotypic interactions
Numerous structural studies revealed that KaiC, purified from S. elongatus PCC 7942 or thermophilic strains, interacts homotypically (Hayashi et al., 2004; Hayashi et al., 2006; Hayashi et al., 2003; Kang et al., 2009; Ming et al., 2007; Mori et al., 2002). KaiC, when purified in the presence of ATP, forms a dumb-bell shape consisting of six KaiC monomers and 12 bound ATP molecules (Hayashi et al., 2003; Mori et al., 2002). A thinner region connects the CI and CII structural domains and a channel exists in the center of the ring structure that narrows near the CI regions of the KaiC monomers (Figure 1). Mutations in each of the Walker A motifs affect the clock phenotype, increase the Km for ATP, and inhibit hexamerization (Hayashi et al., 2003); however, mutants of the KaiC CI domain Walker A motifs and catalytic carboxylate glutamate residues have greater effect on hexamerization than their CII domain counterparts, suggesting that the CI domain of KaiC is important for hexamerization. The CII domain of KaiC was shown to have phosphorylation activity similar to the wild-type levels while the CI domain lacked phosphorylation activity. As such, the CII domain, particularly the last 25-aminoacyl residues, was shown to interact with KaiA, while CI alone did not (Hayashi et al., 2004; Hayashi et al., 2006; Pattanayek et al., 2006).
Structural information obtained for KaiA provided additional insightful hints at its function (Garces et al., 2004; Uzumaki et al., 2004; Vakonakis et al., 2004; Williams et al., 2002). KaiA forms a dimer in solution through interaction of a novel fold in its C-terminus. The C-terminal domain of KaiA is capable of binding a small peptide from the CII domain of KaiC in solution, and is alone responsible for the stimulation of KaiC autophosphorylation (Uzumaki et al., 2004; Vakonakis et al., 2004; Williams et al., 2002). The N-terminal domain of KaiA resembles the receiver domains of proteins typically involved in bacterial two-component signal transduction pathways; however, this domain is considered a pseudo-receiver (PsR) in that it does not contain the conserved aspartate residue of bona fide receivers that accepts a phosphoryl group from a partner kinase (Stock et al., 2000). The hypothesized function of the N-terminal domain of KaiA is to act as a conduit for environmental information to the Kai oscillator to regulate the extent by which KaiA stimulates the autophosphorylation of KaiC (Williams et al., 2002; see Section III.C).
The X-ray crystal structure of KaiB reveals four subunits that are organized into a dimer of two asymmetric dimers that bind to KaiC in the presence of KaiA and ATP (Iwase et al., 2005; Pattanayek et al., 2008). The KaiB C-terminal tail is flexible, negatively charged, and via its interaction with KaiC has been hypothesized to displace ATP and interfere with KaiC subunit exchange by interacting with the CII hexameric barrel (Pattanayek et al., 2008). The same structural studies also suggest that KaiB binding to KaiC sterically inhibits the binding of KaiA.
As suggested by the biochemical relationship among the Kai proteins, KaiA, KaiB, and KaiC physically interact with one another both in vitro and in vivo. Initial yeast two-hybrid assays demonstrated both homotypic and heterotypic interactions among these clock components (Iwasaki et al., 1999). In vitro analysis using GST-Kai protein fusions also showed that KaiA and KaiB only weakly interact, but their interaction is strengthened in the presence of KaiC. Both the CI and CII domains of KaiC interact with the other two Kai proteins, and the interaction between KaiA and KaiB is strengthened more by the CI domain than by full-length KaiC; CII had the least positive effect on KaiA/KaiB binding and further demonstrates the differences in function of the two KaiC domains. Mutational analysis also supports the importance of Kai interactions in maintaining circadian rhythmicity. A mutation in kaiA that causes a 33-h free-running period (FRP) was shown to enhance the interaction of KaiA with KaiB (Iwasaki et al., 1999). Mutations in kaiC that cause enhanced interactions between KaiA and KaiC lengthen the circadian period to 44 h and 60 h, respectively, whereas a KaiC mutant that interacts weakly with KaiA results in a short 16-h period in LL (Taniguchi et al., 2001). These data show that strong interactions between KaiA, KaiB, and KaiC can slow the rate at which the clock proceeds.
Not only do the Kai proteins interact, but they do so dynamically both in vivo and in vitro. KaiA, KaiB, and KaiC have been shown to interact in a ratio of 1:1:4 by weight (Kitayama et al., 2003) with the largest amounts of KaiA and KaiB proteins interacting with KaiC 16–20 h after being released into LL to form a large complex during the early subjective night (Kitayama et al., 2003). In the cell KaiB and KaiC protein levels display robust rhythmicity in either LD or LL conditions with peak levels occurring 4–6 h (~CT15) after the peak in mRNA. KaiA protein levels are thought to be more consistent throughout the circadian cycle (Ivleva et al., 2005; Xu et al., 2000). The result of the dynamic interaction of the Kai proteins is the alteration of the KaiC phosphorylation (KaiC-P) state at various times of day with the highest levels of KaiC-P appearing to accumulate around CT12–16 with reduced levels at CT 0–4 (Iwasaki et al., 2002). Kai protein stability is also rhythmic, as KaiC is most stable with a 20-h half-life at CT16 when accumulation and phosphorylation are at maximum levels; in the early subjective day KaiC stability is decreased to a 6-h half-life (Imai et al., 2004). It has been suggested that the Kai system may resemble an F1-ATPase system, whereby KaiA rotates inside KaiC through phosphorylation and release of ADP, and KaiB association slows down the rotation antagonizing the function of KaiA on KaiC-P (Wang, 2005).
II. KaiC: Breakthroughs into Oscillator Timing and Clock Synchronization
What became clear from early studies into the Kai oscillator was that dynamic interactions among KaiA, KaiB and KaiC lead to KaiC phosphorylation events that are central to the timing mechanism, but questions still remained regarding the biochemical basis for timekeeping. Pivotal genetic and mutational studies led to four major findings that have adjusted previous paradigms established for not only the cyanobacterial circadian system, but for clock mechanisms in general: first, the canonical transcription-translation feedback loop, central to timing models in eukaryotic circadian oscillators, takes a back seat to strictly post-translational oscillations in KaiC phosphorylation; second, ordered KaiC autokinase and autophosphatase activity drives the Kai circadian oscillator; third, shuffling of various KaiC monomer phosphoforms between KaiC hexamers provides a mechanism for stability and synchronization; and fourth, the innate and slow-turnover ATPase activity of KaiC dictates timing in the Kai oscillator.
A. Which came first? Feedback loops and the in vitro oscillator
Clearly, initial studies into the Kai oscillator dictated that the kai genes and their protein products possess autoregulatory properties, similar to those of eukaryotic circadian systems (Ishiura et al., 1998). However, placing kai genes under the control of heterologous promoters was shown to have little effect on timekeeping (Ditty et al., 2005; Xu et al., 2003), indicating that transcriptional control of clock genes is not central to Kai-based timekeeping. Arguably, the biggest breakthrough toward understanding the mechanism of maintaining oscillations came from the realization that transcriptional control is not required for the maintenance of circadian rhythmicity in the prokaryotic model. An in vivo temperature-compensated cycling of KaiC-P was sustained in the absence of nascent rhythmic accumulation of Kai proteins or kai mRNA for 56 hours in DD (Tomita et al., 2005). In addition, the presence of transcription and translation inhibitors had no effect on DD KaiC-P rhythms, suggesting a self-sustainable post-translational rhythm in KaiC-P. The C28a kaiC4 allele, which directs a 28 h period, resulted in a KaiC-P rhythm extended by 4 h in DD (Tomita et al., 2005). Most convincingly, in vitro incubation of KaiA, KaiB and KaiC with ATP leads to a stable, temperature-compensated near 24-h rhythm in KaiC-P for multiple cycles (Figure 2A), and various KaiC variants that cause period changes in vivo had similarly correlated changes in the period of the in vitro KaiC phosphorylation rhythms (Nakajima et al., 2005).
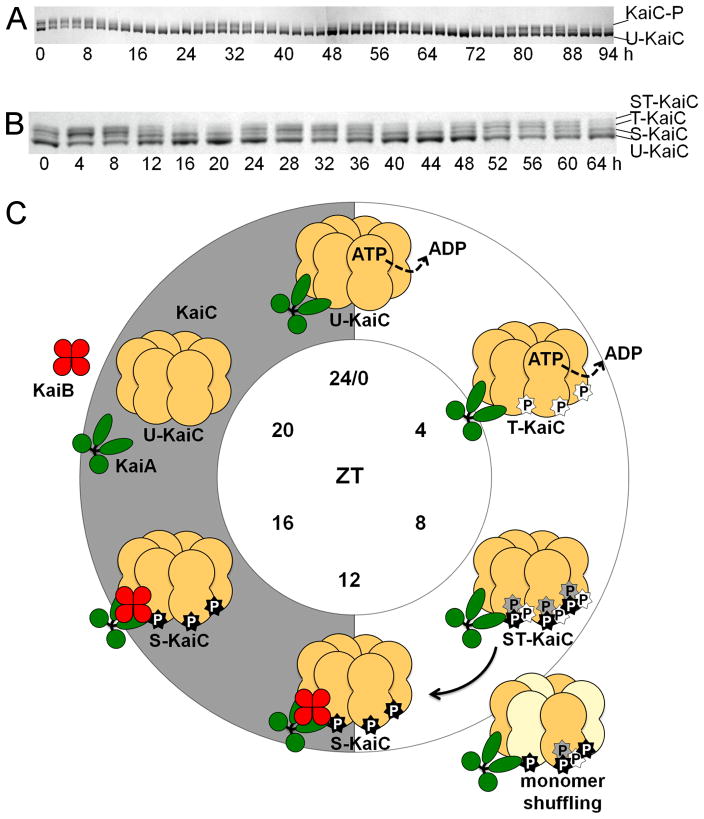
Figure 2.
In vitro KaiC-P oscillation and biochemical model for KaiC-P oscillatory activity. (A) In vitro oscillation of KaiC-P for 4 days taken at 2-h time points. U-KaiC, unphosphorylated KaiC; KaiC-P, phosphorylated KaiC. (B) In vitro KaiC-P oscillation with resolved phosphoforms. In vitro reactions were incubated for 3 days with samples taken at 4-h time points. Four phosphoforms of KaiC exist over the course of a phosphorylation cycle. ST -KaiC, S431 and T432 KaiC double phosphoform; T-KaiC, T432 KaiC phosphoform; S-KaiC, S431 KaiC phosphoform; and U-KaiC, unphosphorylated KaiC. Immunoblot images provided by Yong-Ick Kim, UC-San Diego. (C) Model for ordered phosphorylation and monomer shuffling activity in KaiC. Initially at ZT 0, unphosphorylated KaiC (U-KaiC) associates with KaiA to induce phosphorylation of KaiC (T-KaiC) at T432 (white star) at ZT 4, which is subsequently followed by phosphorylation at S431 (black star) and possibly T426 (gray star) for a fully phosphorylated KaiC (ST-KaiC) by ZT 8. Fully phosphorylated KaiC initiates the dephosphorylation stage and is amenable to monomer shuffling between ZT 8 and 12 to synchronize multiple intracellular KaiC hexamers, leading to the lone S431 phosphoform (S-KaiC) by ZT 12 and association with KaiB. KaiC continues autodephosphorylation through ZT 16, yielding U-KaiC by ZT 20. Note that indicated KaiC phosphoforms at each ZT time represent the most prominent KaiC-P phosphoform relative to the population. P, phosphate.
