Abstract
Background
Hepatitis C virus (HCV) has been reported to replicate in peripheral blood mononuclear cells (PBMCs), particularly in patients coinfected with HCV and human immunodeficiency virus (HIV). However, there are limited data regarding the prevalence of and the factors associated with extrahepatic replication.
Methods
The presence of negative-strand HCV RNA in PBMCs was evaluated by a strand-specific assay for 144 anti-HCV–positive/HIV-infected women enrolled in the Women’s Interagency HIV Study. One to 5 PBMC samples obtained from each woman were tested. Multivariate analyses were used to assess for associations with the clinical and demographic characteristics of the women.
Results
Negative-strand HCV RNA was detected in 78 (25%) of 315 specimens, and, for 61 women (42%), ≥1 specimen was found to have positive results. The presence of negative-strand HCV RNA in PBMCs was significantly positively associated with an HCV RNA plasma level of ≥6.75 log copies/mL (P =.04) and consumption of ≥7 alcoholic drinks per week (P =.02). It was also negatively associated with injection drug use occurring in the past 6 months (P =.03). A negative association with a CD4+CD38+DR+ cell percentage of >10% and a positive association with acquired immunodeficiency syndrome were borderline significant (P =.05).
Conclusions
HCV replication in PBMCs is common among HIV-coinfected women and appears to be a dynamic process related to lifestyle, virologic, and immunologic factors.
Hepatitis C virus (HCV) is a positive-strand RNA virus that replicates through a negative-strand intermediary. Although hepatocytes are the primary sites for HCV replication, there is evidence of negative-strand HCV RNA in peripheral blood mononuclear cells (PBMCs), and the HCV genomic sequences present in PBMCs have been found to differ from those found in serum and the liver [1–6]. HCV RNA has also been detected in PBMCs and hematopoietic progenitor cells by means of in situ hybridization [7]. The presence of HCV replication has been documented in lymph nodes from patients with AIDS [8] and, recently, also in lymph nodes from HIV-negative liver transplant recipients [9]. Importantly, the same minor quasi-species variants of HCV strain H77, which were selected in lymphoblastoid cells in vitro, were found to be replicating in vivo in the PBMCs of chimpanzees inoculated with the same parental strain [10].
HCV is common among persons infected with HIV, because both pathogens share similar routes of transmission. In the United States and Europe, 13%–43% of HIV-infected persons are also infected with HCV [11], and this proportion of persons with HIV/HCV coinfection is even higher among injection drug users. HIV coinfection has important implications for HCV infection. First, it is likely to facilitate the spread of HCV. Mothers who are coinfected with HIV and HCV have been reported to transmit HCV to their infants at a much higher rate than mothers infected with HCV only [12–14]. Similarly, horizontal, possibly sexual, transmission of HCV is more common among HIV/HCV-coinfected persons than among HCV-monoinfected persons [15]. Second, HIV accelerates the development of HCV-associated severe liver disease [16–19]. Paradoxically, the reduction in mortality and morbidity among HIV-infected patients after the introduction of highly active antiretroviral therapy (HAART) may have contributed to the emergence of HCV as a significant pathogen in this population [20].
There is emerging evidence that HIV facilitates HCV replication in vivo, not only in the liver but also at extrahepatic sites [8, 21–23]. However, the prevalence of this phenomenon and the factors associated with its occurrence need further investigation. We addressed these questions in a cohort study of anti-HCV–positive/HIV-infected women enrolled in the Women’s Interagency HIV Study (WIHS).
PATIENTS AND METHODS
Patients
The study included 144 anti-HCV–positive/HIV-infected women enrolled in the WIHS. The WIHS is a prospective, multicenter cohort study established in 1993 to conduct comprehensive investigations of the influence of HIV infection on women in the United States. Participants are seen every 6 months and undergo an extensive interview, physical and gynecologic examinations, and multiple laboratory evaluations. A detailed description of the WIHS has been published elsewhere [24]. Written, informed consent was obtained from all study participants, and human experimentation followed the guidelines of the US Department of Health and Human Services and the institutional review boards of the participating institutions.
Specimen processing and storage
For viral load measurement and cell and plasma storage, blood was collected in sodium citrate cell-preparation tubes (Vacutainer; Becton-Dickinson), which were either processed within 6 h or centrifuged at 1500 g and then were processed, as directed by the manufacturer, and subsequently stored at −80°C at a central repository (BBI Biotech Research Laboratories).
HIV RNA and HCV RNA load determination and HCV serologic testing
Plasma HIV RNA levels were measured using the NASBA/NucliSens HIV RNA assay (bioMérieux), in accordance with the recommendations of the manufacturer, in laboratories that participate in and are certified by the Virology Quality Assurance certification program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health [20]. HCV RNA levels were measured using COBAS Amplicor Monitor 2.0 (Roche Diagnostics) with a linear range of 600–700,000 IU/mL. Because we anticipated that coinfected women would have HCV RNA levels that were higher than the upper limit of the assay, all samples were initially diluted 1:10. If the samples were found to be HCV RNA negative, they were retested in undiluted form by use of a qualitative Amplicor HCV assay, which has a lower limit of detection of 50 IU/mL (Roche Diagnostics); if samples were found to be HCV RNA positive, then they were retested in undiluted form by use of the quantitative assay. All specimens that were nonreactive according to both quantitative and qualitative polymerase chain reaction assays were considered to be HCV RNA negative. Results of HCV serologic tests were determined at study entry by use of the contemporary commercial HCV EIAs (Abbott EIA 2.0 and 3.0). In addition, all women with undetectable HCV RNA had their samples retested using HCV 3.0 EIA (Ortho Diagnostic).
