Abstract
A member of a Plasmodium receptor family for erythrocyte invasion was identified on chromosome 13 from the Plasmodium falciparum genome sequence of the Sanger Centre (Cambridge, U.K.). The protein (named BAEBL) has homology to EBA-175, a P. falciparum receptor that binds specifically to sialic acid and the peptide backbone of glycophorin A on erythrocytes. Both EBA-175 and BAEBL localize to the micronemes, organelles at the invasive ends of the parasites that contain other members of the family. Like EBA-175, the erythrocyte receptor for BAEBL is destroyed by neuraminidase and trypsin, indicating that the erythrocyte receptor is a sialoglycoprotein. Its specificity, however, differs from that of EBA-175 in that BAEBL can bind to erythrocytes that lack glycophorin A, the receptor for EBA-175. It has reduced binding to erythrocytes with the Gerbich mutation found in another erythrocyte, sialoglycoprotein (glycophorin C/D). The interest in BAEBL's reduced binding to Gerbich erythrocytes derives from the high frequency of the Gerbich phenotype in some regions of Papua New Guinea where P. falciparum is hyperendemic.
The erythrocytic stage of Plasmodium falciparum causes several million deaths yearly, primarily in Africa. The parasite lives within the erythrocyte except during the brief period when merozoites, parasites in the invasive stage, are released from infected erythrocytes to invade uninfected erythrocytes. Invasion of erythrocytes by merozoites is a multistep process that includes attachment, reorientation of the merozoite in such a way that its apical end is in contact with the erythrocyte surface, junction formation, and entry into the parasitophorous vacuole (1, 2). The binding of merozoites to erythrocytes requires parasite receptors (3–7).
One family of these parasite receptors is named Duffy-binding-like erythrocyte-binding protein (DBL-EBP) for its similarity to the Plasmodium vivax and Plasmodium knowlesi proteins that bind to the Duffy blood-group antigens (Duffy positive) on human erythrocytes (8). P. vivax does not infect Africans lacking the Duffy blood-group antigens (Duffy negative), and P. knowlesi will not form a junction with or invade Duffy-negative human erythrocytes (9, 10). region 2, a domain of the P. vivax DBL-EBP, has the same specificity as the full-length protein (11).
P. knowlesi has three highly homologous DBL proteins, each with different specificities as defined by region 2 (4, 12). One binds to Duffy blood-group antigens on human and rhesus erythrocytes; a second binds to sialic acid on rhesus erythrocytes; and a third binds to an unidentified receptor on rhesus erythrocytes. Whereas P. knowlesi can only invade Duffy-positive human erythrocytes, it can invade rhesus erythrocytes that have been rendered Duffy negative by protease treatment and by removal of sialic acid with neuraminidase (4, 13). P. knowlesi invades these enzymatically treated erythrocytes at the same rate as the untreated erythrocytes, indicating a highly efficient alternative pathway of invasion.
The Duffy-binding proteins of P. vivax and P. knowlesi are part of a larger family of Plasmodium proteins that includes EBA-175 of P. falciparum. EBA-175 binds to sialic acid and the peptide backbone of glycophorin A on the erythrocyte surface (14). As in the case of P. vivax, the binding domain of EBA-175 is defined by region 2. Unlike P. vivax, which cannot infect Duffy-negative erythrocytes, some strains of P. falciparum parasites have alternative pathways of invasion; they do not require glycophorin A for either invasion or growth in vitro. Thus, other receptors must be involved in these alternative pathways (15).
The P. falciparum genome sequence identifies at least four paralogues of EBA-175. We have studied one of these DBL genes of P. falciparum, named BAEBL (16), to explore its possible role in invasion and its erythrocyte receptor specificity.
Materials and Methods
Structure of BAEBL.
The sequence of BAEBL was identified (16) from cDNA (GenBank Accession No. N97830) deposited by D. Chakrabarti and from the database supplied by the Sanger Centre (Cambridge, U.K.) for chromosome 13 (>MAL13_001500, December 27, 2000).¶ Based on this sequence, we sequenced the P. falciparum clone, Dd2/Nm (15) from genomic DNA (GenBank Accession No. AF332918). The exon/intron boundaries were defined by reverse transcription–PCR of the P. falciparum clone Dd2/Nm (GenBank Accession No. AF332919). Primers used for Dd2/Nm sequencing were f1, 5′-AGACCAATAAATTATATATAATGAAAGGA-3′ and 5′-TTTAAACTTTTCCATTGTTTCTAAACG-3′; f2, 5′-ATAAATTTAATTCACTTTCCGAAAATGA-3′ and 5′-AAAACAATCTCTTCTTTTCCATCAAG-3′; f3, 5′-TTTATAGGTGATGATATGGATTTTGG-3′ and 5′-TCGTAAATGTTCCAGTACAATTCCT-3′; f4, 5′-CAAATGGAGGTTTTGACGAACTTG-3′ and 5′-TAAGTACTGCTGACATTACTTTCCA-3′; f5, 5′-GGAGCTTCAATATATGAGGCGCA-3′ and 5′-ATATCTTCATATTCATTTGGACTCTC-3′; f6, 5′-TGAGTCATTTAAGGTAGAATGTAAGA-3′ and 5′-GGAACTTTCCGAATGTCCATTCGT-3′; f7, 5′-TAAATGAACAACAAAGTGGGAAGGA-3′ and 5′-ATTCTCAATTTGCGTTATATATTGATG-3′; f8, 5′-AGTTCCTTCAGAGGATAATACCCA-3′ and 5′-CTTGATTGACCCTCGCTTTTAA AAC-3′; f9, 5′-ACTAAAAGAGTAAGGGAGGAAATAAT-3′ and 5′-TATAAAATACATTGAATTATTTAAACTATTG-3′. PCR from total RNA untreated with reverse transcriptase never produced PCR-amplified products (data not shown). Oligonucleotides 5′-ATTCCTTATTTTG CTGCTGGAGGT-3′ and 5′-AAGTTGCTTCTATATTAGATTCTCCT-3′ were also used to sequence fragment f9. Only the 3′ region was sequenced for cDNA to determine the precise location of the intron/exon boundaries.