While in vitro oscillation of KaiC-P in the absence of transcriptional-translational feedback shattered the dogma, further investigation into the timing mechanism in vivo revealed that transcription-translation-based oscillation of kai genes is still important. In a KaiA-overexpressing strain, which forces constitutive KaiC-P, temperature-sensitive and phase re-settable rhythms in PkaiBC and KaiB and KaiC are detected, albeit with a shorter period and higher trough (Kitayama et al., 2008). Strains producing unphosphorylated S431A/T432A mutant KaiC proteins were arrhythmic at the level of the PkaiBC promoter and S431E/T432E KaiC mutant proteins that are phospholocked generate damped and 48-h period rhythms. In a K294H KaiC mutant strain that lacks autokinase activity, KaiC is unphosphorylated; however, a weak PkaiBC oscillation with a long period is evident (Kitayama et al., 2008). Therefore, multiple coupled oscillatory systems are important for precise and robust rhythms, and the importance of the transcription-translation feedback loop plays second chair to the primary post-translational KaiC-P oscillation. It is the work of the entire clock system, with its input and output pathways, that helps to coordinate this independent Kai oscillation with the environmental day/night cycle.
B. Ordered KaiC autokinase and autophosphatase activities drive the circadian oscillator
KaiC protein autophosphorylates when incubated with radiolabeled ATP, and crystallographic, mass spectrometric, and mutational analyses demonstrated at least two main, and one additional, phosphorylation sites in KaiC (Nishiwaki et al., 2004; Pattanayek et al., 2009; Pattanayek et al., 2004; Xu et al., 2004; Xu et al., 2009). Kinetic studies divulged an intricate and ordered phosphorylation of KaiC on two adjacent aminoacyl residues in CII, S431 and T432. Four phosphoforms of KaiC exist over the course of a phosphorylation cycle with T432-KaiC-P and S431-KaiC-P phosphoforms predominating during the phosphorylation and dephosphorylation phases, respectively (Figures 2A and 2B). When KaiA is mixed with unphosphorylated KaiC, T432 is initially phosphorylated followed by S431 to result in a fully phosphorylated KaiC. T432 and S431-KaiC double phosphorylation initiates the switch in KaiC from autokinase to autophosphatase activity, resulting in the dephosphorylation of T432 followed by S431. As expected, KaiA association is only seen during the initial KaiC phosphorylation phase. Association of KaiC with KaiA and KaiB results from S431-KaiC-P, and these interactions are important for maintaining the amplitude of KaiC phosphorylation (Nishiwaki et al., 2007; Rust et al., 2007). An additional third phosphorylation site at T426 seems to play a role in KaiC phosphorylation status. Structural evidence suggests that T426 and S431 face each other across a looped region of KaiC (Xu et al., 2004) and that T426 can be phosphorylated in vitro (Pattanayek et al., 2009). In vivo studies of T426A and T426E mutant strains, which are expected to mimic unphosphorylated and phospholocked states, respectively, abolish rhythmicity (Xu et al., 2009). Various substitutions at T426, S431 and T432 cannot support KaiC phosphorylation rhythms, and the T426N mutant KaiC variant specifically hinders the rate of dephosphorylation. When overexpressed with wild-type KaiC, T426N or T426E KaiC variants lengthen the period of the clock to 31 h in vivo, inhibit interactions with KaiA and KaiB, and increase the Q10 value, a measure of temperature compensation, to 1.3. While direct phosphorylation of T426 is debatable, this residue could be transiently phosphorylated or be involved in stabilizing phosphorylation at S431 (Pattanayek et al., 2009; Xu et al., 2009).
Additional structural and mutational analyses of KaiC revealed how the KaiA and KaiB proteins act to shift the active state of KaiC from autokinase to autophosphatase (Kim et al., 2008). Aminoacyl residues 488–497 at the C-terminus of every KaiC monomer constitute a regulatory switch called the “A-loop,” and whether these residues are buried or exposed determines the steady-state level of phosphorylation (Figure 1). The current model depicts KaiA as stabilizing the exposed loop to direct KaiC phosphorylation, as a KaiC487 mutant that lacks the A-loop and additional tail residues (498–519) structurally mimics the exposed state and is constitutively phosphorylated irrespective of the presence of KaiA. A KaiC497 mutant protein is truncated such that it has the A-loop but not the solvent-exposed tail, which mimics an A-loop buried state resulting in unphosphorylated KaiC, either alone or in combination with KaiA. Truncations partway through the A-loop are hyperphosphorylated, but to a lesser level than that of KaiC487. In addition, ectopic expression of KaiC487 or KaiC497 abolishes rhythmicity in wild-type S. elongatus PCC 7942, as measured by the PkaiBC::luxAB reporter strain, demonstrating dominant-negative effects. KaiA directly binds and stabilizes the exposed state of the A-loop, while KaiB indirectly stabilizes the buried state of the A-loop by hindering KaiC interaction with KaiA. It is hypothesized that A-loop displacement moves bound ATP closer to the sites of phosphorylation (Kim et al., 2008).
In addition to Kai protein interactions, concentrations of the Kai proteins affect the in vitro KaiC-P oscillation (Nakajima et al., 2010). In a reaction with 3.5 μM KaiC, a KaiA concentration range from 0.6 to 6.0 μM is necessary for rhythmic KaiC-P and the period and amplitude is continuously influenced by KaiA concentration; outside this concentration range, the KaiC-P rhythm is damped. Although KaiB protein at a concentration of 1.75 μM or higher is necessary for a KaiC-P rhythm, excess KaiB does not affect the period or amplitude. As such, KaiA and KaiB are “parameter-tuning” and “state-switching” regulators of the KaiC-P rhythm, respectively (Nakajima et al., 2010). The ability of KaiA to fine-tune KaiC-P rhythms based on concentration may give future insight to an entrainment mechanism for the clock.
C. Stabilization of the KaiC phosphorylation cycle by monomer shuffling
The Kai circadian system functions as a result of quantitative fluctuations in clock components and KaiC-P levels that ultimately dictate cellular activity. Therefore, the question of how stable oscillations are synchronized among multiple KaiC hexamers in dynamic interaction with all other aspects of the clock drew attention. Studies on the rhythm in single cells revealed that the oscillations in individual cells are extremely resilient to outside perturbations; single S. elongatus cells maintain a robust circadian oscillation with a correlation time of several months (Amdaoud et al., 2007; Mihalcescu et al., 2004). Therefore, stability in the clock is innate to the clock machinery of individual cells without the need for cell-to-cell communication.
The stability phenomenon has provided fodder for many mathematical models predicting how synchronization occurs within the Kai oscillator (Clodong et al., 2007; Eguchi et al., 2008; Emberly and Wingreen, 2006; Kurosawa et al., 2006; Markson and O’Shea, 2009; Mehra et al., 2006; Miyoshi et al., 2007; Mori et al., 2007; Nagai et al., 2010; Rust et al., 2007; Takigawa-Imamura and Mochizuki, 2006; van Zon et al., 2007; Yoda et al., 2007). Stability is based on monomer exchange among KaiC hexamers and is linked to KaiC dephosphorylation. Using native and FLAG-tagged KaiC monomers, the monomers were shown to shuffle among KaiC hexamers, and this shuffling is phosphorylation-dependent as S431A and T432A mutant monomers do not shuffle (Kageyama et al., 2006). KaiA inhibited shuffling and KaiB had no effect, suggesting that monomer shuffling is a potential dephosphorylation mechanism by diluting out KaiC-P with nascent, unphosphorylated KaiC (Kageyama et al., 2006). This model was tested by monitoring resilience of the in vitro KaiC-P rhythm by mixing KaiC hexamers from different phases offset by 4 h. When KaiC hexamers from six different circadian phases are mixed, the overall KaiC-P pattern is immediately rhythmic, suggesting that that individual KaiC hexamers synchronize due to monomer shuffling. KaiC-P cycling persists without damping for 10 days in vitro as long as sufficient ATP is present in the reaction. Unphosphorylated S431A/T432A KaiC proteins do not affect monomer shuffling while the phospholocked S431D/T432E KaiC proteins increase shuffling by almost 2-fold and fix KaiC in the dephosphorylation stage leading to arrhythmia. In addition, KaiC hexamers in the dephosphorylation state are dominant in shuffling as they have been shown to exchange monomers with KaiC proteins in other states. Therefore, shuffling is tightly linked with dephosphorylation and is mediated early in the dephosphorylation phase (ZT 28–32 h) (Ito et al., 2007).
D. KaiC ATPase activity as a basis for oscillator timing and cell division control
While earlier studies on nucleotide binding revealed insights into KaiC function and hexamerization (Hayashi et al., 2004; Hayashi et al., 2006; Hayashi et al., 2003; Ishiura et al., 1998; Nishiwaki et al., 2000), investigations into the rate of ATP consumption by KaiC revealed that KaiC consumes ATP at an extremely low level, as each monomer only requires approximately 15 ATP molecules for one circadian cycle (Terauchi et al., 2007). Peak ATPase activity occurs 4 h before phosphorylation of KaiC in vitro (Terauchi et al., 2007) and in vivo KaiC ATPase is at its peak at CT12 (Dong et al., 2010).
One major finding regarding KaiC ATPase activity is the linear correlation between ATPase activity and circadian frequency. The ATPase activities of short and long period KaiC mutant proteins are higher and lower than wild-type KaiC, respectively, such that timing of the circadian cycle depends directly on the rate of ATP hydrolysis (Terauchi et al., 2007). Per circadian dogma, a characteristic required for period determination is that it must be temperature compensated. Using purified KaiC from Thermosynechococcus elongatus BP-1, ATPase activity was shown to be temperature compensated in the wild-type, but not in a S431A/T432A KaiC-P double mutant, providing a possible mechanism for temperature compensation of the circadian period (Murakami et al., 2008).
Recent studies of the role of KaiC ATPase activity reveals a link between ATPase activity and clock output by providing a checkpoint for cell division (Dong et al., 2010; see Section IV.F). An oscillator state that correlates with elevated KaiC-P under normal conditions is responsible for a daily pause in cytokinesis; however, the correlating activity matches that of ATPase activity rather than that of phosphorylation of KaiC. For example, cells expressing the S431D/T432E phospholocked KaiC variant mimic the peak KaiC-P state, but this phosphomimicry does not inhibit cell division. Conversely, the S431A/T432A unphosphorylated KaiC variant blocks cell division. This conundrum was resolved through the recognition that S431A/T432A, and other variants that cause cell elongation, have elevated ATPase activity, and low ATPase mutants have no effect on cell division (Dong et al., 2010). Thus, although the phosphorylation cycle is the trackable mark of oscillator progression, it only hints at other processes that comprise the oscillator time stamp.
Generating an in vitro Kai circadian oscillation in KaiC-P was a monumental breakthrough that allowed prokaryotic chronobiologists to think outside the box for an endogenous timekeeping mechanism (Figure 2C). The combination of ordered KaiC autokinase and autophosphatase activity drives the Kai circadian oscillator, and ATPase activity dictates timing as the biochemical basis for timekeeping. Shuffling of various KaiC monomer phosphoforms among KaiC hexamers provides a mechanism for stability and synchronization. A complete clock system is more than an oscillator alone, but rather the sum of all of its parts; recent insights into input and output pathways have further shaped the cyanobacterial circadian model system.
III. Input Pathways: Light-Dependent Cellular Metabolism Synchronizes the Clock with Local Time
As an obligate photoautotroph, S. elongatus benefits from its ability to organize its cellular processes in anticipation of “feeding time” that occurs with the rising sun. In contrast to the dedicated photoreceptors of eukaryotic clock systems that transduce light cues to their central oscillator components, no photoreceptors have been identified as essential to the input pathways of the cyanobacterial clock. Multiple genetic screens for phase-resetting mutants as well as directed gene inactivation of each of the seven predicted blue light photoreceptors failed to demonstrate a direct link between these photoreceptors and input to the oscillator (Mackey et al., 2009). Instead, the S. elongatus input pathways rely on the metabolic changes within the cell that result from changes in light quantity to synchronize their circadian cycle to that of the environment.