Detection of negative-strand HCV RNA and HCV genotyping
PBMCs that were viably frozen were used in this study. RNA was extracted as described elsewhere [8]. The specificity of our real-time RT-PCR assay for the detection of negative-strand HCV RNA was ascertained by conducting cDNA synthesis at a high temperature with the thermostable enzyme Tth (Applied Biosystems). Descriptions of the Tth-based assay and its real-time modification have been published elsewhere [25, 26]. The strand-specific assay is capable of detecting ~100 viral genomic eq/mL of the correct negative strand while non-specifically detecting 107 –108 viral genomic eq/mL of the incorrect positive strand. Although the assay is quantitative over a wide range of template, the results were commonly close to the level of detection and, thus, were reported only as being either positive or negative. Amplification of positive-strand HCV RNA from PBMCs was not attempted as in our previous studies [8, 27], as well as in studies conducted by others [6]; positive-strand HCV RNA was found to be almost universally present among viremic patients and can represent nothing more than viral adsorption [2]. For women who were found to be viremic, the HCV genotype was determined using the NC Trugene HCV 5′ NC genotyping kit (Bayer HealthCare), as recommended by the manufacturer and as described elsewhere [28].
Immunofluorescence staining and flow cytometric analysis
Lymphocyte subsets were quantitated using frozen PBMCs. Flow cytometric analysis was performed using a FACSCalibur Flow Cytometer utilizing CELLQuest software (Becton Dickinson). Activation status was defined as simultaneous expression of CD38 and HLA-DR on CD4+ and CD8+ cells.
Data analysis
To investigate the association of demographic, lifestyle, virologic, and immunologic characteristics with the presence of negative-strand HCV RNA (i.e., extrahepatic replication) in PBMCs, we analyzed data collected by structured interview and by laboratory testing, as described above. The independent variables that were evaluated at baseline included only race and education. The independent variables that were evaluated at each study visit included age, status as a current smoker, alcohol use, drug use, number of sex partners, AIDS diagnosis, HAART, and the laboratory variables described above. HAART was defined as (a) ≥2 nucleoside reverse-transcriptase inhibitors (NRTIs) in combination with ≥1 protease inhibitor (PI) or 1 nonnucleoside reverse-transcriptase inhibitor (NNRTI), (b) 1 NRTI in combination with ≥1 PI and ≥1 NNRTI, (c) a regimen containing ritonavir and saquinavir in combination with 1 NRTI and no NNRTIs, or (d) an abacavir-or tenofovir-containing regimen of ≥3 NRTIs in the absence of both PIs and NRTIs, except for the 3 NRTI regimens consisting of abacavir + tenofovir + lamivudine or didanosine + tenofovir + lamivudine. Statistical procedures included logistic regression and Spearman rank correlation (SAS software; version 9; SAS Institute). For the logistic regression models, the dependent variable was the presence/absence of negative-strand HCV RNA. To account for the fact that data from multiple visits were evaluated for the same women, the regression models used generalized estimating equations with a logit link function (i.e., a logistic regression model) [29]. An exchangeable correlation matrix was specified. Because the unit of analysis was a study visit, not a subject, differential degrees of follow-up among subjects would not affect the analysis of associations. The analytic approach further accounted for correlated outcomes, over study visits, within subjects.
The association of each independent variable with the presence of negative-strand HCV RNA was first analyzed univariately and then was adjusted for the HCV RNA level. The subsequent multivariate model included independent variables with P values of <.10 after adjusting for HCV RNA level. Results are reported as odds ratios and 95% confidence intervals.
RESULTS
Characteristics at baseline
The demographic, lifestyle, and clinical characteristics at enrollment into the WIHS for the 144 anti-HCV–positive/HIV-infected women included in this analysis are presented in table 1. Seventy-four percent were >35 years of age at study entry. The majority of women were black (58%) or Hispanic (22%). More than one-half (56%) had exchanged sex for money, drugs, or shelter, and 66% had been involved with >10 sex partners. Almost one-quarter (23%) drank ≥7 alcoholic drinks per week, and 69% were current cigarette smokers. Ninety-one percent (91%) had a history of drug use, including injection drug use (IDU) (85%). Fifty-seven percent (57%) of the women had initiated antiretroviral therapy for HIV infection, but none were receiving HAART at the time of enrollment. One woman had received a diagnosis of AIDS. None of the women had been treated for their HCV infection.
Table 1.
Characteristics, at baseline, of 144 anti–hepatitis C virus (HCV)–positive/HIV-infected women who were evaluated for the presence of negative-strand HCV RNA in their peripheral blood mononuclear cells.
| Characteristic | No. (%) of women |
|---|---|
| Age, years | |
| ≤35 | 37 (26) |
| >35 | 107 (74) |
|
| |
| Race/ethnicity | |
| White | 26 (18) |
| Black | 83 (58) |
| Hispanic | 31 (22) |
| Other | 4 (3) |
|
| |
| Education completed, no. of years | |
| <12 | 65 (45) |
| ≥12 | 79 (55) |
|
| |
| Received transfusion | |
| Never | 117 (81) |
| Ever | 26 (18) |
| NA | 1 (1) |
|
| |
| Traded sexa | |
| Never | 64 (44) |
| Ever | 80 (56) |
|
| |
| Sex partners in lifetime, no. | |
| 1–4 | 20 (14) |
| 5–10 | 28 (19) |
| >10 | 95 (66) |
| NA | 1 (1) |
|
| |
| Alcohol use, drinks/week | |
| 0 | 61 (42) |
| 1–6 | 45 (31) |
| >7 | 33 (23) |
| NA | 5 (3) |
|
| |
| Current smoker | |
| No | 44 (31) |
| Yes | 100 (69) |
|
| |
| Drug use history | |
| Injection drug(s) | |
| Never | 22 (15) |
| Ever | 122 (85) |
|
| |
| Marijuana/hash | |
| Never | 109 (76) |
| Ever | 34 (24) |
|
| |
| Crack/freebase cocaine | |
| Never | 105 (73) |
| Ever | 39 (27) |
|
| |
| Cocaine | |
| Never | 115 (80) |
| Ever | 29 (20) |
|
| |
| Heroin | |
| Never | 114 (79) |
| Ever | 30 (21) |
|
| |
| Methadone | |
| Never | 140 (97) |
| Ever | 4 (3) |
|
| |
| Amphetamine | |
| Never | 140 (97) |
| Ever | 4 (3) |
|
| |
| Any drug use | |
| Never | 13 (9) |
| Ever | 131 (91) |
|
| |
| HIV therapy received | |
| None | 60 (42) |
| Monotherapy | 45 (31) |
| Combination therapy | 38 (26) |
| HAARTb | 0 |
| NA | 1 (1) |
|
| |
| AIDS diagnosis | |
| No | 143 (99) |
| Yes | 1 (1) |
NOTE. HAART, highly active antiretroviral therapy; NA, not available.