Antisera.
Antisera to BAEBL region 2 and region 6 of Dd2/Nm were generated by immunization of rats with a DNA vaccine with the vector VR1050 (kindly supplied by S. Hoffman, Naval Medical Research Center, Silver Spring, MD) that contains the T cell epitopes P2P30 from tetanus toxoid. Region 2 and region 6 gene fragments of BAEBL were amplified from P. falciparum clone Dd2/Nm and cloned into the VR1050 vector, described as VR1012tPAp2p30 by Becker et al. (17) but now renamed VR1050 (W. O. Rogers, personal communication). The inserts for regions 2 and 6 of Dd2/Nm spanned from amino acid Q141 to I756 and K1046 to S1132, respectively (GenBank Accession No. AF332918). Rats were immunized intradermally with 500 μg of DNA at 3-week intervals for a total of four immunizations. Sera were obtained from the rats a week after the fourth immunization.
Rabbit anti-region 2 of EBA-175 (KLS14) was a kind gift of D. Narum and K. L. Sim (EntreMed, Rockville, MD). Mouse anti-RAP-1 monoclonal antibody 7H8/50 [MRA-79, Malaria Research and Reference Reagent Resource (MR4) Center, Manassas, VA] was a kind gift of A. Saul (Queensland Institute of Medical Research, Brisbane, Australia).
Erythrocytes Used in the Studies.
Blood was collected in 10% (vol/vol) citrate-phosphate-dextrose and stored for up to 4 weeks at 4°C. At the time of study, the erythrocytes were washed three times in incomplete medium (RPMI medium 1640; Life Technologies, Rockville, MD) with 25 mM Hepes and 0.36 mM hypoxanthine (Sigma). For neuraminidase treatment, 5.5 ml of a 5% (vol/vol) suspension of the washed human erythrocytes in incomplete medium was incubated twice with 3 milliunits of neuraminidase (Vibrio cholerae; Calbiochem) for 2 h at 37°C each time. For trypsin treatment, washed human erythrocytes were incubated with 1 mg/ml tosyl-phenylalanine-chloromethyl-ketone-treated trypsin (Sigma) for 2 h at 37°C. After trypsin treatment, the cells were washed once in incomplete medium and incubated with 2 mg/ml soybean trypsin inhibitor (Sigma) for 10 min at room temperature. The cells were washed twice before use in a study.
The glycophorin A and glycophorin B null erythrocytes [En(a−) and S−s−U−, respectively] and the glycophorin D null/glycophorin C modified erythrocytes (Gerbich cells) were frozen within a few days of receipt and thawed by the Red Cross method (18). Blood from a Gerbich donor was collected in 10% (vol/vol) anticoagulant citrate-phosphate-dextrose.
Other glycophorin C/D mutant cells (Leach, Gerbich, and Yus cells) had been stored in liquid nitrogen as frozen pellets (19) and thawed directly into PBS at 37°C.
Metabolic Labeling of Parasite Proteins.
Soluble, metabolically labeled parasite proteins were obtained from culture supernatant of schizont-infected erythrocytes that released merozoites in the absence of uninfected erythrocytes. The parasites were left to lyse and release proteins into the culture supernatant. The Dd2/Nm clone of P. falciparum was cultured as described (20) with the following exceptions. Schizont-infected erythrocytes (5 × 107 per ml of culture medium) were used during the metabolic labeling. The culture supernatant was ultracentrifuged in a Beckman Optima TLX Ultracentrifuge (Beckman Coulter) at 40,000 rpm (98,600 × g) for 10 min at 4°C before storage at −70°C.
Immunoprecipitation.
Proteins in the supernatant and in the diluted eluate were immunoprecipitated as described (20) with the following exceptions. The supernatant (50 μl) was diluted into 250 μl of NETT (50 mM Tris, pH 7.4/150 mM NaCl/1 mM EDTA/0.5% Triton X-100) supplemented with 0.5% BSA (ICN). To determine whether the proteins immunoprecipitated by anti-BAEBL region 2 and anti-BAEBL region 6 are identical, we preabsorbed with one antiserum and immunoprecipitated with the other. Radiolabeled supernatant (50 μl) was preabsorbed with protein A-Sepharose as described elsewhere (20). The supernatant was incubated with 10 μl of anti-BAEBL region 2 or 10 μl of anti-BAEBL region 6 for 2 h at 4°C. Protein G-Sepharose (40 μl; 50%, vol/vol) was added to remove the immune complexes. Supernatant was split into two equal volumes and immunoprecipitated with 5 μl of anti-BAEBL region 2 and 5 μl of anti-BAEBL region 6, as described above.