Mutations have been identified in three S. elongatus genes – pex (period extender), cikA (circadian input kinase), and ldpA (light dependent period) – that alter the ability of S. elongatus to effectively synchronize the phasing of behaviors with repetitive LD cues, reset phase in response to changes in light cues, and adjust period in response to subtle differences in light intensity, respectively. Inactivation of any of these three genes results in a short circadian period as compared to that of wild-type cells (Katayama et al., 2003; Kutsuna et al., 1998; Kutsuna et al., 2007; Schmitz et al., 2000); these data suggest that the S. elongatus input pathway(s) is used to delay the internal Kai-based oscillation to better match daily time. In addition to these components, KaiA has been implicated as being a key player in the coordination between environmental stimuli and the timing of the KaiC-P rhythm (Wood et al., 2010). Given the central role that KaiC plays in the circadian system, it is not surprising that a mutation of kaiC exists that allows cells to maintain near 24-h time, but prevents the ability to synchronize that timing with the solar day (Kiyohara et al., 2005).
A. Pex protein synchronizes phase with LD cycles
Originally identified through its apparent complementation of a 22-h short period kaiC mutant, further analysis showed that the resulting wild-type rhythm was due to the presence of an ectopic copy of the pex gene, which resulted in a 2-h extension of the 22-h mutant period (Kutsuna et al., 1998). The pex gene encodes a 148 amino acid protein whose structure has a winged-helix motif similar to DNA-binding domains of the PadR family (Arita et al., 2007). Overexpression of the pex gene by an inducible promoter demonstrated a dose-dependent lengthening of the period of the circadian rhythm in both wild-type and kai circadian period mutant backgrounds, while inactivation of pex results in a 1-h period shortening in LL conditions (Kutsuna et al., 1998). Cells that lack pex do not fully synchronize to the entraining LD cycles, such that the resulting phase of the rhythm in LL is advanced as compared to wild type (Takai et al., 2006). This defect in synchronization likely results from a lack of Pex protein accumulation during the dark (subjective night) phase of the cycle, during which wild-type pex mRNA and Pex protein levels peak at ZT16 and ZT20, respectively, during LD12:12 cycles (Takai et al., 2006).
Pex protein is predicted to function as a dimer and binds preferentially to the upstream promoter region of the kaiA gene; individual alanine substitution of arginine residues in the wing region abolishes the DNA-binding activity of Pex (Arita et al., 2007). Cells that lack pex display a substantial increase in kaiA promoter activity and mRNA accumulation while overexpression of Pex protein reduces kaiA expression (Kutsuna et al., 2007). The period length that results from pex inactivation or overexpression can be recapitulated by direct manipulation of kaiA levels using an inducible promoter to overexpress wild-type or antisense kaiA messages, respectively. Pex is predicted to function during the dark phase of an LD12:12 cycle, where its accumulation likely leads to the repression of kaiA through direct binding of the kaiA promoter region; the resulting decrease in KaiA protein relieves the positive effect of KaiA on KaiC-P and delays the internal oscillation such that it more closely matches that of the environment (Kutsuna et al., 2007).
B. CikA protein bridges input and output pathways
Unlike pex mutants that display a subtle phase difference as compared to that of wild type, cells that lack the cikA gene are unable to effectively reset the phase of their circadian rhythm to abrupt changes in environmental stimuli, such as pulses of darkness. The cikA gene was originally identified as altering expression of a PpsbAII luciferase reporter by causing a shortened circadian period (~ 22 h), altered phase angle and low amplitude rhythmicity in LL conditions (Schmitz et al., 2000). Subsequent analyses revealed a cell division defect in the cikA mutant (Miyagishima et al., 2005). The role of CikA in circadian periodicity appears to be non-overlapping relative to those of the Kai proteins because inactivation of cikA in kai mutant backgrounds results in additional period shortening and adjustments to the phase angle as compared to the kai mutation alone (Schmitz et al., 2000). Overexpression of cikA in vivo results in arrhythmia (Mutsuda et al., 2003). CikA protein accumulates in a circadian fashion with lower levels during the subjective day and an increase during the subjective night (Ivleva et al., 2006). In the absence of kaiC, CikA levels remain low and constant in LL, but not LD conditions, which suggests that the clock system can regulate CikA protein levels in the absence of external cues. Thus, the level of CikA protein is modulated both by the environment and by the circadian system.
Bioinformatic analysis of the CikA protein predicted an N-terminal GAF domain, which is typically involved in bilin chromophore attachment in bacterial photoreceptors. The presence of this domain initially suggested a role for CikA in phototransduction, which led to the testing of CikA as a component on the input pathway of the clock. As predicted, inactivation of cikA prevents S. elongatus cells from resetting the phase of their circadian rhythm by more than 2 h in response to pulses of darkness that would otherwise cause wild-type cells to adjust their phase by up to 8–10 h (Schmitz et al., 2000). This phenotype is not likely attributed to the GAF domain, which lacks the conserved cysteine amino acid residue responsible for bilin adduct formation and phototransduction; attempts to purify CikA with an attached bilin in vivo were unsuccessful (Mutsuda et al., 2003). Rather, this GAF domain regulates the phosphorylation state of the CikA central histidine protein kinase (HPK) domain, which is similar to that of sensor protein kinases involved in bacterial two-component regulatory systems (Stock et al., 2000). Removal of the GAF domain results in severely decreased CikA auto-phosphorylation at its conserved histidine residue (H393) in vitro (Figure 3A) (Mutsuda et al., 2003). A CikA variant that lacks the GAF domain is unable to complement a cikA null in vivo (Zhang et al., 2006), which is likely due to the decreased autophosphorylation of CikA, an essential post-translational modification for normal CikA function.
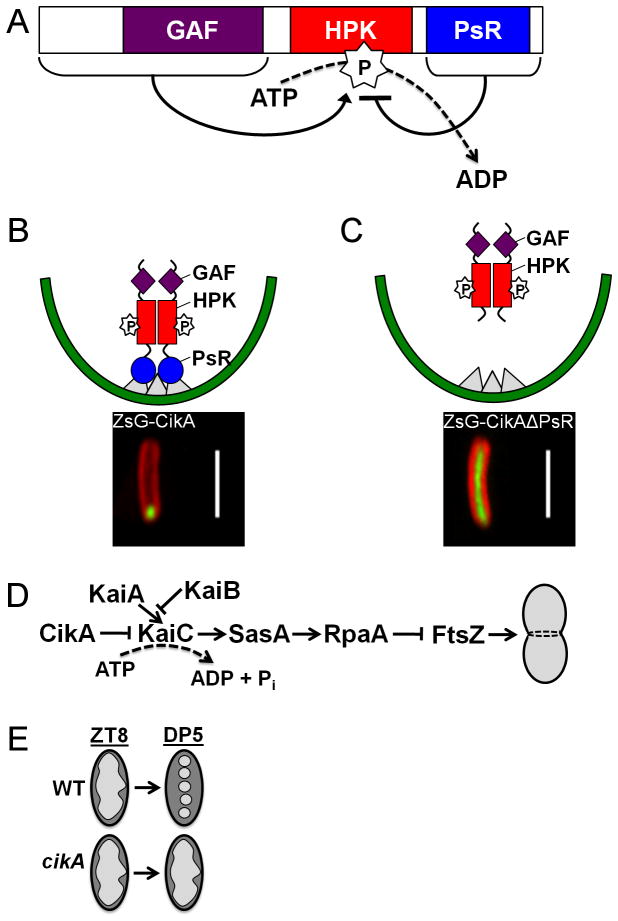
Figure 3.
Multiple roles for CikA in the S. elongatus circadian system. (A) Autophosphorylation of CikA at its histidine protein kinase (HPK) domain is positively influenced by the N-terminal GAF domain, but inhibited by the C-terminal pseudo-receiver (PsR) domain. (B) The PsR domain (circle) of CikA is proposed to interact with as-yet-to-be-identified proteins (triangles) at the pole of the cyanobacterial cell. Micrograph image of an S. elongatus cell harboring a ZsGreen-CikA fusion protein exhibits polar localization. Scale bar = 5μm. (C) Absence of the CikA-PsR domain results in the delocalization of this CikA variant to exist throughout the cytoplasm of the cell as shown in the cartoon and micrograph image, which visualizes the ZsGreen-CikAΔPsR fusion protein. Scale bar = 5μm. ZsGreen fusion constructs from Zhang et al., 1999; micrograph images courtesy of Julie Bordowitz, UC-San Diego. (D) CikA inhibits the ATPase activity of KaiC, such that in the absence of CikA, the ATPase activity rises above a maximum threshold to activate the SasA/RpaA two-component system of the circadian output pathway. RpaA is predicted to indirectly repress the localization of FtsZ protein that is involved in determining the midline of the cell to allow for division to proceed. (E) At ZT8, the nucleoid region of the wild-type (WT) and cikA null is diffuse. After being subjected to a 5-h dark pulse (DP5) beginning at ZT8, the cikA null does not display complete chromosome compaction like that of a wild-type cell.
The C-terminal end of the CikA protein contains a PsR domain that is receiver-like in sequence and structure (Gao et al., 2007), with the exception of the conserved aspartic acid residue that is involved in phosphotransfer from the HPK partner protein of a true two-component system. No phosphorylation event could be detected in experiments designed to allow the transfer of a phosphate group from the HPK of CikA to its PsR domain (Mutsuda et al., 2003). The PsR is involved in multiple roles that provide CikA with its fundamental phenotypic characteristics. It is predicted that the PsR domain physically blocks the H393 residue through direct interaction with the CikA HPK domain (Gao et al., 2007); this interaction would repress autophosphorylation and is consistent with in vitro experiments that show a 10-fold increase in phosphorylated CikA when the PsR domain is removed (Mutsuda et al., 2003). Additionally, the PsR domain is necessary for CikA localization (Figure 3B). A fluorescent-tagged CikA variant that lacks the PsR domain is found throughout the cell and no longer localizes to the pole of the cell like that of the wild-type CikA protein (Figure 3C) (Zhang et al., 2006). Overexpression of the CikA-PsR alone in a wild-type background produces a phenotype similar to that of the cikA null, which suggests that the free PsR constructs may be competing with wild-type CikA for binding sites at the pole to allow for proper CikA function (Zhang et al., 2006) or preventing autophosphorylation of HPK domains on full-length CikA protein.
The role of CikA in resetting of the circadian phase appears to derive from its effect on KaiC activity. In response to a 5-h dark pulse that resets the phase of bioluminescent reporter gene expression by 8 h, CikA protein levels increase (Ivleva et al., 2006). Pex protein levels also increase in the dark to repress kaiA expression (Takai et al., 2006). CikA inhibits the ATPase activity of KaiC protein (Figure 3D) (Dong et al., 2010), and the Pex-induced decrease in KaiA prevents KaiA-stimulated autophosphorylation of KaiC. Together, these input components lead to a decrease in the population of phosphorylated KaiC molecules (Ivleva et al., 2006). The combined data suggest that resetting of the circadian phase is dependent upon changes in KaiC-P status.
Consistent with a role for KaiC-P in resetting the circadian phase rather than in maintenance of circadian period, two mutants that lack the ability to reset their rhythm do not display robust fluctuations in the ratio of KaiC-P throughout the circadian cycle. Both a cikA null and a cyanobacterial strain that harbors a point mutation in the kaiC gene, named kaiC (pr1), are unable to reset the phase of their rhythm in response to external dark pulses, yet produce the rhythmic output of bioluminescence in LL conditions (Kiyohara et al., 2005; Schmitz et al., 2000). Neither of these phase-resetting mutants undergoes complete chromosome compaction when placed in 5-h darkness that would otherwise result in tightly condensed chromosomes in a wild-type cell (Figure 3E) (Smith and Williams, 2009). Taken together, these data suggest a relationship between discrete entrainment, KaiC activity, and nucleoid topology, although the pathway that unites them is unknown.