For drugs, money, and/or shelter.
HAART was not available at the time of enrollment in the Women’s Interagency HIV Study.
Negative-strand HCV RNA in PBMCs
PBMC specimens obtained during the 315 WIHS visits made by the 144 women were tested for the presence of negative-strand HCV RNA. In 78 (25%) of the 315 specimens obtained, negative-strand HCV RNA was detected.
The number of visits evaluated per woman ranged from 1 to 5. For the 94 women who had samples tested at ≥2 visits, the interval between the first and last visits at which samples were tested ranged from 0.3 to 6.5 years (median, 1.0 year). Table 2 shows the number of visits at which negative-strand HCV RNA was detected in relation to the total number of visits at which samples were tested per women. Negative-strand HCV RNA was detected in ≥1 specimen obtained from 61 (42%) of the 144 women. Of the 94 women (65%) who had specimens tested at ≥1 visit, 14 (15%) had negative-strand HCV RNA repeatedly detected.
Table 2.
No. of study visits at which negative-strand hepatitis C virus (HCV) RNA was detected in peripheral blood mononuclear cells (PBMCs) in relation to the total no. of study visits at which samples were tested for each woman.
| Total study visitsa | No. of women (n = 144) | Women with negative-strand HCV RNA in PBMCs, no. (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| At ≥ 1 visit | By study visit | |||||||
| 0 | 1 | 2 | 3 | 4 | 5 | |||
| 1 | 50 | 12 (24) | 38 (76) | 12 (24) | … | … | … | … |
| 2 | 33 | 11 (33) | 22 (67) | 11 (33) | 0 (0) | … | … | … |
| 3 | 48 | 29 (60) | 19 (40) | 20 (42) | 8 (17) | 1 (2) | … | … |
| 4 | 10 | 9 (90) | 1 (10) | 4 (40) | 3 (30) | 2 (20) | 0 (0) | … |
| 5 | 3 | 0 (0) | 3 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
No. of visits at which samples were evaluated per woman.
Seven specimens obtained from 7 different women had positive test results despite the women having had an undetectable HCV RNA level in plasma. Six of these women had evaluations performed at 2–4 visits; at all visits, HCV RNA was not detectable in plasma, and at only 1 visit was negative-strand HCV RNA detectable in PBMCs. The seventh woman had evaluations performed at 4 visits. At 3 visits, HCV RNA was detectable in plasma, and, at 2 of these 3 visits, negative-strand HCV RNA was detectable in PBMCs. At a fourth visit, no HCV RNA was detectable in plasma, but negative-strand HCV RNA was detectable in PBMCs.
Univariate analysis of factors associated with negative-strand HCV RNA in PBMCs
The associations of negative-strand HCV RNA in PBMCs with demographic, lifestyle, virologic, clinical, and immunologic characteristics were initially investigated univariately and after adjustment for HCV RNA (table 3). Negative-strand HCV RNA detection was positively associated with alcohol use and negatively associated with a high level of the CD8 activation marker CD8+CD38+DR+.
Table 3.
Characteristics associated with the presence of negative-strand hepatitis C virus (HCV) RNA in peripheral blood mononuclear cells among 315 samples obtained from 144 anti-HCV–positive/HIV-infected women.
| Characteristic | Samples analyzed, no. | Samples positive for negative-strand HCV RNA, no. (%) | OR (P) |
|
|---|---|---|---|---|
| Unadjusted | Adjusteda | |||
| Age, years | ||||
| <35 | 36 | 12 (33) | 1.0 | 1.0 |
| ≥35 | 279 | 66 (24) | 0.6 (.13) | 0.6 (.10) |
|
| ||||
| Race/ethnicity | ||||
| White | 48 | 15 (31) | 1.0 | 1.0 |
| Hispanic | 85 | 23 (27) | 0.8 (.64) | 0.9 (.85) |
| Black | 174 | 39 (22) | 0.6 (.20) | 0.7 (.34) |
|
| ||||
| Education completed, no. of years | ||||
| <12 | 142 | 42 (30) | 1.0 | 1.0 |
| ≥12 | 173 | 36 (21) | 0.6 (.08) | 0.6 (.06) |
|
| ||||
| Current smoker | ||||
| No | 111 | 26 (23) | 1.0 | 1.0 |
| Yes | 202 | 51 (25) | 1.1 (.68) | 0.9 (.73) |
|
| ||||
| Alcohol use, drinks/week | ||||
| 0 | 184 | 41 (22) | 1.0 | 1.0 |