Modified Erythrocyte-Binding Assay.
Erythrocyte-binding assays were developed for metabolically labeled proteins (as described above) that bind erythrocytes. The original assay required that parasite proteins be bound and eluted from some erythrocytes and not from others. In the original study (3), the major protein eluted from the erythrocytes was EBA-175. Lower molecular mass proteins could be proteolytic fragments of EBA-175 or other proteins. Furthermore, this assay is insensitive for less abundant proteins. Therefore, we have developed an assay that depends on the identification of BAEBL with two antisera against different regions of BAEBL and its removal from the culture supernatant by human erythrocytes. The parasite protein can also be identified and quantified by elution of bound protein from erythrocytes followed by immunoprecipitation. It is then possible to study its specificity for erythrocyte receptors with erythrocytes lacking various proteins or with enzymatically modified erythrocytes. First, we determined the quantity of erythrocytes that would remove the majority of BAEBL and used this quantity with erythrocytes of various types (enzyme-modified erythrocytes and erythrocytes that were genetically deficient in membrane proteins) to determine the erythrocyte specificity of BAEBL. We found that one or two absorptions with a volume of packed erythrocytes equal to the volume of metabolically labeled supernatant were required to remove BAEBL from the supernatant, depending on the concentration of BAEBL in the supernatant.
Elution from erythrocytes of bound parasite proteins was performed as described (20). Parasite proteins were eluted only from the erythrocytes of the first adsorption. The parasite proteins were eluted as described above. Because of the adverse effect of high salt on immunoprecipitation, the eluate was diluted 5-fold (vol/vol) in NETT with 0.5% BSA before immunoprecipitation.
Competitive Inhibition Assay.
An inhibition assay was conducted in the presence of Neu5Ac(α2–3) lactosialic acid or Neu5Ac(α2–6) lactosialic acid (Sigma). Metabolically labeled parasite supernatant (50 μl) was preincubated with 1, 10, 100, or 1,000 μM in 15 μl of the aforementioned carbohydrates for 1 h at room temperature. Packed erythrocytes (50 μl) were added to the mixture. The erythrocyte-binding assay was conducted as described above.
Immunolocalization of BAEBL.
The methods for immunolocalization of BAEBL by confocal microscopy were performed as described (20) with the following modifications. The blocking buffer consisted of PBS (pH 7.4), containing 0.1% Triton X-100 (Bio-Rad), and 2.5 mg/ml normal goat serum (Jackson ImmunoResearch). The secondary antisera consisted of Alexa 488-conjugated goat anti-rat IgG and Alexa 594-conjugated goat anti-rabbit IgG (Molecular Probes) diluted 1:500 in blocking buffer. For antiquenching, we mounted labeled parasites in Prolong Antifade (Molecular Probes).
Invasion Assay.
Prewashed A+ human erythrocytes treated with neuraminidase and A+ human erythrocytes of the Gerbich type (−2, −3, −4) were tested for invasion by P. falciparum clones Dd2 and Dd2/Nm (15) as described (21). Rhesus (Macaca mulatta) erythrocytes that are resistant to invasion by P. falciparum were used as a control for normal erythrocytes introduced with the parasitized erythrocytes.
Results
Structure of the baebl Gene.
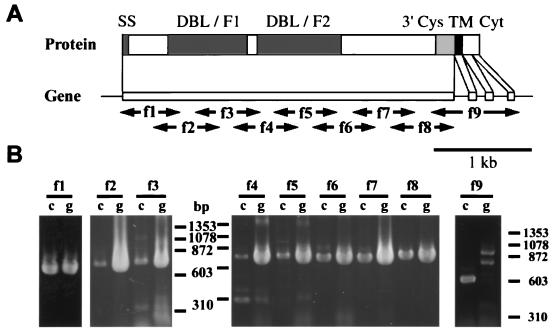
BAEBL is predicted to have two cysteine-rich domains (regions 2 and 6), a transmembrane region, and a cytoplasmic region. This structure is characteristic of all DBL-EBP genes (8). The sequence was obtained from the Sanger Centre chromosome 13 genomic sequence of P. falciparum clone 3D7. The sequence of the gene was also determined from genomic and cDNA sequences of Dd2/Nm. The exon/intron structure for Dd2/Nm was identical to that of EBA-175 in that it had four exons, one for the extracellular domain, one for the transmembrane domain, and two encoding the cytoplasmic region (Fig. 1). The extracellular exon 1 of Dd2/Nm was identical to the genomic sequence of 3D7 from the Sanger Centre except for three changes in region 2 (I185V, N239S, and K261T; Dd2/Nm amino acid number from GenBank is AF332918/3D7 Sanger Centre chromosome 13). By using microsatellites and a genetic cross between Dd2 and HB3 (22), baebl was localized to the end of chromosome 13 close to marker C13 M51 (data not shown).