C. LdpA regulates continuous entrainment of the clock
The ldpA locus was initially identified in a screen for light input mutants that disrupted the ability to reset to pulses of darkness throughout the circadian cycle. Further analyses demonstrated that the true role of the ldpA gene and its protein product is in continuous entrainment of the circadian rhythm through modulation of the period of the internal rhythm in response to changes in light intensity (Katayama et al., 2003). Like other diurnal organisms that possess intrinsic biological timing mechanisms, the circadian period decreases (indicative of a faster clock) as light intensity increases, a phenomenon named Aschoff’s rule (Aschoff, 1981); however, mutants that lack ldpA maintain the short period (22–23 h) phenotype associated with high light intensity regardless of the actual environmental light intensity (Katayama et al., 2003). This light-dependent period mutant phenotype also affects the enhanced susceptibility of the kaiC22a mutation to changing light intensity. Inactivation of ldpA in the C22a mutant showed no substantial change in period across light intensities that would otherwise display up to 3-h difference in period; the ldpA null displayed the high light intensity phenotype across the light spectrum (Katayama et al., 2003). These data indicate that ldpA is epistatic to kaiC22a with regards to light input.
In an ldpA mutant background, CikA protein is maintained at its trough level throughout the circadian cycle; this level corresponds to the amount of CikA that is present in high light conditions in a wild-type strain (Ivleva et al., 2005). In contrast, KaiA protein levels are elevated as compared to wild type, which may lead to the short period phenotype of the ldpA null, as kaiA induction is associated with a 1-h period shortening in vivo (Kutsuna et al., 2007). Despite this increase in KaiA protein and decrease in CikA protein, both of which would be predicted to result in increased expression from the PkaiBC promoter (Ishiura et al., 1998; Taniguchi et al., 2010), there is no noticeable change in the level of bioluminescence from a PkaiBC::luc reporter in the absence of ldpA.
The ldpA gene encodes a 352 amino acid protein that carries two redox-active Fe4S4 clusters (Ivleva et al., 2005). The presence of these Fe4S4 clusters, coupled with the circadian phenotype of the ldpA mutant, suggested that LdpA is involved in carrying redox signals from the photosynthetic apparatus to the circadian oscillator to align the circadian rhythm with the changing external environment. Addition of DCMU (3-(3,4-dichlorophenyl)-1,1-dimethylurea), which inhibits electron transport from the photosystem II reaction center to the plastoquinone (PQ) pool, was able to partially rescue the ldpA phenotype; in the presence of DCMU, the ldpA mutant displayed a small change in period length across light intensities (Katayama et al., 2003). These data suggest that photosynthetic activity contributes to continuous entrainment of the circadian oscillator, but that LdpA is not the sole component in this pathway.
The addition of the quinone analog DBMIB (2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone) results in a saturation of electrons (overall reduction) of the PQ pool by preventing electron transport from PQ to cytochrome b6f; introduction of sub-lethal concentrations of DBMIB leads to the rapid degradation of both the LdpA and CikA proteins (Ivleva et al., 2005; Ivleva et al., 2006). DBMIB-dependent degradation of CikA occurs more slowly in the absence of LdpA. The PsR domain of CikA binds directly the DBMIB molecule and is necessary and sufficient for DBMIB-dependent degradation (Ivleva et al., 2006); thus, the environmental input that is received by CikA and transduced to the central oscillator reflects that of the metabolic state of the cell. These data provide a molecular mechanism for the phenotypic phenomenon of Aschoff’s rule.
In contrast to the eukaryotic clock systems that require compartmentalization of clock components to allow for proper circadian rhythmicity, the cyanobacterial clock proteins of the oscillator, input, and output pathways interact closely with one another to dictate the internal timing mechanisms. Using a 6xHis-tagged LdpA variant, KaiA, CikA and a component of the output pathway (Synechococcus adaptive sensor, SasA) were shown to form a complex in vivo (Ivleva et al., 2005). Interaction with KaiA peaked during LL8–12, despite overall soluble KaiA protein accumulation peaking between LL16–20. CikA co-purification with LdpA remained relatively constant across the circadian cycle. The interaction between KaiA and CikA is likely indirect and possibly mediated through LdpA because co-purification of LdpA and CikA was uninterrupted in a kaiA mutant, and the interaction between LdpA and KaiA is maintained in a cikA mutant background (Ivleva et al., 2005). The path by which CikA transduces input information to the oscillator does not appear to require LdpA as an ldpA mutant, unlike a cikA null, can reset its phase in response to dark pulses. CikA co-purifies with KaiA and KaiC in large multimeric complexes in vivo that change in their overall molecular weight throughout the circadian cycle (Ivleva et al., 2006); these interactions place CikA in the heart of the clock complex.
The interactions between components of the input pathways and the central oscillator suggest direct transduction of environmental information among protein partners. The current model places KaiA as the conduit for environmental information to the Kai-based oscillator. The N-terminus of KaiA contains a PsR, similar to that of CikA, which lacks the necessary aspartic acid residue necessary for phosphoryl transfer (Williams et al., 2002). This PsR domain binds directly the oxidized, but not reduced, form of the DBMIB quinone analog (Wood et al., 2010). The bound oxidized DBMIB results in irreversible aggregation of KaiA protein, such that KaiA is no longer able to stimulate the autokinase activity of KaiC in vitro (Wood et al., 2010). Although the electron donor for KaiA is not yet known, the co-purification of CikA and LdpA with the Kai proteins, the direct binding of DBMIB to the CikA-PsR, and the rapid degradation of CikA and LdpA in response to DBMIB together implicate the possibility of direct electron transfer from the photosynthesis machinery via known input proteins to the redox-sensitive KaiA protein as a mechanism of entrainment of the cyanobacterial clock.
IV. Output Pathways: Multiple Independent Pathways Merge to Coordinate Cellular and Physiological Processes
The presence of circadian rhythms in a diverse range of organisms – including cyanobacteria, fungi, insects, plant and mammals – has led most chronobiologists to hypothesize that these internal oscillations beget an adaptive advantage to those organisms through behaviors anticipatory of the changing LD cycle (Dunlap et al., 2004). Microarray analyses demonstrate that 89% of the S. elongatus genes that encode proteins involved in photosynthesis display peak expression near subjective dawn, which would allow for their photosynthesis-related products to be most prominent during the daytime (Vijayan et al., 2009). The enhanced fitness of mammals has been demonstrated in the decreased survival rates of clock mutants as compared to those organisms with circadian oscillations that are consonant with the external LD cycles (DeCoursey et al., 2000). The natural variation in free-running periods of Arabidopsis thaliana plants that live at different latitudes correlates with the day length to which those plants are subjected (Dodd et al., 2005; Michael et al., 2003), which suggests a selective pressure to possess an internal rhythm that matches that of the environment.
Because S. elongatus does not fix nitrogen (Herrero et al., 2001) and has no physiological requirement for temporally separating this oxygen-sensitive process from that of photosynthesis as do diazotrophic cyanobacteria, the evolutionary advantage for maintaining internal time was not initially clear. To test the evolutionary advantage that the clock may bestow upon S. elongatus, strains that harbor mutations in one of the three genes – kaiA, kaiB or kaiC – that encode the core oscillator proteins were placed in direct competition with wild-type cells in a variety of environmental conditions (Ouyang et al., 1998; Woelfle et al., 2004). When grown independently as pure cultures, the mutant cultures that produce long period, short period or arrhythmic phenotypes from bioluminescent reporters grew at a rate indistinguishable from that of wild type in either LL or LD conditions. When two cultures were grown together in a continuously-diluted culture, which allows for growth to continue for 30–45 generations, the strain whose FRP most closely matched that of the given LD cycle would out-compete the other strain (Ouyang et al., 1998; Woelfle et al., 2004). If cells are subjected to an LD12:12 cycle, a wild-type strain that produces a FRP of 25 h outcompetes a 23-h mutant, yet this mutant will quickly dominate the population if provided an LD11:11 cycle (Ouyang et al., 1998). This advantage occurs only when the competing cultures are subjected to LD cycles and not in LL conditions, and only when strains are co-cultured, which suggests that the internal timekeeping system confers an adaptive advantage to S. elongatus only when in a competitive environment. Notably, nature is a competitive environment, and the laboratory setting is likely the only location in which cyanobacteria would be in pure culture and LL conditions.
A. Global clock-controlled regulation of gene expression
Even before the kai genes were identified, a screen was conducted to determine the extent by which the circadian system regulates the timing of S. elongatus gene expression; the strategy used a “promoter trap” that inserted promoterless luxAB throughout the genome to randomly sample promoter activity via bioluminescence (Liu et al., 1995). Two major classes of peak expression patterns emerged that were 12 h out of phase with one another. Class 1 appears to be the “default” phasing for promoters because the majority of promoter activities peak at subjective dusk, including heterologous E. coli promoters (PconII and Ptrc) that have been introduced into the cyanobacterial chromosome (Tsinoremas et al., 1996; Xu et al., 2003). Taken together, these data pinpoint the circadian system as a global regulator for gene expression in S. elongatus.
The widespread circadian regulation of transcription is not necessarily indicative of rhythmic mRNA and/or protein accumulation. Microarray analyses show that only 30–64% of S. elongatus transcripts accumulate in a rhythmic fashion in LL (Ito et al., 2009; Vijayan et al., 2009); similar to those of the promoter trap assay, transcript rhythms fall into the two major categories of peaking near subjective dusk (CT8–12) or subjective dawn (CT20–24). Interestingly, not all of the known clock components display rhythmic mRNA levels throughout the day. The kaiBC and cikA transcripts accumulate with a near 24-h rhythm, yet those of other known clock components, including sasA, ldpA and pex, do not (Ito et al., 2009). The timing of the peak KaiB, KaiC and CikA protein levels lag their respective mRNA rhythms by 6–8 h; this fact implies that post-transcriptional and post-translational modifications play an important role in the functionality of those proteins, especially in the dark where mRNA levels are quickly reduced within 4 h, yet KaiC-P rhythms persist for 3 days in DD (Ito et al., 2009; Tomita et al., 2005).
The inhibitory role that KaiC plays on its own expression is paralleled throughout most of the genome, such that constitutive overexpression of kaiC leads to repression of nearly every promoter tested (Ito et al., 2009; Nakahira et al., 2004). For class 1 genes, the level of promoter activity after KaiC-induced repression is at a level consistent with the trough of the activity level in a wild-type background. These promoters are divided into two main types based on the level of expression that remained after the KaiC-induced repression occurred. The “clock-dominated” promoters display high amplitude rhythms in the presence of the clock, but almost no expression occurred when the clock was disrupted by excess KaiC. Other “clock-modulated” promoters maintained relatively higher levels of arrhythmic expression with the nonfunctional clock. The mRNA levels that result from KaiC overexpression are similar to those seen at subjective dawn with a functional clock, such that class 1 genes produce mRNA levels that mirror the trough level in LL and class 2 genes escalate mRNA production to reach peak levels (Ito et al., 2009).
B. Chromosome topology as a mediator for rhythmic gene expression
Multiple lines of evidence have hinted that the pervasiveness of clock control of gene expression in S. elongatus is not necessarily a direct result of the transcription-translation feedback loop of the kai genes and their protein products (Ito et al., 2009; Kutsuna et al., 2005; Nakahira et al., 2004; Nakajima et al., 2005; Terauchi et al., 2007; Tomita et al., 2005; Xu et al., 2003). Negative regulation of kaiBC by excess KaiC is not due to specific cis-acting elements within the promoter. Although a negative regulatory region exists within the kaiBC promoter region, removal of this negative element does not prevent the promoter from responding to KaiA or KaiC overexpression. Furthermore, there do not appear to be specific cis- or trans-acting elements that contribute to the phasing of expression for class 1 or 2 genes during LL conditions (Min and Golden, 2000; Min et al., 2004). Rather, the local DNA topology likely determines the times at which a promoter is accessible to the transcriptional machinery. Early evidence for this “oscilloid model” (Mori and Johnson, 2001a) stems from introduction of E. coli promoters (Pfis and PtyrT), whose expression patterns in E. coli are dependent upon the surrounding DNA topology. These promoters drive luciferase reporter genes with a near 24-h rhythm and specific phasing to suggest the importance of DNA superhelicity and arrangement within the cell in the maintenance of circadian-regulated gene expression (Min et al., 2004).