| 1–6 | 87 | 19 (22) | 1.0 (.97) | 1.0 (.82) |
| ≥7 | 42 | 17 (40) | 2.4 (.012) | 2.1 (.05) |
|
| ||||
| History of IDU | ||||
| Never | 44 | 8 (18) | 1.0 | 1.0 |
| Ever | 271 | 70 (26) | 1.6 (.28) | 1.5 (.32) |
|
| ||||
| Drug use in past 6 months | ||||
| IDU | ||||
| No | 292 | 75 (26) | 1.0 | 1.0 |
| Yes | 21 | 2 (10) | 0.3 (.09) | 0.3 (.07) |
|
| ||||
| Marijuana | ||||
| No | 257 | 68 (26) | 1.0 | 1.0 |
| Yes | 56 | 9 (16) | 0.6 (.17) | 0.6 (.20) |
|
| ||||
| Crack/freebase cocaine | ||||
| No | 267 | 67 (25) | 1.0 | 1.0 |
| Yes | 46 | 10 (22) | 0.9 (.75) | 0.8 (.60) |
|
| ||||
| Cocaine | ||||
| No | 297 | 76 (26) | 1.0 | 1.0 |
| Yes | 15 | 1 (7) | 0.2 (.12) | 0.2 (.11) |
|
| ||||
| Heroin | ||||
| No | 290 | 74 (26) | 1.0 | 1.0 |
| Yes | 23 | 3 (13) | 0.5 (.31) | 0.4 (.28) |
|
| ||||
| Sex partners, no. | ||||
| At baseline | ||||
| 1–4 | 144 | 32 (22) | 1.0 | 1.0 |
| 5–10 | 68 | 20 (29) | 1.5 (.25) | 1.6 (.18) |
| >10 | 103 | 26 (25) | 1.2 (.56) | 1.4 (.32) |
|
| ||||
| In past 6 months | ||||
| 0 | 119 | 33 (28) | 1.0 | 1.0 |
| 1 | 170 | 38 (22) | 0.8 (.29) | 0.9 (.66) |
| ≥2 | 24 | 6 (25) | 0.9 (.88) | 1.0 (1.00) |
|
| ||||
| Plasma HCV RNA level, log copies/mL | ||||
| >0 to <6.0 | 48 | 10 (21) | 1.0 | … |
| 6.0 to <6.75 | 174 | 49 (28) | 1.5 (.31) | … |
| ≥6.75 | 22 | 11 (50) | 3.8 (.01) | … |
| HCV RNA negative | 60 | 7 (12) | 0.5 (.18) | … |
|
| ||||
| HCV genotype | ||||
| 1b | 72 | 22 (31) | 1.0 (1.0) | … |
| Other | 186 | 50 (27) | 0.8 (.63) | 0.7 (.37) |
|
| ||||
| AIDS diagnosis | ||||
| No | 228 | 50 (22) | 1.0 | 1.0 |
| Yes | 87 | 28 (32) | 1.7 (.07) | 1.6 (.10) |
|
| ||||
| Plasma HIV RNA level, copies/mL | ||||
| ≤400 | 103 | 27 (26) | 1.0 | 1.0 |
| 401–50,000 | 155 | 38 (25) | 0.9 (.79) | 1.0 (.99) |
| >50,000 | 53 | 12 (23) | 0.8 (.62) | 0.8 (.49) |
|
| ||||
| Received HAART | ||||
| Pre-HAART period | 130 | 30 (23) | 1.0 | 1.0 |
| Post-HAART period | 181 | 48 (27) | 1.2 (.52) | 1.2 (.50) |
|
| ||||
| Duration of HAART, no. of months | ||||
| Did not receive HAART | 129 | 30 (23) | 1.0 | 1.0 |
| <6 | 85 | 20 (24) | 1.0 (.98) | 1.0 (.91) |
| 6 to <12 | 81 | 23 (28) | 1.3 (.44) | 1.4 (.37) |
| ≥12 | 15 | 5 (33) | 1.7 (.39) | 2.1 (.24) |
|
| ||||
| CD4 cell count, cells/mm3 | ||||
| ≥200 | 241 | 58 (24) | 1.0 | 1.0 |
| <200 | 72 | 20 (28) | 1.2 (.61) | 1.2 (.62) |
|
| ||||
| CD8 cell count, cells/mm3 | ||||
| <800 | 146 | 35 (24) | 1.0 | 1.0 |
| ≥800 | 167 | 43 (26) | 1.1 (.80) | 1.0 (.91) |
|
| ||||
| Percentage of cells | ||||
| CD8+CD38+DR+ | ||||
| <55% | 226 | 63 (28) | 1.0 | 1.0 |
| ≥55% | 31 | 4 (13) | 0.4 (.08) | 0.4 (.05) |
|
| ||||
| CD4+CD38+DR+ | ||||
| <10% | 142 | 43 (30) | 1.0 | 1.0 |
| ≥10% | 115 | 24 (21) | 0.6 (.08) | 0.6 (.06) |
NOTE. Analyses used logistic regression with generalized estimating equations for repeated measures within subjects. HAART, highly active antiretroviral therapy; IDU, injection drug use; OR, odds ratio.
For HCV RNA level.
Figure 1 illustrates the association between negative-strand HCV RNA in PBMCs and plasma HIV RNA and HCV RNA loads. Although no association with HIV load is apparent, HCV replication in PBMCs is progressively more common at higher plasma HCV RNA loads. Parenthetically, there was no correlation between plasma levels of HCV and HIV RNA (Spearman correlation coefficient,−.009; P =.87).
Figure 1.
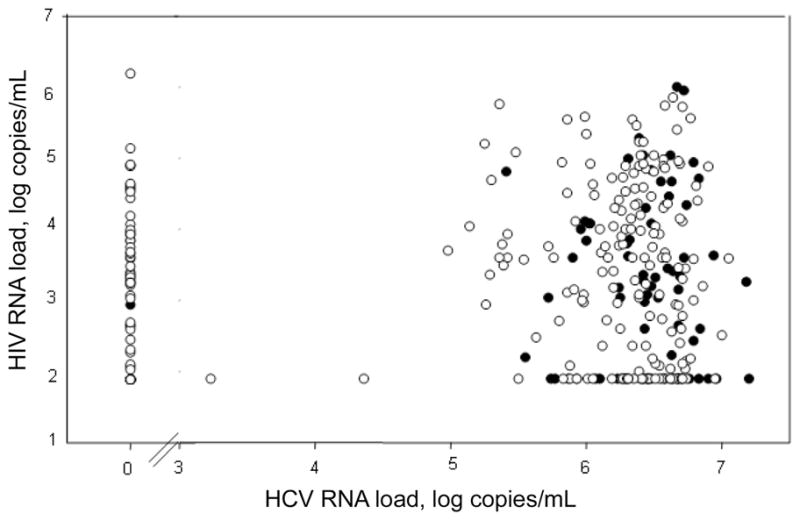
Presence of negative-strand hepatitis C virus (HCV) RNA in 315 peripheral blood mononuclear cell samples obtained from anti-HCV–positive/HIV-infected women, in relation to plasma levels of HIV and HCV RNA. Filled circles, samples that were positive for negative-strand HCV RNA; open circles, samples that were negative for negative-strand HCV RNA. The 7 filled circles denoting an HCV RNA load of 0 log copies/mL are partly obscured; HIV RNA loads are 1.9, 2.7, 2.9, 3.8, 3.9, 4.6, and 4.9 log copies/mL.