Figure 1.
Sequencing strategy and exon/intron structure of baebl. Oligonucleotides were designed based on the genomic sequence obtained from the P. falciparum genome project (Sanger Centre) and used for the sequencing of genomic DNA (GenBank Accession No. AF332918) and reverse transcription–PCR of mRNA (GenBank Accession No. AF332919) to determine the intron/exon structure in P. falciparum Dd2/Nm strain. (A) Schematic representation of the gene and predicted protein structure of baebl. Predicted protein structure has strong similarity with EBA-175, containing the putative signal sequence (SS, amino acids 1–21) predicted by SIGNALP V2.0; region 2 (two DBL domains, F1 and F2); region 6 (3′Cys), the transmembrane domain (TM, amino acids 1,134–1,153) predicted by TMHMM V2.0, followed by the putative cytoplasmic domain (Cyt). (B) f1 to f9 primers (see Materials and Methods) are used for reverse transcription–PCR of mRNA (lanes marked c) and PCR of genomic DNA (lanes marked g). (Bar = 1 kb.)
Localization and Expression of BAEBL.
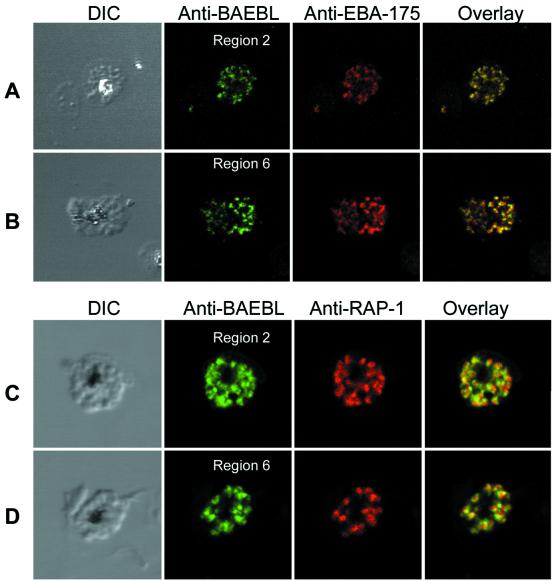
Antibodies to the two cysteine-rich domains (regions 2 and 6) for BAEBL of Dd2/Nm were used to determine localization and expression of the protein. Antibodies to regions 2 and 6 localize to the same organelle as EBA-175 (Fig. 2 A and B), which has been shown to localize to the micronemes (23). Furthermore, immunolocalization of RAP-1, a protein found in rhoptries, other apical organelles of merozoites, shows that BAEBL is adjacent to but not overlapping RAP-1 (Fig. 2 C and D). This distribution is consistent with the localization of BAEBL within micronemes, a distribution identical to that of EBA-175 and the Duffy-binding proteins of P. knowlesi (5, 23). The fact that antisera against two different regions of BAEBL showed identical localization within the parasite indicates that the antisera are not crossreacting with another protein.
Figure 2.
Confocal microscopy demonstrates the localization of BAEBL in micronemes. (A) Dd2/Nm schizonts were double labeled with anti-BAEBL region 2 and anti-EBA-175. Schizonts immunolabeled with anti-BAEBL region 2 were stained with Alexa 488 secondary antibody (green). Schizonts labeled with anti-EBA-175 were stained with Alexa 594 secondary antibody (red). (B) Dd2/Nm schizonts were double labeled with anti-BAEBL region 6 and anti-EBA-175. Schizonts immunolabeled with anti-BAEBL region 6 were stained with Alexa 488 secondary antibody (green). Schizonts labeled with anti-EBA-175 were stained with Alexa 594 secondary antibody (red). (C) Dd2/Nm schizonts were double labeled with anti-BAEBL region 2 and anti-RAP-1 monoclonal antibody. Schizonts immunolabeled anti-BAEBL region 2 were stained with Alexa 488 secondary antibody (green). Schizonts labeled with anti-RAP-1 were stained with tetramethyl rhodamine isothiocyanate (TRITC) secondary antibody (red). (D) Dd2/Nm schizonts were double labeled with anti-BAEBL region 6 and anti-RAP-1 monoclonal antibody. Schizonts immunolabeled with anti-BAEBL region 6 were stained with Alexa 488 secondary antibody (green). Schizonts labeled with anti-RAP-1 were stained with TRITC secondary antibody (red).