When S. elongatus cells are sampled over time and stained with DAPI (4′, 6-diamidino-2-phenylindole) dye to visualize the nucleoid, a rhythm in the compaction/decompaction of the chromosome can be observed (Smith and Williams, 2006). This rhythm occurs in a circadian pattern in LD12:12 cycles, and continues with a period of approximately 24 h when cells are transferred to LL. The chromosome slowly condenses during the light (or subjective day) with full compaction occurring just before the anticipated light to dark transfer, and decompaction proceeds during the (subjective) night (Smith and Williams, 2006). The predictable changes in DNA topology over time are not limited to the chromosome; endogenous plasmids of S. elongatus exhibit the rhythmic fluctuations in superhelicity, and luciferase reporters display similar patterns of bioluminescence when expressed from the endogenous pANS plasmid as they do from the chromosome (Woelfle et al., 2007).
By comparing microarray analysis and the superhelical conformation of the pANS plasmid of S. elongatus, each topological state of the DNA can be associated with a particular state of gene expression (Vijayan et al., 2009). The activation or repression of genes results directly from response to the change in superhelicity of the chromosome. These responses were tested by adding novobiocin, a DNA gyrase inhibitor that results in a more relaxed chromosome, to cells at sub-lethal concentrations. Almost 80% of the tested promoters responded in a predictable manner: those genes that are highly expressed when the plasmid is supercoiled DNA were repressed, while other genes had increased expression upon novobiocin addition to suggest that they are normally repressed when the DNA is supercoiled (Vijayan et al., 2009).
The link between the Kai oscillator and changes in DNA topology likely lies in KaiC (Figure 4). The structure of KaiC resembles the hexameric proteins of the RecA/DnaB family – recombinase and helicase proteins that can bind DNA – and KaiC can interact with forked, double-stranded DNA molecules (Mori et al., 2002). In a kaiC null background, the rhythms in chromosome condensation are lost; however, the rhythm is not dependent solely upon the presence of KaiC, as the inactivation of kaiA produces a phenotype indistinguishable from that of the kaiC mutant (Smith and Williams, 2006). The period of the genome compaction rhythm is dependent upon the timing of the Kai oscillator. The 14-h periodicity of a kaiC14 mutant is paralleled in the compaction rhythm. An inactivation of sasA still allows the chromosome to condense in a rhythmic fashion, but the transfer of information is lost without SasA present, and the result is arrhythmic gene expression (Smith and Williams, 2006). Additionally, unlike its wild-type counterpart, the kaiC (pr1) allele cannot induce repression globally when overexpressed (Kiyohara et al., 2005) and cannot undergo dark-induced chromosome compaction. The kaiC (pr1) mutant will allow for wild-type rhythms to occur in LL conditions. These data suggest that the kaiC (pr1) allele is defective in its ability to regulate stimulus-induced chromosome compaction. Pulsed kaiC (pr1) overexpression at different times throughout the cycle cannot induce phase shifts like that of wild type, which suggests that there is a role for KaiC feedback to input pathways or compaction as an input to the cell (Smith and Williams, 2006; Smith and Williams, 2009).
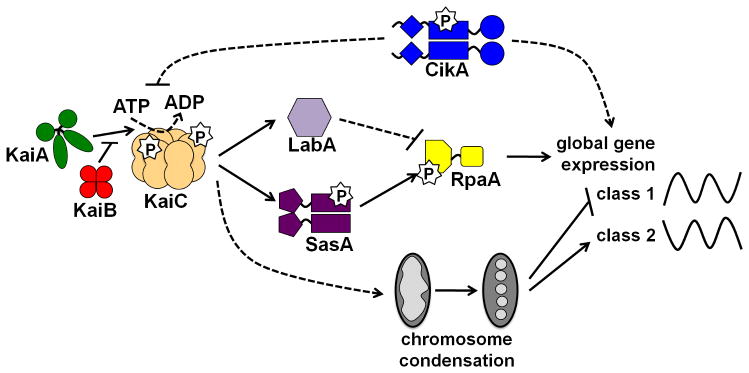
Figure 4.
Model of S. elongatus circadian output pathways. Internal timekeeping is mediated via regulation of KaiC ATPase and phosphorylation activities, where KaiA enhances KaiC autophosphorylation and KaiB averts the KaiA-induced stimulation. KaiC ATPase activity is repressed (indirectly) by CikA in a KaiA-independent pathway. Temporal information from the Kai complex is transduced through at least three protein-based pathways that regulate gene expression. KaiC stimulates SasA autophosphorylation; phosphorylated SasA subsequently transfers its phosphoryl group to RpaA, which leads to RpaA activation. This phosphotransfer event is predicted to result in changes in genome-wide gene expression patterns. LabA is involved in the negative regulation of RpaA function through indirect mechanisms. Residual rhythmic output in the absence of RpaA results from CikA-mediated gene expression pathways. KaiC activity also regulates chromosome compaction; a compact chromosome turns off expression of class 1 genes, while class 2 genes peak at times when compaction is at its highest. The exact mechanism by which KaiC influences chromosome topology is not yet known. Solid lines and dashed lines represent direct and indirect mechanisms, respectively. Arrows represent positive influence, while blunt lines represent inhibition. P, phosphate.
C. The SasA-RpaA two-component system as a positive limb of output
The positive limb of the S. elongatus output pathway is comprised of the two-component regulatory system proteins SasA and RpaA (Regulator of phycobiliosome associated). The importance of SasA in the S. elongatus circadian timing mechanism was recognized through its direct interaction with KaiC using a yeast two-hybrid screen (Iwasaki et al., 2000). Immunoprecipitation experiments showed that either full-length or the N-terminal domain (residues 1–97) of SasA is capable of interaction with KaiC; either the CI or CII domain of KaiC is sufficient for this binding. No direct interactions between SasA and either KaiA or KaiB were detected in vivo or in vitro (Iwasaki et al., 2000). The sasA gene encodes a 387 amino acid protein that belongs to the HPK family of proteins involved in His-to-Asp two-component signal transduction (Stock et al., 2000). Further bioinformatic analyses revealed the N-terminal domain of SasA has 60% amino acid similarity to the full-length KaiB protein (Iwasaki et al., 2000); however, the tetrameric structure of KaiB (Iwase et al., 2005) differs from the thioredoxin-like fold of the KaiC-interacting N-terminus of SasA (Klewer et al., 2002).
Removal of sasA shortens the circadian period and severely decreases the amplitude of circadian gene expression to near-baseline levels (Iwasaki et al., 2000). The clock is still running and can be reset by light and temperature cues, but the timekeeping mechanism can no longer effectively transduce that information to clock-controlled processes. Rhythmic accumulation of kaiA and kaiBC mRNA does not persist in a sasA mutant background; KaiA protein levels are reduced to approximately 70% compared to those of wild type, while KaiB and KaiC protein levels decrease to levels that are nearly undetectable using standard protocols. Constitutive overexpression of SasA causes arrhythmia as measured by bioluminescent reporters. Short pulses of SasA overexpression shift the phase of the rhythm with advances in the oscillation when sasA levels are decreasing and delays when sasA levels are on the rise (Iwasaki et al., 2000). The direction of the phase shifts that result from short pulses of SasA overexpression are contradictory to those seen when KaiC is overexpressed for short durations. This discrepancy further demonstrates that SasA is important for the functionality of the clock system, but that its function is distinct and separate from that of the Kai complex.
The interaction between the N-terminal sensor domain of SasA and KaiC allows for KaiC to increase the rate by which SasA autophosphorylates, though the reciprocal experiment does not show a SasA-dependent change in KaiC autokinase activity (Smith and Williams, 2006). The autokinase activity of SasA is crucial to its function, as an amino acid substitution at the conserved histidine residue (H162Q) results in a sasA null phenotype (Iwasaki et al., 2000). This KaiC-induced activation of SasA is predicted to be limited during the late subjective night/early subjective day of the circadian cycle in which SasA is part of a large multimeric complex that contains at least the KaiA, KaiB, and KaiC proteins (Kageyama et al., 2003), and likely also CikA (Ivleva et al., 2006) and LdpA (Ivleva et al., 2005). SasA transfers its phosphate group to RpaA (Figure 4), which is predicted to activate the protein to serve as a transcription factor through its DNA-binding domain (Takai et al., 2006); the exact target(s) of RpaA has not yet been elucidated.
RpaA in S. elongatus was identified using bioinformatic analyses to identify sequences that encode receiver domains commonly found in RR proteins (Takai et al., 2006). Of 24 identified sequences, only disruption of rpaA resulted in the attenuated expression from the kaiBC promoter that would be expected of the partner protein of SasA. The phenotype of the rpaA null is more severe than that of a sasA mutant in that elimination of rpaA results in arrhythmia from nearly all promoters regardless of light intensity; however, the residual rhythmicity from a PpsbAI reporter in the absence of rpaA shows that the oscillator continues to function in the absence of this output pathway (Takai et al., 2006). The overall decrease in the level of gene expression in the absence of either sasA or rpaA suggests that the SasA-RpaA system acts as a positive regulator of the output pathway.
D. LabA and CikA as negative regulators of output
The labA (low-amplitude and bright) gene was identified because its absence lessened the inhibitory effects of excess KaiC protein on the kaiBC promoter (Taniguchi et al., 2007). In an otherwise wild-type background, a labA null mutant displays decreased amplitude that results from elevated trough levels from bioluminescence reporters, but maintains a wild-type circadian period. These phenotypes are paralleled in the overall increased levels of kaiBC mRNA and KaiC protein with a maintained 24-h rhythmic accumulation and KaiC-P rhythm (Taniguchi et al., 2007). Overexpression of labA reduces the overall level of bioluminescence and corresponding KaiC protein levels, but a wild-type period is maintained by the luxAB reporter and KaiC-P status. These data support the role of LabA as a negative limb of the output pathway through which KaiC-P acts to repress gene expression.
LabA and SasA are predicted to function in separate pathways that converge at RpaA (Figure 4). The labA/sasA double mutant displays an intermediate phenotype of overall low amplitude rhythms, but overall higher bioluminescence levels than that of the sasA mutant alone; inactivation of labA can attenuate the bioluminescence repression that results from SasA overexpression (Taniguchi et al., 2007). The rpaA gene is epistatic to labA because inactivation of both labA and rpaA results in the phenotype that is similar to that of rpaA alone (Taniguchi et al., 2007). Thus, RpaA likely receives circadian output information through (at least) two independent pathways: SasA positively influences RpaA function via phosphotransfer, and LabA negatively regulates RpaA function by indirect mechanisms.
The residual rhythmicity of the labA/sasA double mutant may be the result of the CikA protein, which is most notable for its role in the S. elongatus input pathway for discrete entrainment. A transposon insertion in the cikA gene was identified in the same screen that identified labA; inactivation of cikA resulted in decreased repression of kaiBC promoter by excess KaiC (Taniguchi et al., 2010). Inactivation of labA, sasA, and cikA in the same cell resulted in arrhythmic bioluminescence from luciferase reporters; however, the oscillator itself appears to be intact as the pattern of KaiC-P over the circadian cycle was similar to that of wild type (Taniguchi et al., 2010).
Genetic analyses demonstrated that LabA and CikA work through independent output pathways to negatively regulate expression of kaiBC. The cikA/labA double mutant displays the additive phenotype of a short period and low amplitude rhythm associated with the removal of cikA, but with an overall increase in bioluminescence as seen in the labA null (Taniguchi et al., 2010). In each single mutant and the double mutant, KaiC protein levels were higher than those of wild type; however, only the cikA null showed a change in the phase of the KaiC-P pattern to suggest that CikA-mediated repression of kaiBC expression is separate from that of the KaiC-P-mediated feedback in which LabA participates.