Multivariate analysis of factors associated with negative-strand HCV RNA in PBMCs
The association between the presence of negative-strand HCV RNA and the factors that were found to be marginally significant (P < .10) in the univariate analyses adjusted for HCV RNA level were further evaluated in a multivariate logistic regression model that was adjusted for each of these parameters. The model included age, education, alcohol use, IDU in the past 6 months, plasma HCV RNA load, AIDS diagnosis, and the percentage of CD4+CD38+DR+ and CD8+CD38+DR cells. Only alcohol use (≥7 drinks/week) and the plasma HCV RNA level (≥6.75 log copies/mL) had significant positive associations with the presence of negative-strand HCV RNA (table 4), and IDU in the past six months had a significant negative association. The association with diagnosis of AIDS remained borderline positive, and the association with the percentage of CD4+CD38+DR+ cells (>10%) remained borderline negative (P =.05).
Table 4.
Multivariate analysis of characteristics associated with the presence of negative-strand hepatitis C virus (HCV) RNA in peripheral blood mononuclear cells in 315 samples, controlling for repeated measures in 144 anti-HCV–positive/HIV-infected women.
| Characteristic | OR (95% CI) | P |
|---|---|---|
| Alcohol use, drinks/week | ||
| 0 | 1.0 | |
| 1–6 | 1.2 (0.6–2.5) | .54 |
| ≥7 | 2.9 (1.2–7.1) | .02 |
|
| ||
| IDU in past 6 months | ||
| No | 1.0 | |
| Yes | 0.2 (0.1–0.9) | .03 |
|
| ||
| Plasma HCV RNA level, log copies/mL | ||
| >0 to <6.0 | 1.0 | |
| 6.0 to <6.75 | 1.4 (0.5–4.1) | .52 |
| ≥6.75 | 4.6 (1.1–19.9) | .04 |
| HCV RNA negative | 0.4 (0.1–1.7) | .22 |
|
| ||
| CD4+CD38+DR+ cells, % | ||
| <10 | 1.0 | |
| ≥10 | 0.5 (0.3–1.0) | .05 |
|
| ||
| AIDS diagnosis | ||
| No | 1.0 | |
| Yes | 2.0 (1.0–3.9) | .05 |
NOTE. Analyses used logistic regression with generalized estimating equations for repeated measures within subjects. CI, confidence interval; IDU, injection drug use; OR, odds ratio.
DISCUSSION
This is the largest report to date demonstrating that extrahepatic replication of HCV in PBMCs is frequent among HIV-infected women. More than 40% of women in our study had negative-strand HCV RNA detectable at some time during follow-up, but replication in PBMCs was not constant among women evaluated at multiple visits that spanned from 0.3 to 6.5 years. We found that high HCV RNA levels, an AIDS diagnosis, and alcohol consumption were predictors of HCV replication in PBMCs. On the other hand, recent IDU and an increased percentage of the activation marker CD4+CD38+DR+ cells were negatively associated with HCV replication in PBMCs.
The association between the presence of negative-strand HCV RNA in PBMCs and the plasma HCV RNA load might reflect the overall high replication fitness of viral strain(s) present in a particular host. Furthermore, high viral turnover might facilitate the development of mutations, some of which may be advantageous to colonization of different cells. Such changes could be relatively minor. For example, it has been demonstrated for lymphocytic choriomeningitis virus that strains differing by a single amino acid substitution, when inoculated together into a mouse, are competitively selected either by the liver and spleen or by neurons [30]. Although the extent of changes necessary for successful colonization of extrahepatic sites by HCV is unclear, it is intriguing that many sequences identified at sites other than the liver share common mutations [27, 31–35]. This association with viral load is unlikely to be the result of nonspecific detection of the positive strand because the strand specificity of our assays was ~7–8 logs and because PBMC pellets were extensively washed after separation, reducing any contamination by circulating virions.
There was also a statistically significant association between extrahepatic replication and alcohol use: women who had ≥7 drinks per week were 3 times more likely to have negative-strand HCV RNA present in their PBMCs than were their nondrinking counterparts. This phenomenon could represent the well-known immunosuppressive effect of alcohol [36]. However, there is also evidence that interactions between alcohol and HCV infection are more complex, as is suggested by the cumulative effect of both on progression of liver disease [37].
Because all of the women in the present cohort were infected with HIV, we could not evaluate the influence that HIV status by itself might have on extrahepatic replication of HCV in PBMCs. However, we did investigate whether there was an association with the plasma HIV RNA level, and we found none. The mechanisms by which HIV could enhance extrahepatic HCV infection are still speculative, with one possibility being that this effect is related to general immunosuppression. Accordingly, in one small study, negative-strand viral RNA was more common in PBMC samples obtained from patients after liver transplantation than in those obtained from patients before liver transplantation [38], and HCV replication was enhanced by the presence of immunodeficiency in a mouse model [39]. It has also been reported that removal of CD8+ T cells results in increased HCV replication in PBMC cultures, which suggests that the adaptive immune system does exert some level of control over extrahepatic replication [40]. We found only a marginal positive association with AIDS diagnosis, and neither in this study nor in 2 previous studies was extrahepatic replication associated with a low CD4+ cell count [8, 23].
Although previous studies have shown that coinfection with HIV may facilitate HCV replication in vitro, either by rendering cells more susceptible to HCV infection or by increasing HCV replication [26], we found no association of HCV replication in PBMCs with either the HIV RNA level or receipt of HAART. Because we found no correlation between plasma HIV RNA and HCV RNA levels, no correlation would be expected at the extrahepatic sites. The absence of a correlation between HIV load and the presence of extrahepatic replication would mitigate against a significant direct effect of HIV in vivo. However, the HIV RNA load in PBMCs, which would be more relevant than that in plasma, was not evaluated in this study.