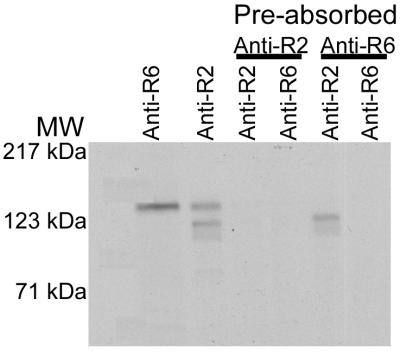
To study the molecular characteristics of BAEBL, we used methods developed for the production of soluble, metabolically labeled erythrocyte-binding proteins (see Materials and Methods). Antisera to regions 2 and 6 immunoprecipitated a protein of ≈148 kDa; antibodies to region 2 also immunoprecipitated two lower-molecular-mass proteins (129 kDa and 117 kDa). The proof that the two 135-kDa proteins were identical derived from studies of immunoabsorption with one serum followed by immunoprecipitation with the second serum (Fig. 3). The same 135-kDa protein was removed by both sera, indicating that the antisera to the two regions of BAEBL were not crossreacting with another protein. The two lower-molecular-mass proteins identified by anti-region 2, but not by anti-region 6, probably resulted from immunoprecipitation of proteolytic products of BAEBL that contained region 2 but not region 6.
Figure 3.
Evidence that anti-region 2 (Anti-R2) and anti-region 6 (Anti-R6) sera immunoprecipitate the same protein. The supernatant was preabsorbed with either anti-region 2 or anti-region 6 followed by immunoprecipitation by the two sera. BAEBL was removed by both sera. MW, molecular mass.
Erythrocyte Binding Specificity.
We have developed a new assay for measuring the binding of BAEBL to erythrocytes. Previously, EBA-175 was identified in parasite proteins bound and eluted from human erythrocytes. Its identification was based on the fact that it was the most abundant and the highest-molecular-mass protein eluted from these erythrocytes. Lower-molecular-mass proteins may be proteolytic fragments of EBA-175 or products of different genes. To identify BAEBL positively, we immunoprecipitated BAEBL from proteins eluted from erythrocytes with anti-region 2 and anti-region 6.
We also modified the protocol; we identified proteins removed from the supernatant by erythrocyte absorption. We determined the different quantities of erythrocytes required to remove BAEBL from the parasite supernatant. It was found that 25 μl of packed erythrocytes slightly reduced the quantity of immunoprecipitated BAEBL from 50 μl of parasite supernatant. The protein was largely removed by absorbing twice with 50 μl of packed erythrocytes for some culture supernatants and by only one absorption with 50 μl for other supernatants (data not shown). Therefore, with some samples, we absorbed with 25 μl once, 50 μl once, and 50 μl twice for comparison between normal erythrocytes and mutant erythrocytes or enzyme-treated erythrocytes. The protein also was eluted from the first 50 μl of packed erythrocytes used for absorption, which set the conditions for absorbing and eluting BAEBL and demonstrated that BAEBL may be a parasite receptor for binding erythrocytes.
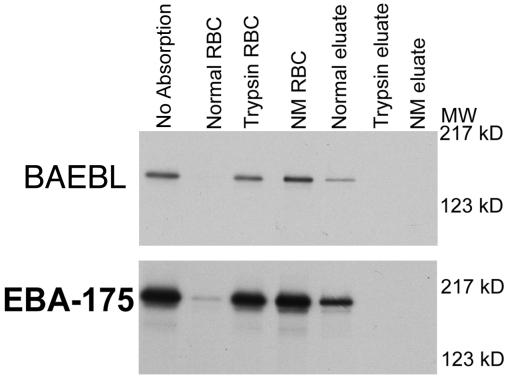
To determine the specificity of binding, we studied binding to neuraminidase- and trypsin-treated human erythrocytes and human erythrocytes with various genetically modified blood groups. Both enzymes eliminated the binding of BAEBL to human erythrocytes (Fig. 4), indicating that the erythrocyte receptor required sialic acid attached to a peptide backbone and must therefore be a sialoglycoprotein. This failure of neuraminidase- and trypsin-treated erythrocytes to bind was identical to that of EBA-175 (Fig. 4). To determine whether BAEBL was binding to the carbohydrates on the erythrocyte receptor, we performed competitive inhibition with Neu5Ac(α2–3) lactosialic and Neu5Ac(α2–6) lactosialic acid at 1, 10, 100, and 1,000 μM. We determined that neither Neu5Ac(α2–3) nor Neu5Ac(α2–6) lactosialic acid inhibited the binding of BAEBL to human erythrocytes (data not shown). These results indicate that BAEBL is binding either to a more complex polysaccharide or to a combination of sialic acid and a peptide backbone of an erythrocyte sialoglycoprotein.
Figure 4.
BAEBL and EBA-175 did not bind to neuraminidase (NM eluate) or trypsin-treated erythrocytes (Trypsin RBC). Eluates of BAEBL and EBA-175 were seen only from normal erythrocytes.
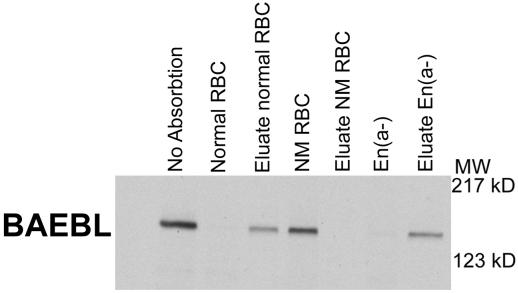
To define further the binding specificity of BAEBL, we studied En(a−) erythrocytes, which lack glycophorin A. We found that EBA-175 failed to bind En(a−) erythrocytes as described in ref. 14 (data not shown). BAEBL, however, bound to these erythrocytes in a manner similar to normal erythrocytes (Fig. 5), thus demonstrating that the binding specificity of BAEBL differed from that of EBA-175. S−s−U− erythrocytes that lacked glycophorin B bound both EBA-175 and BAEBL (data not shown). Thus, neither glycophorin A nor glycophorin B is the sole receptor for BAEBL.