E. CpmA and Group 2 sigma factors affect subsets of genes
The circadian phase modifier (cpmA) gene has also been implicated as a component in output from the S. elongatus clock. Although overexpression of the cpmA gene does not alter the circadian rhythm, its inactivation results in a drastic change in the relative phase angle of a small subset of cyanobacterial reporters (Katayama et al., 1999). In a cpmA background, rhythms from the kaiBC promoter are unaffected, but the phase of expression from a PkaiA::luxAB reporter is shifted by 10 h, such that its expression peaks almost in anti-phase to that of PkaiBC (Katayama et al., 1999). Despite this discrepancy in timing from the kai promoters, robust rhythms of gene expression continue with wild-type period lengths. This fact suggests that cpmA is not part of the oscillator, or a general output pathway, but rather specifically relays temporal information to a subset of genes.
The basic transcriptional machinery is implicated in contributing to circadian output pathways. In bacteria the RNA polymerase holoenzyme can include different sigma factors, which are the subunits that recognize promoter elements (Gross et al., 1998). The Group 2 sigma factors in cyanobacteria are a family of proteins that have sequence similarity to the housekeeping sigma-70 RpoD1 protein, but are not essential for viability of the cells (Tanaka et al., 1992). Inactivation of any of the Group 2 sigma factor genes (rpoD2, rpoD3, rpoD4, or sigC) either alone or in pairs alters the rhythm from a PpsbAI::luxAB reporter by affecting the phase, period, or amplitude of expression (Nair et al., 2002; Tsinoremas et al., 1996). The inactivation of sigC lengthens the period of expression from the PpsbAI promoter but has little effect on the expression patterns from either the PkaiBC or PpurF promoters. PkaiBC promoter is only slightly perturbed as a result of the rpoD2 single mutant, or the rpoD3/rpoD4 and rpoD2/rpoD3 double mutants, which suggests that there is greater buffering of the kaiBC promoter compared to others. The current model proposes that RNA polymerase forms holoenzymes with different Group 2 sigma factors throughout the circadian cycle. Durations of activity for some sigma factors likely overlap; redundancy in their roles is indicated by discrepancies in their phenotypes. The connection between the SasA-RpaA pathway and the regulation by the Group 2 sigma factors is still unclear.
F. Cell Division
Circadian oscillations in gene expression persist in S. elongatus cells with generation times much shorter (as fast as 6 h) than a full circadian cycle (Kondo et al., 1993; Mori et al., 1996). The Kai-based system regulates cell division such that the number of cells in a continuously diluted culture displays a circadian rhythm in LL that matches the phase of its synchronizing LD cycle (Mori et al., 1996). In an arrhythmic kaiC mutant, the rhythm in cell division can be driven by external LD cycles, but does not continue in LL (Mori and Johnson, 2001b). The circadian clock gates cell division, such that there are times in the circadian cycle where cell division is forbidden. For S. elongatus, this forbidden phase occurs early to mid-subjective night (Mori et al., 1996). The regulation is unidirectional, as the cell division cycle does not influence the timing of circadian oscillation (Mori and Johnson, 2001b). Cells maintain wild-type rhythms in bioluminescence from transcriptional fusion reporters regardless of doubling time, which varies in response to light intensity, as well as during stationary phase where cells do not undergo cell division, and during chemically-induced inhibition of cell division (Mori and Johnson, 2001b).
Because DNA synthesis occurs at a constant rate in free-running cells (Mori et al., 1996), the checkpoint for the cell division cycle is likely cytokinesis. Coordination of protein components from the three intermingled sectors of the clock – input, oscillator, and output – has been implicated in the transduction pathway leading to the cell cycle checkpoint. This gating results from elevated ATPase activity of KaiC, rather than overall abundance or phosphorylation state of this central oscillator component (Dong et al., 2010); the two latter properties play important roles in the maintenance of circadian period and global repression of S. elongatus promoters.
Of the known clock genes, only the cikA and kaiB individual mutants produce elongated cells indicative of altered circadian control of cell division (Dong et al., 2010; Miyagishima et al., 2005), although deletion of cikA does not eliminate the gating phenomenon. The elongated cell phenotype in the absence of either CikA or KaiB is suppressed when kaiC, sasA, or rpaA is removed from the cell (Dong et al., 2010). The current model predicts that when KaiC ATPase activity rises above a necessary threshold, cell division is not allowed to proceed because the gate is closed (Figure 3D). Conformational changes in KaiC that are predicted to occur when ATPase activity is above the threshold allow for its stimulation of SasA autophosphorylation and subsequent phosphotransfer to RpaA. Phosphorylated RpaA likely inhibits the formation and localization of FtsZ rings that function to denote the midline of the cell where cell division would occur. Support for this model includes mutants with elevated KaiC ATPase activity that display mislocalized FtsZ proteins (Dong et al., 2010).
Interestingly, the forbidden phase of cell division is coincident with the formation of the clock complex and with the transition from a decondensed to condensed nucleoid. It is possible that cell division is tightly regulated by the circadian system in order to protect the clock itself from disruptions that would result in an otherwise desynchronized population. Mathematical models that describe the coupling of the cell division cycle with that of the circadian rhythm (Yang et al., 2010) help to explain how a population of cyanobacterial cells is able to maintain a similar circadian phase with one another after multiple generations (Mihalcescu et al., 2004).
CONCLUSIONS
The benefits of the genetic and mutational exploitation of the S. elongatus PCC 7942 model for a circadian oscillator have been multifold. The ease of working with this single-celled system was monumental to unveiling the nuts and bolts of the clock and continues to provoke new ideas about circadian mechanisms. The idea of a self-sustained oscillation in the absence of a traditional feedback loop (Nakajima et al., 2005; Nakajima et al., 2010; Tomita et al., 2005) flew in the face of canonical circadian models for timing (Cheng et al., 2003; Cyran et al., 2003; Dunlap, 1999; Harmer et al., 2001; Okamura et al., 2002; Van Gelder et al., 2003) and has subsequently transformed the way, or at least opened the possibilities as to how, eukaryotic-centered chronobiologists think about circadian mechanisms. Recent work in the unicellular picoeukaryotic alga Ostreococcus tauri and in human red blood cells have shown that circadian rhythms in peroxiredoxin oxidation and oligomerization, respectively, exist in the absence of transcription and in eukaryotic cells lacking a nucleus (O’Neill and Reddy, 2011; O’Neill et al., 2011). With redox sensing as an essential part of LdpA, CikA and KaiA function, oscillators in cellular metabolism and redox status are emerging as interesting avenues for further investigation to the understanding of timekeeping mechanisms.
While the lack of a need for a transcriptional-translational feedback oscillation is not required, the fact does not negate its existence. The theory of, and evidence for, multiple oscillators in circadian systems are not new (Bell-Pedersen et al., 2005; de Paula et al., 2007; Indic et al., 2007). There are multiple lines of evidence for the presence of at least a second oscillator in the S. elongatus model. Inactivation of sigC causes period changes in some, but not all reporters leading to the idea that there might be more than one oscillator in the S. elongatus system that become uncoupled from one another when SigC is missing (Nair et al., 2002). In microarray analyses using a kaiABC null strain, 17 genes were found to continue to display rhythmic mRNA accumulation, again suggestive that there is a weak secondary oscillator that controls those transcripts in the absence of the Kai-based oscillator (Vijayan et al., 2009). In addition, genome-wide proteomic studies to determine which clock components (or other proteins) fluctuate their abundance in a circadian manner suggest that only 30–64% of mRNA levels are rhythmic, some of which are known clock components. A more thorough proteomic analysis would allow for a determination of the physiological relevance of these proteins in the circadian system. While there is evidence to predict multiple oscillator(s), their mechanism(s) have not yet been described. And, more importantly, the mechanism by which these multiple oscillators communicate with one another to drive precise rhythms has yet to be elucidated.
In addition to the oscillator mechanism itself, there are many questions still remaining with regards to the prokaryotic clock system as a whole. The current data suggest that KaiA is the conduit for relaying input information to the oscillator as a result of direct expression regulation by Pex (Arita et al., 2007) and interactions with CikA (Ivleva et al., 2006) and LdpA (Ivleva et al., 2005), but the story is not complete. While sensing redox state via the quinone pool seems to be the cue, questions still remain regarding how CikA and LdpA relay this environmental information to the central oscillator. The existence of a cognate RR for CikA that relays redox information to the clock directly is currently elusive. In terms of output, how KaiC-P, KaiC ATPase activity, the SasA/RpaA two-component regulatory system, LabA, Group 2 factors and chromosome compaction coordinate to dictate precise rhythmic activity is not understood. KaiC overexpression has been shown to have differential effects on the promoters that exist on the chromosome and on plasmids. On the chromosome, KaiC overexpression eliminates rhythmic gene expression to the trough levels of all promoters tested and leads to complete chromosome compaction. On the other hand, gene expression from the same reporter construct on the pANS plasmid results in elevated, arhythmic expression (Woelfle et al., 2007). How the clock controls DNA topology and gene expression, how it differentially controls gene expression based on chromosome or plasmid status, and how topology changes feed back on the clock itself are also still in question.
We have also learned to be careful of our own biases. Since its discovery and as dictated by its name, CikA is known to function in the input pathway for the Kai oscillator (Ivleva et al., 2006; Schmitz et al., 2000; Zhang et al., 2006). However, recent lines of evidence elucidated by genetic studies have shown that CikA also functions as an inhibitor of KaiC ATPase activity (Dong et al., 2010) and as a component of one of three independent output pathways to negatively regulate expression of kaiBC (Taniguchi et al., 2010). Regardless of its multiple personalities, the activity of CikA in input, the oscillator, or output is currently placed in an indirect role; exactly how CikA functions in each of these parts of the oscillator is unclear.
No circadian system is understood to completion; however, insights from classic circadian studies and dogma-shattering breakthroughs have shaped the pathway for understanding the Kai oscillator, illustrating the importance of multiple model systems and basic science. What we have learned thus far is that it is important to keep an eye on the past but an open mind to future possibilities that may make up the overall clock processes. Undoubtedly, the ease of this single-celled prokaryotic model will aid in teasing out the intricacies of a circadian clock model, which will lead to advances in other systems as well.
Acknowledgments
The authors thank Dr. Yong-Ick Kim for structural and immunoblot images used in Figures 1 and 2, respectively, Dr. Julie Bordowitz for micrograph images used in Figure 3, and Dr. Michael Vitalini for careful reading of the manuscript. The Synechococcus elongatus PCC 7942 sequence data were produced by the US Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) in collaboration with the user community.
ABBREVIATIONS
- LL
constant light
- DD
constant dark
- LD
light/dark
- FRP
free-running period
- KaiC-P
KaiC phosphorylation
- CT
circadian time
- RR
response regulator
- PsR
pseudo-receiver
- HPK
histidine protein kinase
DEFINITIONS
- Aschoff’s rule
a phenomenon originally described by Jürgen Aschoff in which the circadian period of diurnal organisms decreases with increasing light intensity (nocturnal organisms display the opposite response)
- Continuous entrainment
the modulation of the internal period as a result of gradual changes in the environment, including the changes in light intensity that arise from the rotation of the Earth on its axis
- Discrete entrainment
the effect of the phase of the internal circadian rhythm in response to abrupt changes in environmental cues, such as a brief dark pulse during the maintenance of the organism in constant light conditions
- Q10 value
the ratio of the rate of a given process at one temperature to the rate at a temperature 10°C lower; Q10 values of rhythms maintained by a circadian clock are near that of 1.0
- Subjective day/night
the time during constant conditions that the organism would ordinarily be in the light or dark, respectively, of a light/dark cycle
- Two-component signal transduction system
a regulatory system common in prokaryotic organisms that consists of two proteins – a histidine protein kinase that autophosphorylates in response to some stimulus, and subsequently transfers its phosphoryl group to a cognate response regulator protein to elicit a response to the initial stimulus
Contributor Information
Shannon R. Mackey, Email: MackeyShannonR@sau.edu.