Negative-strand HCV RNA was detected in PBMC samples obtained from 42% of women evaluated in this study, which is similar to the detection rate of 36% found in a previous smaller study [8], as well as the rates of 32% and 55% noted among HCV-monoinfected and HIV/HCV-coinfected women, respectively, as recently reported by Blackard et al. [23]. This number was likely to be influenced by repeated testing, as well as by the high overall prevalence of a history of IDU (85%) among the women in our study. IDU has been reported to be a risk factor for extrahepatic replication, possibly because of associated immunosuppression or repeated exposure to HCV [41].
Superinfection with a new HCV strain leading to an “overtake phenomenon,” in which the original strains are supplanted by the new strain, is a well-recognized phenomenon among high-risk patients, including persons with hemophilia [42], active injection drug users [43], and blood transfusion [44] and liver transplant recipients [45]. A recent report supports the role of superinfection in extrahepatic colonization, finding that HCV sequence differences between plasma and PBMC compartments were 3 times higher in multiply than in singly transfused patients and were twice as high in injection drug users than in patients infected through an unknown source [5]. Although there was a trend toward a higher prevalence of HCV replication in PBMCs among women with a history of IDU, this difference was not significant either by univariate analysis (P =.28) or after adjustment for the HCV RNA level (P = .32), perhaps because of the small number of women with no history of IDU (15% of women at baseline). The significant negative association of HCV replication in PBMCs with recent IDU (i.e., in the past 6 months, as evaluated at each study visit) is puzzling in view of previous reports that found a positive association with a history of IDU. Perhaps immune activation, which may be common in active IDUs, is protective with respect to extrahepatic replication. This is consistent with our observation that an increase in the percentage of activated CD4+ cells (CD38+DR+) was associated with protection against HCV replication in PBMCs. The latter may seem contradictory to reports that pokeweed and phytohemagglutinin mitogens enhance HCV replication in PBMC cultures [27, 46]. However, such in vitro activation by mitogens results in massive cytokine secretion and cell proliferation and might not necessarily reflect events in vivo.
Thus, extrahepatic replication may be a highly dynamic process influenced by many factors, including current IDU and alcohol use and changing immune activation status. Such multiple and interacting factors could be the reason for the inconsistent presence of negative-strand HCV RNA in samples collected from the same women for up to 6.5 years. However, detection of negative-strand HCV RNA is also expected to be influenced by the typical low HCV RNA load noted at extra-hepatic sites and the limited sensitivity of strand-specific assays [47].
The consequences of extrahepatic replication of HCV remain unclear. In one report, HCV RNA was found in the livers of 57% of patients who had chronic liver disease of unclear etiology [48] and were negative for both anti-HCV and HCV RNA in serum. Of these patients, 84% had negative-strand HCV RNA detectable in the liver and 70% had HCV RNA detectable in PBMCs. Persistence of low-level extrahepatic replication may be common after spontaneous and treatment-induced resolution of infection [46, 47], and, in our study, 7 women had detectable HCV replication in PBMCs despite having undetectable HCV RNA in plasma. It is unclear whether persistence of virus at extrahepatic sites could be responsible for treatment failures, which are particularly common among HIV-positive patients. Interestingly, in a recently published case report, an HCV-infected patient cleared the infection and subsequently experienced viral reactivation twice over 8.5 years. Throughout that time, the infecting viral strain remained virtually identical [49]. Interestingly, in a recent study, Di Liberto et al. [6] found that the presence of compartmentalization in PBMCs was strongly predictive of sustained virologic response. The reasons for this phenomenon remain unclear.
In summary, our study revealed a high prevalence of HCV replication in PBMCs obtained from a large cohort of HIV-positive women. A high HCV RNA load in plasma, an AIDS diagnosis, and alcohol use were positively associated with an increased prevalence of extrahepatic replication, whereas an increase in the percentage of CD4+CD38+DR+ cells and recent IDU were associated with lower prevalence. The mechanisms for these associations, as well as their clinical implications, require further investigation.
Acknowledgments
Financial support: National Institute of Allergy and Infectious Diseases (NIAID; grant RO1 A1052065 to A.K.); NIAID, with supplemental funding from the National Cancer Institute and the National Institute on Drug Abuse (grants UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590), the National Institute of Child Health and Human Development (grant UO1-HD-23632), the National Center for Research Resources (grants MO1-RR-00071, MO1-RR-00079, and MO1-RR-00083), and the Institute of Mental Health (grant 1R21 MH073422-01A1 to T.L.).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Laskus T, Radkowski M, Wang LF, Jang SJ, Vargas H, Rakela J. Hepatitis C virus quasispecies in patients infected with HIV-1: correlation with extrahepatic viral replication. Virology. 1998;248:164–71. doi: 10.1006/viro.1998.9269. [DOI] [PubMed] [Google Scholar]
- 2.Laskus T, Radkowski M, Wang LF, Nowicki M, Rakela J. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J Virol. 2000;74:1014–7. doi: 10.1128/jvi.74.2.1014-1017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerat H, Rumin S, Habersetzer F, et al. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood. 1998;91:3841–9. [PubMed] [Google Scholar]