Figure 5.
BAEBL binds and is eluted from En(a−) erythrocytes that lack glycophorin A. NM RBC are neuraminidase-treated normal erythrocytes. MW, molecular mass; kD, kilodalton.
Abnormal Binding to Glycophorin C/D Mutant Erythrocytes.
Another characterized sialoglycoprotein on the surface of human erythrocytes is glycophorin C/D (for review, see refs. 24 and 25). Its peptide backbone is completely unrelated to glycophorins A and B; like these, however, it has a mucin-like region of serines and threonines for O-linked sugars at the N terminus of the protein. Both glycophorin C and glycophorin D are encoded by the same gene with use of alternative start codons. Glycophorin C, the full-length protein, contains one N-linked glycan. There are three mutations of the glycophorin C/D gene that lack high-incidence antigens (25). Leach erythrocytes are null for these proteins; Gerbich and Yus erythrocytes contain exon 3 and 2 deletions, respectively, that lead to a shortened glycophorin C and missing glycophorin D. Both Gerbich and Yus cells have abnormal N-linked glycosylation of the truncated form of glycophorin C (24).
We screened for the binding of BAEBL to erythrocytes of the Gerbich (−2, −3, −4) and Yus (−2, −3, −4) phenotype that had been frozen as pellets in liquid nitrogen. BAEBL had reduced binding to Gerbich and Yus erythrocytes (data not shown). These differences were consistent for pellet-frozen erythrocytes from different donors. EBA-175 bound normally to these erythrocytes (data not shown).
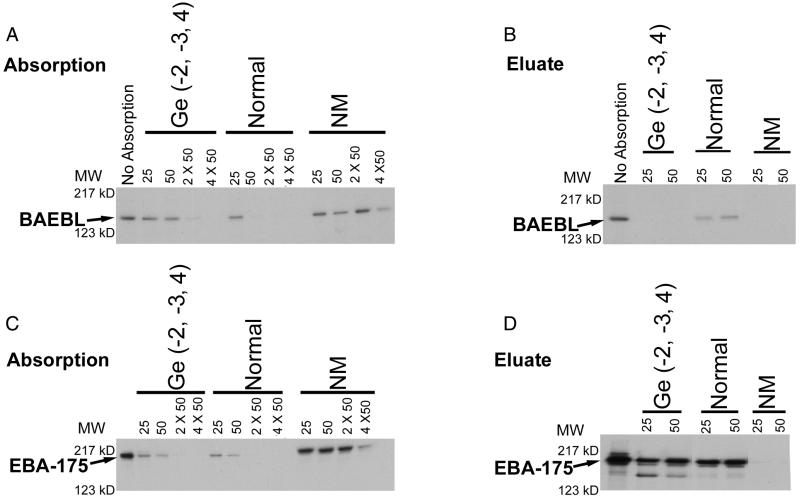
Because the quality of the pellet-frozen erythrocytes was unpredictable, we obtained fresh blood from a person with the Gerbich mutation. In two separate experiments, we found that it required twice as many Gerbich cells to remove BAEBL from the culture supernatant compared with that of normal erythrocytes (Fig. 6A). In contrast to BAEBL, EBA-175 bound equally well to Gerbich and normal erythrocytes (Fig. 6C). This difference between normal and Gerbich erythrocytes for absorption of BAEBL was similar to the results obtained with pellet-frozen erythrocytes as described above.
Figure 6.
Absorption and elution of BAEBL (A and B) and EBA-175 (C and D) with various amounts (25, 50, 2 × 50, and 4 × 50 μl of packed erythrocytes) of Gerbich [Ge(−2, −3, 4)], normal, and neuraminidase (NM)-treated erythrocytes. For elution, 25 and 50 μl of packed erythrocytes were used. MW, molecular mass; kD, kilodalton.
BAEBL was eluted from normal but not Gerbich erythrocytes, indicative of its poor binding to Gerbich erythrocytes (Fig. 6B). BAEBL also did not elute from neuraminidase-treated normal erythrocytes. These results are indicative of the poor binding of BAEBL to Gerbich erythrocytes. In contrast to BAEBL, EBA-175 was eluted from both Gerbich and normal erythrocytes (Fig. 6D).
Invasion of Gerbich Erythrocytes.
P. falciparum clones Dd2 and Dd2/Nm invaded Gerbich erythrocytes at the same rate as normal erythrocytes (Table 1). Dd2, but not Dd2/Nm, had markedly reduced invasion into neuraminidase-treated erythrocytes as described (15).
Table 1.
Invasion rate of P. falciparum into Gerbich erythrocytes
| Red-cell type |
P. falciparum clones
|
|
|---|---|---|
| Dd2, % | Dd2/Nm, % | |
| Normal | 3.7* | 1.6 |
| Gerbich | 3.0 | 1.8 |
| Neuraminidase-treated normal | 0 | 1.8 |
| Rhesus | 0 | 0 |
Percentage of ring-infected erythrocytes.