Susan S. Golden, Email: sgolden@ucsd.edu.
Jayna L. Ditty, Email: jlditty@stthomas.edu.
References
- Amdaoud M, Vallade M, Weiss-Schaber C, Mihalcescu I. Cyanobacterial clock, a stable phase oscillator with negligible intercellular coupling. Proc Natl Acad Sci USA. 2007;104:7051–6. doi: 10.1073/pnas.0609315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K, Hashimoto H, Igari K, Akaboshi M, Kutsuna S, Sato M, Shimizu T. Structural and biochemical characterization of a cyanobacterium circadian clock-modifier protein. J Biol Chem. 2007;282:1128–35. doi: 10.1074/jbc.M608148200. [DOI] [PubMed] [Google Scholar]
- Aschoff J. Freerunning and entrained circadian rhythms. Handbook of behavioral neurobiology: Biological rhythms. 1981:81–93. [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet. 2005;6:544–56. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TH, Chen TL, Hung LM, Huang TC. Circadian rhythm in amino acid uptake by Synechococcus RF-1. Plant Physiol. 1991;97:55–9. doi: 10.1104/pp.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, Yang Y, Wang L, He Q, Liu Y. WHITE COLLAR-1, a multifunctional neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J Biol Chem. 2003;278:3801–8. doi: 10.1074/jbc.M209592200. [DOI] [PubMed] [Google Scholar]
- Clodong S, Duhring U, Kronk L, Wilde A, Axmann I, Herzel H, Kollmann M. Functioning and robustness of a bacterial circadian clock. Mol Syst Biol. 2007;3:90. doi: 10.1038/msb4100128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell. 2003;112:329–41. doi: 10.1016/s0092-8674(03)00074-6. [DOI] [PubMed] [Google Scholar]
- de Paula RM, Vitalini MW, Gomer RH, Bell-Pedersen D. Complexity of the Neurospora crassa circadian clock system: multiple loops and oscillators. Cold Spring Harb Symp Quant Biol. 2007;72:345–51. doi: 10.1101/sqb.2007.72.002. [DOI] [PubMed] [Google Scholar]
- DeCoursey PJ. Effect of light on the circadian activity rhythm of the flying squirrel, Glaucomys volans. Z Vgl Physiol. 1961;44:331–354. [Google Scholar]
- DeCoursey PJ, Walker JK, Smith SA. A circadian pacemaker in free-living chipmunks: essential for survival? J Comp Physiol A. 2000;186:169–80. doi: 10.1007/s003590050017. [DOI] [PubMed] [Google Scholar]
- Ditty JL, Canales SR, Anderson BE, Williams SB, Golden SS. Stability of the Synechococcus elongatus PCC 7942 circadian clock under directed anti-phase expression of the kai genes. Microbiology. 2005;151:2605–13. doi: 10.1099/mic.0.28030-0. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–3. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Dong G, Yang Q, Wang Q, Kim YI, Wood TL, Osteryoung KW, van Oudenaarden A, Golden SS. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 2010;140:529–39. doi: 10.1016/j.cell.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–90. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: biological timekeeping. Sinauer Associates; Sunderland, Mass: 2004. [Google Scholar]
- Edmunds LN., Jr Chronobiology at the cellular and molecular levels: models and mechanisms for circadian timekeeping. Am J Anat. 1983;168:389–431. doi: 10.1002/aja.1001680404. [DOI] [PubMed] [Google Scholar]
- Eguchi K, Yoda M, Terada TP, Sasai M. Mechanism of robust circadian oscillation of KaiC phosphorylation in vitro. Biophys J. 2008;95:1773–84. doi: 10.1529/biophysj.107.127555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai J, Wolk CP. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–54. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- Emberly E, Wingreen NS. Hourglass model for a protein-based circadian oscillator. Phys Rev Lett. 2006;96:038303. doi: 10.1103/PhysRevLett.96.038303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Zhang X, Ivleva NB, Golden SS, LiWang A. NMR structure of the pseudo-receiver domain of CikA. Protein Sci. 2007;16:465–75. doi: 10.1110/ps.062532007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces RG, Wu N, Gillon W, Pai EF. Anabaena circadian clock proteins KaiA and KaiB reveal a potential common binding site to their partner KaiC. EMBO J. 2004;23:1688–98. doi: 10.1038/sj.emboj.7600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden SS. Mutagenesis of cyanobacteria by classical and gene-transfer-based methods. Methods Enzymol. 1988;167:714–27. doi: 10.1016/0076-6879(88)67083-2. [DOI] [PubMed] [Google Scholar]
- Golden SS, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–31. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- Golden SS, Sherman LA. A hybrid plasmid is a stable cloning vector for the cyanobacterium Anacystis nidulans R2. J Bacteriol. 1983;155:966–72. doi: 10.1128/jb.155.3.966-972.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden SS, Sherman LA. Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J Bacteriol. 1984;158:36–42. doi: 10.1128/jb.158.1.36-42.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobbelaar N, Huang TC. Effect of oxygen and temperature on the induction of a circadian nitrogenase activity rhythm in Synechococcus RF-1. Plant Physiol. 1992;140:391–394. [Google Scholar]
- Grobbelaar N, Huang TC, Lin HY, Chow TJ. Dinitrogen fixing endogenous rhythm is Synechococcus RF-1. FEMS Microbiol Lett. 1986;37:173–177. [Google Scholar]
- Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb Symp Quant Biol. 1998;63:141–55. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annu Rev Cell Dev Biol. 2001;17:215–53. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Ito H, Fujita M, Iwase R, Uzumaki T, Ishiura M. Stoichiometric interactions between cyanobacterial clock proteins KaiA and KaiC. Biochem Biophys Res Commun. 2004;316:195–202. doi: 10.1016/j.bbrc.2004.02.034. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Iwase R, Uzumaki T, Ishiura M. Hexamerization by the N-terminal domain and intersubunit phosphorylation by the C-terminal domain of cyanobacterial circadian clock protein KaiC. Biochem Biophys Res Commun. 2006;348:864–72. doi: 10.1016/j.bbrc.2006.07.143. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Suzuki H, Iwase R, Uzumaki T, Miyake A, Shen JR, Imada K, Furukawa Y, Yonekura K, Namba K, Ishiura M. ATP-induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells. 2003;8:287–96. doi: 10.1046/j.1365-2443.2003.00633.x. [DOI] [PubMed] [Google Scholar]
- Herrero A, Muro-Pastor AM, Flores E. Nitrogen control in cyanobacteria. J Bacteriol. 2001;183:411–25. doi: 10.1128/JB.183.2.411-425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtman CK, Chen Y, Sandoval P, Gonzales A, Nalty MS, Thomas TL, Youderian P, Golden SS. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 2005;12:103–15. doi: 10.1093/dnares/12.2.103. [DOI] [PubMed] [Google Scholar]
- Huang TC, Tu J, Chow TJ, Chen TH. Circadian rhythm of the prokaryote Synechococcus sp RF-1. Plant Physiol. 1990;92:531–3. doi: 10.1104/pp.92.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Nishiwaki T, Kondo T, Iwasaki H. Circadian rhythms in the synthesis and degradation of a master clock protein KaiC in cyanobacteria. J Biol Chem. 2004;279:36534–9. doi: 10.1074/jbc.M405861200. [DOI] [PubMed] [Google Scholar]
- Indic P, Schwartz WJ, Herzog ED, Foley NC, Antle MC. Modeling the behavior of coupled cellular circadian oscillators in the suprachiasmatic nucleus. J Biol Rhythms. 2007;22:211–9. doi: 10.1177/0748730407301238. [DOI] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–23. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- Ito H, Kageyama H, Mutsuda M, Nakajima M, Oyama T, Kondo T. Autonomous synchronization of the circadian KaiC phosphorylation rhythm. Nat Struct Mol Biol. 2007;14:1084–8. doi: 10.1038/nsmb1312. [DOI] [PubMed] [Google Scholar]
- Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci USA. 2009;106:14168–73. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Bramlett MR, Lindahl PA, Golden SS. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 2005;24:1202–10. doi: 10.1038/sj.emboj.7600606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva NB, Gao T, LiWang AC, Golden SS. Quinone sensing by the circadian input kinase of the cyanobacterial circadian clock. Proc Natl Acad Sci USA. 2006;103:17468–73. doi: 10.1073/pnas.0606639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA. 2002;99:15788–93. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Taniguchi Y, Ishiura M, Kondo T. Physical interactions among circadian clock proteins KaiA, KaiB and KaiC in cyanobacteria. EMBO J. 1999;18:1137–45. doi: 10.1093/emboj/18.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 2000;101:223–33. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- Iwase R, Imada K, Hayashi F, Uzumaki T, Morishita M, Onai K, Furukawa Y, Namba K, Ishiura M. Functionally important substructures of circadian clock protein KaiB in a unique tetramer complex. J Biol Chem. 2005;280:43141–9. doi: 10.1074/jbc.M503360200. [DOI] [PubMed] [Google Scholar]
- Johnson CH. Testing the adaptive value of circadian systems. Methods Enzymol. 2005;393:818–37. doi: 10.1016/S0076-6879(05)93043-7. [DOI] [PubMed] [Google Scholar]
- Kageyama H, Kondo T, Iwasaki H. Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J Biol Chem. 2003;278:2388–95. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- Kageyama H, Nishiwaki T, Nakajima M, Iwasaki H, Oyama T, Kondo T. Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell. 2006;23:161–71. doi: 10.1016/j.molcel.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kubota K, Ming H, Miyazono K, Tanokura M. Crystal structure of KaiC-like protein PH0186 from hyperthermophilic archaea Pyrococcus horikoshii OT3. Proteins. 2009;75:1035–9. doi: 10.1002/prot.22367. [DOI] [PubMed] [Google Scholar]
- Katayama M, Kondo T, Xiong J, Golden SS. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J Bacteriol. 2003;185:1415–22. doi: 10.1128/JB.185.4.1415-1422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama M, Tsinoremas NF, Kondo T, Golden SS. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181:3516–24. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Dong G, Carruthers CW, Jr, Golden SS, LiWang A. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2008;105:12825–30. doi: 10.1073/pnas.0800526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippert F. Endocytobiotic coordination, intracellular calcium signaling, and the origin of endogenous rhythms. Ann NY Acad Sci. 1987;503:476–95. doi: 10.1111/j.1749-6632.1987.tb40631.x. [DOI] [PubMed] [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–34. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–21. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara YB, Katayama M, Kondo T. A novel mutation in kaiC affects resetting of the cyanobacterial circadian clock. J Bacteriol. 2005;187:2559–64. doi: 10.1128/JB.187.8.2559-2564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klewer DA, Williams SB, Golden SS, LiWang AC. Sequence-specific resonance assignments of the N-terminal, 105-residue KaiC-interacting domain of SasA, a protein necessary for a robust circadian rhythm in Synechococcus elongatus. J Biomol NMR. 2002;24:77–8. doi: 10.1023/a:1020649703380. [DOI] [PubMed] [Google Scholar]
- Kondo T, Ishiura M. Circadian rhythms of cyanobacteria: monitoring the biological clocks of individual colonies by bioluminescence. J Bacteriol. 1994;176:1881–5. doi: 10.1128/jb.176.7.1881-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–6. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa G, Aihara K, Iwasa Y. A model for the circadian rhythm of cyanobacteria that maintains oscillation without gene expression. Biophys J. 2006;91:2015–23. doi: 10.1529/biophysj.105.076554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsuna S, Kondo T, Aoki S, Ishiura M. A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp strain PCC 7942. J Bacteriol. 1998;180:2167–74. doi: 10.1128/jb.180.8.2167-2174.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsuna S, Kondo T, Ikegami H, Uzumaki T, Katayama M, Ishiura M. The circadian clock-related gene pex regulates a negative cis element in the kaiA promoter region. J Bacteriol. 2007;189:7690–6. doi: 10.1128/JB.00835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsuna S, Nakahira Y, Katayama M, Ishiura M, Kondo T. Transcriptional regulation of the circadian clock operon kaiBC by upstream regions in cyanobacteria. Mol Microbiol. 2005;57:1474–84. doi: 10.1111/j.1365-2958.2005.04781.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Golden SS, Kondo T, Ishiura M, Johnson CH. Bacterial luciferase as a reporter of circadian gene expression in cyanobacteria. J Bacteriol. 1995;177:2080–6. doi: 10.1128/jb.177.8.2080-2086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackey SR, Ditty JL, Zeidner G, Chen Y, Golden SS. Mechanisms for entraining the cyanobacterial circadian clock system with the environment. In: Ditty JL, Mackey SR, Johnson CH, editors. Bacterial Circadian Programs. Springer-Verlag; Berlin: 2009. pp. 141–156. [Google Scholar]