- 4.Navas S, Martin J, Quiroga JA, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–6. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roque-Afonso AM, Ducoulombier D, Di Liberto G, et al. Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells. J Virol. 2005;79:6349–57. doi: 10.1128/JVI.79.10.6349-6357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Liberto G, Roque-Afonso AM, Kara R, et al. Clinical and therapeutic implications of hepatitis C virus compartmentalization. Gastroenterology. 2006;131:76–84. doi: 10.1053/j.gastro.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Sansonno D, Iacobelli AR, Cornacchiulo V, Iodice G, Dammacco F. Detection of hepatitis C virus (HCV) proteins by immunofluorescence and HCV RNA genomic sequences by non-isotopic in situ hybridization in bone marrow and peripheral blood mononuclear cells of chronically HCV- infected patients. Clin Exp Immunol. 1996;103:414–21. doi: 10.1111/j.1365-2249.1996.tb08296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. The presence of active hepatitis C virus replication in lymphoid tissue in patients coinfected with human immunodeficiency virus type 1. J Infect Dis. 1998;178:1189–92. doi: 10.1086/515682. [DOI] [PubMed] [Google Scholar]
- 9.Pal S, Sullivan DG, Kim S, et al. Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology. 2006;130:1107–16. doi: 10.1053/j.gastro.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 10.Shimizu YK, Igarashi H, Kanematu T, et al. Sequence analysis of the hepatitis C virus genome recovered from serum, liver, and peripheral blood mononuclear cells of infected chimpanzees. J Virol. 1997;71:5769–73. doi: 10.1128/jvi.71.8.5769-5773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winnock M, Salmon-Ceron D, Dabis F, Chene G. Interaction between HIV-1 and HCV infections: towards a new entity? J Antimicrob Chemother. 2004;53:936–46. doi: 10.1093/jac/dkh200. [DOI] [PubMed] [Google Scholar]
- 12.Thomas SL, Newell ML, Peckham CS, Ades AE, Hall AJ. A review of hepatitis C virus (HCV) vertical transmission: risks of transmission to infants born to mothers with and without HCV viraemia or human immunodeficiency virus infection. Int J Epidemiol. 1998;27:108–17. doi: 10.1093/ije/27.1.108. [DOI] [PubMed] [Google Scholar]
- 13.Granovsky MO, Minkoff HL, Tess BH, et al. Hepatitis C virus infection in the mothers and infants cohort study. Pediatrics. 1998;102:355–9. doi: 10.1542/peds.102.2.355. [DOI] [PubMed] [Google Scholar]
- 14.Tovo PA, Palomba E, Ferraris G, et al. Increased risk of maternal-infant hepatitis C virus transmission for women coinfected with human immunodeficiency virus type 1. Italian Study Group for HCV Infection in Children. Clin Infect Dis. 1997;25:1121–4. doi: 10.1086/516102. [DOI] [PubMed] [Google Scholar]
- 15.Eyster ME, Alter HJ, Aledort LM, Quan S, Hatzakis A, Goedert JJ. Heterosexual co-transmission of hepatitis C virus (HCV) and human immunodeficiency virus (HIV) Ann Intern Med. 1991;115:764–8. doi: 10.7326/0003-4819-115-10-764. [DOI] [PubMed] [Google Scholar]
- 16.Ridzon R, Gallagher K, Ciesielski C, et al. Simultaneous transmission of human immunodeficiency virus and hepatitis C virus from a needle-stick injury [see comments] N Engl J Med. 1997;336:919–22. doi: 10.1056/NEJM199703273361304. [DOI] [PubMed] [Google Scholar]
- 17.Eyster ME, Diamondstone LS, Lien JM, Ehmann WC, Quan S, Goedert JJ. Natural history of hepatitis C virus infection in multitransfused hemophiliacs: effect of coinfection with human immunodeficiency virus. The Multicenter Hemophilia Cohort Study. J Acquir Immune Defic Syndr. 1993;6:602–10. [PubMed] [Google Scholar]
- 18.Garcia-Samaniego J, Soriano V, Castilla J, et al. Influence of hepatitis C virus genotypes and HIV infection on histological severity of chronic hepatitis C. The Hepatitis/HIV Spanish Study Group. Am J Gastroenterol. 1997;92:1130–4. [PubMed] [Google Scholar]
- 19.Hanley JP, Jarvis LM, Andrews J, et al. Investigation of chronic hepatitis C infection in individuals with haemophilia: assessment of invasive and non-invasive methods. Br J Haematol. 1996;94:159–65. doi: 10.1046/j.1365-2141.1996.6192064.x. [DOI] [PubMed] [Google Scholar]
- 20.Monga HK, Breauz K, Rodrigues-Barradas MC, Yoffe B. Increased HCV-related morbidity and mortality in HIV patients. Hepatology. 1998;28:565A. [Google Scholar]
- 21.Sherman KE, O’Brien J, Gutierrez AG, et al. Quantitative evaluation of hepatitis C virus RNA in patients with concurrent human immunodeficiency virus infections. J Clin Microbiol. 1993;31:2679–82. doi: 10.1128/jcm.31.10.2679-2682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daar ES, Lynn H, Donfield S, et al. Relation between HIV-1 and hepatitis C viral load in patients with hemophilia. J Acquir Immune Defic Syndr. 2001;26:466–72. doi: 10.1097/00126334-200104150-00011. [DOI] [PubMed] [Google Scholar]
- 23.Blackard JT, Smeaton L, Hiasa Y, et al. Detection of hepatitis C virus (HCV) in serum and peripheral-blood mononuclear cells from HCV-monoinfected and HIV/HCV-coinfected persons. J Infect Dis. 2005;192:258–65. doi: 10.1086/430949. [DOI] [PubMed] [Google Scholar]
- 24.Barkan SE, Melnick SL, Preston-Martin S, et al. The Women’s Inter-agency HIV Study. WIHS Collaborative Study Group. Epidemiology. 1998;9:117–25. [PubMed] [Google Scholar]
- 25.Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Lack of evidence for hepatitis G virus replication in the livers of patients coinfected with hepatitis C and G viruses. J Virol. 1997;71:7804–6. doi: 10.1128/jvi.71.10.7804-7806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskus T, Radkowski M, Jablonska J, et al. Human immunodeficiency virus facilitates infection/replication of hepatitis C virus in native human macrophages. Blood. 2004;103:3854–9. doi: 10.1182/blood-2003-08-2923. [DOI] [PubMed] [Google Scholar]