Discussion
A gene (baebl) of the DBL-EBP family of Plasmodium-receptor proteins with erythrocyte specificity different from that of EBA-175 has been studied in P. falciparum.
The exon/intron structure of BAEBL is similar to that of other DBL-EBP (8). The difference between two DBL-EBP from P. falciparum and P. vivax or P. knowlesi is the duplication of region 2 (8). Despite the similarity of BAEBL and EBA-175 in their requirement for sialic acid on erythrocyte proteins, the specificity of BAEBL for receptors on the erythrocyte surface differs from that of EBA-175. The differences are 2-fold. First, BAEBL, but not EBA-175, binds to En(a−) erythrocytes that lack glycophorin A. Second, Gerbich erythrocytes (that have an altered glycophorin C and are missing glycophorin D) bind BAEBL much more weakly than normal erythrocytes but bind EBA-175 normally. Thus, the specificity of these two parasite receptor molecules differs, suggesting alternative pathways for invasion. These data indicate at least two different sialic acid-dependent pathways for invasion.
In preliminary experiments, BAEBL was not detected in the parasite supernatant absorbed with glycophorin C/D null erythrocytes of the Leach phenotype (D.C.G.M. and L.H.M., unpublished data). BAEBL, however, was never detected in the eluate of these erythrocytes. In addition, 2 and 8 μl of Leach erythrocytes, but not normal erythrocytes, removed BAEBL from parasite supernatant. Because of the unknown quality of these pellet-frozen erythrocytes, we tried to obtain fresh Leach erythrocytes but failed. More definitive studies testing the binding of BAEBL to Leach erythrocytes must await the availability of fresh Leach erythrocytes, although the failure to elute BAEBL and its reduction with small numbers of Leach erythrocytes suggest proteolysis.
The Gerbich phenotype is found at high allele frequencies (50%) in some regions of Papua New Guinea (26). The mutation consists of a deletion of exon 3 in glycophorin C that leads to truncated glycophorin C and missing glycophorin D (27). Of interest to our study is the fact that these areas colocalize to hyperendemic areas of malaria. Serjeantson (28) found reduced frequency of heavy infections with P. falciparum and P. vivax with the Gerbich phenotype. The common mutation in erythrocyte band 3, resulting in ovalocytosis, was not described at the time of the study by Serjeantson (28), and this omission may have influenced the results. Pasvol et al. (29) found reduced invasion of Gerbich erythrocytes, but these cells were also ovalocytic (30). It is possible that hereditary ovalocytosis caused by band-3 mutations influenced the invasion and frequency of infection. It is critical to restudy these groups now that the two mutations can be separated by molecular techniques. We were unable to find any reduction in invasion of Gerbich erythrocytes by the two P. falciparum clones Dd2 and Dd2/Nm, but other parasite clones could be affected. It is known that Dd2 switched to the sialic acid-independent pathway (Dd2/Nm) after modification of the eba-175 gene (15), suggesting that Dd2 lacking EBA-175 could not invade by using BAEBL as a pathway. Alternatively, the effect on invasion may be too subtle to be detected by the invasion assay as performed. The tantalizing possibility remains that the high frequency of the Gerbich phenotype is selected for as a result of reduced invasion by P. falciparum mediated by the BAEBL receptor. The Gerbich mutation, like Duffy negativity in West Africa, seems to be going to fixation in these communities. The difference between the Gerbich mutation and Duffy negativity is that the Gerbich negative erythrocytes are still infected by P. falciparum, whereas Duffy-negative erythrocytes are refractory to P. vivax. Still, the parasite receptors for these two blood groups become potential vaccine candidates, because mutation in the host molecules leads to either no infection (P. vivax and Duffy negativity) or potentially reduced infection (P. falciparum and Gerbich mutation).
Acknowledgments
We thank the following for generous donations to the study: Drs. Kim Lee Sim and David Narum (EntreMed) for rabbit anti-EBA-175; Dr. Allan Saul for RAP-1 antibody to MR4; Ms. Marylyn Moulds (GammaImmucor, Houston) for frozen erythrocytes with glycophorin C/D mutations; Ms. Martha Berringer (Virginia Blood Center, Richmond, VA), who obtained a donor of the Gerbich phenotype; and JoLynn Proctor (National Institutes of Health Blood Center) for the S−s−U− erythrocytes. We thank Dr. Xinzhuan Su for the localization of BAEBL by using microsatellites in the cross between Dd2 and HB3. We thank Ms. Andrea Weissberg for scanning the x-ray films. We thank Dr. Owen Schwartz for help with confocal microscopy and Adobe photoshop. We also thank Dr. Peter Zimmerman for molecular typing of the Gerbich donor from the Virginia Blood Center. Sequencing of P. falciparum chromosome 13 was accomplished as part of the Malaria Genome Project with support by the Wellcome Trust.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF332918 and AF332919).
Preliminary sequence data for P. falciparum chromosome 13 were obtained from the Sanger Centre website at http://www.sanger.ac.uk/.