- Markson JS, O’Shea EK. The molecular clockwork of a protein-based circadian oscillator. FEBS Lett. 2009;583:3938–47. doi: 10.1016/j.febslet.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Hong CI, Shi M, Loros JJ, Dunlap JC, Ruoff P. Circadian rhythmicity by autocatalysis. PLoS Comput Biol. 2006;2:e96. doi: 10.1371/journal.pcbi.0020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–53. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–5. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- Min H, Golden SS. A new circadian class 2 gene, opcA, whose product is important for reductant production at night in Synechococcus elongatus PCC 7942. J Bacteriol. 2000;182:6214–21. doi: 10.1128/jb.182.21.6214-6221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Liu Y, Johnson CH, Golden SS. Phase determination of circadian gene expression in Synechococcus elongatus PCC 7942. J Biol Rhythms. 2004;19:103–12. doi: 10.1177/0748730403262056. [DOI] [PubMed] [Google Scholar]
- Ming H, Miyazono K, Tanokura M. Cloning, expression, purification, crystallization and preliminary crystallographic analysis of selenomethionine-labelled KaiC-like protein PH0186 from Pyrococcus horikoshii OT3. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:327–9. doi: 10.1107/S1744309107011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui A, Kumazawa S, Takahashi A, Ikemoto H, Arai T. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature. 1986;323:720–722. [Google Scholar]
- Miyagishima SY, Wolk CP, Osteryoung KW. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol Microbiol. 2005;56:126–43. doi: 10.1111/j.1365-2958.2005.04548.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi F, Nakayama Y, Kaizu K, Iwasaki H, Tomita M. A mathematical model for the Kai-protein-based chemical oscillator and clock gene expression rhythms in cyanobacteria. J Biol Rhythms. 2007;22:69–80. doi: 10.1177/0748730406295749. [DOI] [PubMed] [Google Scholar]
- Mori T, Binder B, Johnson CH. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci USA. 1996;93:10183–8. doi: 10.1073/pnas.93.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Johnson CH. Circadian programming in cyanobacteria. Semin Cell Dev Biol. 2001a;12:271–8. doi: 10.1006/scdb.2001.0254. [DOI] [PubMed] [Google Scholar]
- Mori T, Johnson CH. Independence of circadian timing from cell division in cyanobacteria. J Bacteriol. 2001b;183:2439–44. doi: 10.1128/JB.183.8.2439-2444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH. Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA. 2002;99:17203–8. doi: 10.1073/pnas.262578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Williams DR, Byrne MO, Qin X, Egli M, McHaourab HS, Stewart PL, Johnson CH. Elucidating the ticking of an in vitro circadian clockwork. PLoS Biol. 2007;5:e93. doi: 10.1371/journal.pbio.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R, Miyake A, Iwase R, Hayashi F, Uzumaki T, Ishiura M. ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells. 2008;13:387–95. doi: 10.1111/j.1365-2443.2008.01174.x. [DOI] [PubMed] [Google Scholar]
- Murayama Y, Oyama T, Kondo T. Regulation of circadian clock gene expression by phosphorylation states of KaiC in cyanobacteria. J Bacteriol. 2008;190:1691–8. doi: 10.1128/JB.01693-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuda M, Michel KP, Zhang X, Montgomery BL, Golden SS. Biochemical properties of CikA, an unusual phytochrome-like histidine protein kinase that resets the circadian clock in Synechococcus elongatus PCC 7942. J Biol Chem. 2003;278:19102–10. doi: 10.1074/jbc.M213255200. [DOI] [PubMed] [Google Scholar]
- Nagai T, Terada TP, Sasai M. Synchronization of circadian oscillation of phosphorylation level of KaiC in vitro. Biophys J. 2010;98:2469–77. doi: 10.1016/j.bpj.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Ditty JL, Min H, Golden SS. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J Bacteriol. 2002;184:3530–8. doi: 10.1128/JB.184.13.3530-3538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc Natl Acad Sci USA. 2004;101:881–5. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–5. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- Nakajima M, Ito H, Kondo T. In vitro regulation of circadian phosphorylation rhythm of cyanobacterial clock protein KaiC by KaiA and KaiB. FEBS Lett. 2010;584:898–902. doi: 10.1016/j.febslet.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA. 2000;97:495–9. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T. A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 2007;26:4029–37. doi: 10.1038/sj.emboj.7601832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Takao T, Kondo T. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA. 2004;101:13927–32. doi: 10.1073/pnas.0403906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–8. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura H, Yamaguchi S, Yagita K. Molecular machinery of the circadian clock in mammals. Cell Tissue Res. 2002;309:47–56. doi: 10.1007/s00441-002-0572-5. [DOI] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc Natl Acad Sci USA. 1998;95:8660–4. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Mori T, Xu Y, Pattanayek S, Johnson CH, Egli M. Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLoS One. 2009;4:e7529. doi: 10.1371/journal.pone.0007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M. Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell. 2004;15:375–88. doi: 10.1016/j.molcel.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Williams DR, Pattanayek S, Mori T, Johnson CH, Stewart PL, Egli M. Structural model of the circadian clock KaiB-KaiC complex and mechanism for modulation of KaiC phosphorylation. EMBO J. 2008;27:1767–78. doi: 10.1038/emboj.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, Stewart PL, Egli M. Analysis of KaiA-KaiC protein interactions in the cyanobacterial circadian clock using hybrid structural methods. EMBO J. 2006;25:2017–28. doi: 10.1038/sj.emboj.7601086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–12. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz O, Katayama M, Williams SB, Kondo T, Golden SS. CikA, a bacteriophytochrome that resets the cyanobacterial circadian clock. Science. 2000;289:765–8. doi: 10.1126/science.289.5480.765. [DOI] [PubMed] [Google Scholar]
- Schneegurt MA, Sherman DM, Nayar S, Sherman LA. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp strain ATCC 51142. J Bacteriol. 1994;176:1586–97. doi: 10.1128/jb.176.6.1586-1597.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc Natl Acad Sci USA. 2006;103:8564–9. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Williams SB. Chromosome compaction: Output and phase. In: Ditty JL, Mackey SR, Johnson CH, editors. Bacterial Circadian Programs. Springer-Verlag; Berlin: 2009. pp. 169–182. [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Sweeney BM, Borgese MB. A circadian rhythm in cell division in a prokaryote, the cyanobacterium Syncechococcus WH7803. J Phycol. 1989;25:183–186. [Google Scholar]
- Takai N, Ikeuchi S, Manabe K, Kutsuna S. Expression of the circadian clock-related gene pex in cyanobacteria increases in darkness and is required to delay the clock. J Biol Rhythms. 2006;21:235–44. doi: 10.1177/0748730406289400. [DOI] [PubMed] [Google Scholar]
- Takigawa-Imamura H, Mochizuki A. Predicting regulation of the phosphorylation cycle of KaiC clock protein using mathematical analysis. J Biol Rhythms. 2006;21:405–16. doi: 10.1177/0748730406291329. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Masuda S, Takahashi H. Multiple rpoD-related genes of cyanobacteria. Biosci Biotechnol Biochem. 1992;56:1113–7. doi: 10.1271/bbb.56.1113. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Katayama M, Ito R, Takai N, Kondo T, Oyama T. labA: a novel gene required for negative feedback regulation of the cyanobacterial circadian clock protein KaiC. Genes Dev. 2007;21:60–70. doi: 10.1101/gad.1488107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc Natl Acad Sci USA. 2010;107:3263–8. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Yamaguchi A, Hijikata A, Iwasaki H, Kamagata K, Ishiura M, Go M, Kondo T. Two KaiA-binding domains of cyanobacterial circadian clock protein KaiC. FEBS Lett. 2001;496:86–90. doi: 10.1016/s0014-5793(01)02408-5. [DOI] [PubMed] [Google Scholar]
- Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T. ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA. 2007;104:16377–81. doi: 10.1073/pnas.0706292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–4. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- Tsinoremas NF, Ishiura M, Kondo T, Andersson CR, Tanaka K, Takahashi H, Johnson CH, Golden SS. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;15:2488–95. [PMC free article] [PubMed] [Google Scholar]
- Uzumaki T, Fujita M, Nakatsu T, Hayashi F, Shibata H, Itoh N, Kato H, Ishiura M. Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein. Nat Struct Mol Biol. 2004;11:623–31. doi: 10.1038/nsmb781. [DOI] [PubMed] [Google Scholar]
- Vakonakis I, Klewer DA, Williams SB, Golden SS, LiWang AC. Structure of the N-terminal domain of the circadian clock-associated histidine kinase SasA. J Mol Biol. 2004;342:9–17. doi: 10.1016/j.jmb.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Van Gelder RN, Herzog ED, Schwartz WJ, Taghert PH. Circadian rhythms: in the loop at last. Science. 2003;300:1534–5. doi: 10.1126/science.1085446. [DOI] [PubMed] [Google Scholar]
- van Zon JS, Lubensky DK, Altena PR, ten Wolde PR. An allosteric model of circadian KaiC phosphorylation. Proc Natl Acad Sci USA. 2007;104:7420–5. doi: 10.1073/pnas.0608665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 2009;106:22564–8. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Recent cyanobacterial Kai protein structures suggest a rotary clock. Structure. 2005;13:735–41. doi: 10.1016/j.str.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus: a potential clock input mechanism. Proc Natl Acad Sci USA. 2002;99:15357–62. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–6. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc Natl Acad Sci USA. 2007;104:18819–24. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TL, Bridwell-Rabb J, Kim YI, Gao T, Chang YG, LiWang A, Barondeau DP, Golden SS. The KaiA protein of the cyanobacterial circadian oscillator is modulated by a redox-active cofactor. Proc Natl Acad Sci USA. 2010;107:5804–9. doi: 10.1073/pnas.0910141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J. 2000;19:3349–57. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH. Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 2003;22:2117–26. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH. Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA. 2004;101:13933–8. doi: 10.1073/pnas.0404768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Qin X, Yan H, Egli M, Johnson CH. Intramolecular regulation of phosphorylation status of the circadian clock protein KaiC. PLoS One. 2009;4:e7509. doi: 10.1371/journal.pone.0007509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Pando BF, Dong G, Golden SS, van Oudenaarden A. Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science. 2010;327:1522–6. doi: 10.1126/science.1181759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Eguchi K, Terada TP, Sasai M. Monomer-shuffling and allosteric transition in KaiC circadian oscillation. PLoS One. 2007;2:e408. doi: 10.1371/journal.pone.0000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dong G, Golden SS. The pseudo-receiver domain of CikA regulates the cyanobacterial circadian input pathway. Mol Microbiol. 2006;60:658–68. doi: 10.1111/j.1365-2958.2006.05138.x. [DOI] [PubMed] [Google Scholar]