- 27.Laskus T, Radkowski M, Piasek A, et al. Hepatitis C virus in lymphoid cells of patients coinfected with human immunodeficiency virus type 1: evidence of active replication in monocytes/macrophages and lymphocytes. J Infect Dis. 2000;181:442–8. doi: 10.1086/315283. [DOI] [PubMed] [Google Scholar]
- 28.Germer JJ, Majewski DW, Rosser M, et al. Evaluation of the TRUGENE HCV 5′NC genotyping kit with the new GeneLibrarian module 3.1. 2 for genotyping of hepatitis C virus from clinical specimens. J Clin Microbiol. 2003;41:4855–7. doi: 10.1128/JCM.41.10.4855-4857.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44:1049–60. [PubMed] [Google Scholar]
- 30.Dockter J, Evans CF, Tishon A, Oldstone MB. Competitive selection in vivo by a cell for one variant over another: implications for RNA virus quasispecies in vivo. J Virol. 1996;70:1799–803. doi: 10.1128/jvi.70.3.1799-1803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laporte J, Malet I, Andrieu T, et al. Comparative analysis of translation efficiencies of hepatitis C virus 5′ untranslated regions among intraindividual quasispecies present in chronic infection: opposite behaviors depending on cell type. J Virol. 2000;74:10827–33. doi: 10.1128/jvi.74.22.10827-10833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forton DM, Karayiannis P, Mahmud N, Taylor-Robinson SD, Thomas HC. Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants. J Virol. 2004;78:5170–83. doi: 10.1128/JVI.78.10.5170-5183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laporte J, Bain C, Maurel P, Inchauspe G, Agut H, Cahour A. Differential distribution and internal translation efficiency of hepatitis C virus quasispecies present in dendritic and liver cells. Blood. 2003;101:52–7. doi: 10.1182/blood-2002-03-0818. [DOI] [PubMed] [Google Scholar]
- 34.Lerat H, Shimizu YK, Lemon SM. Cell type-specific enhancement of hepatitis C virus internal ribosome entry site-directed translation due to 5′ nontranslated region substitutions selected during passage of virus in lymphoblastoid cells. J Virol. 2000;74:7024–31. doi: 10.1128/jvi.74.15.7024-7031.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskus T, Radkowski M, Bednarska A, et al. Detection and analysis of hepatitis C virus sequences in cerebrospinal fluid. J Virol. 2002;76:10064–8. doi: 10.1128/JVI.76.19.10064-10068.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson S, Kolls JK. Alcohol, host defence and society. Nat Rev Immunol. 2002;2:205–9. doi: 10.1038/nri744. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharya R, Shuhart MC. Hepatitis C and alcohol: interactions, outcomes, and implications. J Clin Gastroenterol. 2003;36:242–52. doi: 10.1097/00004836-200303000-00012. [DOI] [PubMed] [Google Scholar]
- 38.Radkowski M, Wang LF, Vargas HE, Rakela J, Laskus T. Detection of hepatitis C virus replication in peripheral blood mononuclear cells after orthotopic liver transplantation. Transplantation. 1998;66:664–6. doi: 10.1097/00007890-199809150-00022. [DOI] [PubMed] [Google Scholar]
- 39.Bronowicki JP, Loriot MA, Thiers V, Grignon Y, Zignego AL, Brechot C. Hepatitis C virus persistence in human hematopoietic cells injected into SCID mice. Hepatology. 1998;28:211–8. doi: 10.1002/hep.510280127. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, Wang X, Douglas SD, et al. CD8+ T cell depletion amplifies hepatitis C virus replication in peripheral blood mononuclear cells. J Infect Dis. 2005;192:1093–101. doi: 10.1086/432957. [DOI] [PubMed] [Google Scholar]
- 41.Resti M, Azzari C, Moriondo M, et al. Injection drug use facilitates hepatitis C virus infection of peripheral blood mononuclear cells. Clin Infect Dis. 2002;35:236–9. doi: 10.1086/341302. [DOI] [PubMed] [Google Scholar]
- 42.Eyster ME, Sherman KE, Goedert JJ, Katsoulidou A, Hatzakis A. Prevalence and changes in hepatitis C virus genotypes among multitransfused persons with hemophilia. The Multicenter Hemophilia Cohort Study. J Infect Dis. 1999;179:1062–9. doi: 10.1086/314708. [DOI] [PubMed] [Google Scholar]
- 43.Herring BL, Page-Shafer K, Tobler LH, Delwart EL. Frequent hepatitis C virus superinfection in injection drug users. J Infect Dis. 2004;190:1396–403. doi: 10.1086/424491. [DOI] [PubMed] [Google Scholar]
- 44.Laskus T, Wang LF, Radkowski M, et al. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J Virol. 2001;75:2059–66. doi: 10.1128/JVI.75.5.2059-2066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laskus T, Wang LF, Rakela J, et al. Dynamic behavior of hepatitis C virus in chronically infected patients receiving liver graft from infected donors. Virology. 1996;220:171–6. doi: 10.1006/viro.1996.0297. [DOI] [PubMed] [Google Scholar]
- 46.Pham TN, MacParland SA, Mulrooney PM, Cooksley H, Naoumov NV, Michalak TI. Hepatitis C virus persistence after spontaneous or treatment-induced resolution of hepatitis C. J Virol. 2004;78:5867–74. doi: 10.1128/JVI.78.11.5867-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radkowski M, Gallegos-Orozco JF, Jablonska J, et al. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology. 2005;41:106–14. doi: 10.1002/hep.20518. [DOI] [PubMed] [Google Scholar]
- 48.Castillo I, Pardo M, Bartolome J, et al. Occult hepatitis C virus infection in patients in whom the etiology of persistently abnormal results of liver-function tests is unknown. J Infect Dis. 2004;189:7–14. doi: 10.1086/380202. [DOI] [PubMed] [Google Scholar]
- 49.Lee WM, Polson JE, Carney DS, Sahin B, Gale M., Jr Reemergence of hepatitis C virus after 8. 5 years in a patient with hypogammaglobulinemia: evidence for an occult viral reservoir. J Infect Dis. 2005;192:1088–92. doi: 10.1086/432917. [DOI] [PubMed] [Google Scholar]