References
- 1.Dvorak J A, Miller L H, Whitehouse W C, Shiroishi T. Science. 1975;187:748–750. doi: 10.1126/science.803712. [DOI] [PubMed] [Google Scholar]
- 2.Aikawa M, Miller L H, Johnson J, Rabbege J. J Cell Biol. 1978;77:72–82. doi: 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camus D, Hadley T H. Science. 1985;230:553–556. doi: 10.1126/science.3901257. [DOI] [PubMed] [Google Scholar]
- 4.Haynes J D, Dalton J P, Klotz F W, McGinnis M H, Hadley T J, Hudson D E. J Exp Med. 1988;167:1873–1881. doi: 10.1084/jem.167.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams J H, Hudson D H, Torii M, Ward G E, Wellems T E, Aikawa M. Cell. 1990;63:142–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- 6.Sim B K L, Orlandi P A, Haynes J D, Klotz F W, Carter J M, Camus D. J Cell Biol. 1990;111:1877–1884. doi: 10.1083/jcb.111.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galinski M R, Medina C C, Ingravallo P, Barnwell J W. Cell. 1992;69:1213–1226. doi: 10.1016/0092-8674(92)90642-p. [DOI] [PubMed] [Google Scholar]
- 8.Adams J H, Sim B K L, Dolan S A, Fang X, Kaslow D C, Miller L H. Proc Natl Acad Sci USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller L H, Mason S J, Clyde D F, McGinniss M H. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 10.Miller L H, Aikawa M, Johnson J G, Shiroishi T. J Exp Med. 1979;149:172–184. doi: 10.1084/jem.149.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chitnis C E, Miller L H. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ranjan A, Chitnis C E. Proc Natl Acad Sci USA. 1999;96:14067–14072. doi: 10.1073/pnas.96.24.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller L H, Dvorak J A, Shiroishi T, Durocher J R. J Exp Med. 1973;138:1597–1601. doi: 10.1084/jem.138.6.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sim B K L, Chitnis C E, Wasniowska K, Hadley T J, Miller L H. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 15.Dolan S A, Miller L H, Wellems T E. J Clin Invest. 1990;86:618–624. doi: 10.1172/JCI114753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams, J. H., Kaneko, O., Peterson, D. S. & Blair, P. L. (2001) Trends Parasitol., in press. [DOI] [PubMed]
- 17.Becker S I, Wang R, Hedstrom R C, Aguiar J C, Jones T R, Hoffman S L, Gardner M J. Infect Immun. 1998;66:3457–3461. doi: 10.1128/iai.66.7.3457-3461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallory D, editor. Immunohematology Methods and Procedures. National Reference Laboratory, Rockville, MD: American Red Cross; 1993. pp. 125-1–125-2. [Google Scholar]
- 19.Judd W J. In: Methods in Immunohematology. Judd W J, editor. Durham, NC: Montgomery Scientific; 1994. pp. 188–190. [Google Scholar]
- 20.Kaneko O, Fidock D A, Schwartz O M, Miller L H. Mol Biochem Parasitol. 2000;110:135–146. doi: 10.1016/s0166-6851(00)00263-2. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko O, Soubes S C, Miller L H. Exp Parasitol. 1999;93:116–119. doi: 10.1006/expr.1999.4441. [DOI] [PubMed] [Google Scholar]
- 22.Su X, Ferdig M T, Huang Y, Huynh C Q, Liu A, You J, Wootton J C, Wellems T E. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 23.Sim B K L, Toyoshima T, Haynes J D, Aikawa M. Mol Biochem Parasitol. 1992;51:157–160. doi: 10.1016/0166-6851(92)90211-2. [DOI] [PubMed] [Google Scholar]
- 24.Reid M E, Spring F A. Transfus Med (Oxford) 1994;4:139–146. doi: 10.1111/j.1365-3148.1994.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 25.Colin Y, Le Van Kim C. In: Blood Cell Biochemistry. Cartron J P, Rouger P, editors. New York: Plenum; 1995. pp. 331–350. [Google Scholar]
- 26.Booth P B, Tills D, Warlow A, Kopec A C, Mourant A E, Teesdale P, Hornabrook R W. Hum Hered. 1982;32:385–403. doi: 10.1159/000153328. [DOI] [PubMed] [Google Scholar]
- 27.Serjeantson S W, White B S, Bhatia K, Trent R J. Immunol Cell Biol. 1994;72:23–27. doi: 10.1038/icb.1994.4. [DOI] [PubMed] [Google Scholar]
- 28.Serjeantson S W. Papua New Guinea Med J. 1989;32:5–9. [PubMed] [Google Scholar]
- 29.Pasvol G, Anstee D, Tanner M J A. Lancet. 1984;i:907–908. doi: 10.1016/s0140-6736(84)91366-7. [DOI] [PubMed] [Google Scholar]
- 30.Aanstee D J, Parsons S F, Ridgwell K, Tanner M J A, Merry A H, Thomson E E, Judson P A, Johnson P, Bates S, Fraser I D. Biochem J. 1984;218:615–619. doi: 10.1042/bj2180615. [DOI] [PMC free article] [PubMed] [Google Scholar]