Abstract
Super-resolution techniques provide a route to studying fine scale anatomical detail using multiple lower resolution acquisitions. In particular, techniques that do not depend on regular sampling can be used in medical imaging situations where imaging time and resolution are limited by subject motion. We investigate in this work the use of super-resolution technique for anisotropic fetal brain MR data reconstruction without modifying the data acquisition protocol. The approach, which consists of iterative motion correction and high resolution image estimation, is compared with a previously used scattered data interpolation-based reconstruction method. To optimize acquisition time, an evaluation of the influence of the number of input images and image noise is also performed. Evaluation on simulated MR images and real data show significant improvements in performance provided by the super-resolution approach.
1 Introduction
Imaging moving subjects remains an open issue for Magnetic Resonance Imaging (MRI). Although the development of ultrafast 2D acquisition sequences has led to significant improvements for clinical studies (see for instance [1] for fetal studies), the slice acquisition time is still critical and has to be as short as possible to reduce the impact of the motion. As a result, sets of thick 2D slices are generally acquired in clinical studies and interpretation remains limited by visual inspection.
Several clinical imaging protocols make use of multiple orthogonal 2D multi-planar acquisitions with non-isotropic voxel size for brain studies. In the context of fetal imaging4, Rousseau et al. in [4] have proposed a registration-based method to compound multiple orthogonal sets of 2D fetal MRI slices into a single isotropic high resolution (HR) volume. This algorithm has been shown to be capable of reconstructing 3D geometrically consistent images from challenging clinically acquired data. The image reconstruction process is based on a local neighbourhood approach with a Gaussian kernel. Although this local interpolation approach is relatively computationally efficient for use during iterative refinement of slice to volume alignment, the final reconstruction can introduce additional blurring and does not make use of oversampling of the anatomy provided by the acquisition of multiple datasets. Jiang et al. in [5] have proposed a very similar approach for volume reconstruction based on a slice-to-volume registration and B-spline interpolation. They have applied their method on various MR brain images: awake neonates, deliberately moved adults and foetuses. To form 3D images of fetal brain, Kim et al. in [6] have recently proposed a new registration approach which considers the collective alignment of all slices directly, via shared structure in their intersections, rather than to an estimated 3D volume. After slice alignment a final volume is reconstructed using sparse data interpolation. All these approaches consist of two steps (motion estimation and image reconstruction) and make use of interpolation techniques to reconstruct a final 3D MR images.
In this paper we begin by assuming that fetal motion can be reliably estimated [4], and focus on the image reconstruction step and we investigate the use of a super resolution (SR) approach for this specific issue. SR in MRI has been mostly investigated using a specialized protocol to acquire shifted images, where the motion between slices is known (translation in the slice direction [7,8,9] or rotation around a common frequency encoding axis [10]). In this paper, we study reconstruction techniques with a commonly used fetal imaging protocol: multiple orthogonal stacks of thick slices where the slice thickness is usually around three times the dimension of the in-plane resolution. Since the limits of SR algorithms (maximum magnification factor, minimum number of low-resolution (LR) inputs) is still an open question [11,12], here we also experimentally investigate the number of LR images required to form a given image resolution in order to optimize the overall imaging time.
2 Problem Statement
2.1 Super-Resolution
The principle of super-resolution (SR) is to combine low resolution images to produce an image that has a higher spatial resolution than the original images [13]. This is a large research field encompassing many applications. However the majority of the work has focused on using lower resolution data acquired on a regular grid and often assuming simple translational motion between the lower resolution sample grids, unlike multislice brain MR data which is corrupted by full 3D rigid motion on a slice by slice basis (see Figure 1). Please note that this data specificity does not affect the way the SR problem is modeled. However, it introduces additional challenges because SR becomes a reconstruction problem with scattered anisotropic data.
Fig. 1.
Acquired fetal MR image data with anisotropic resolution. It can be noted that the LR images provide complementary views of the entire brain. From left to right: 1) axial LR image, 2) sagittal LR image, 3) coronal LR image.
As in most of common SR approaches, we model the physical problem and then compute a solution by inverting this observation model:
| (1) |
where n is the number of LR images and mr is the number of slices of the LR image r, yr,s denotes the slice s of the LR image r, x is the HR image, nr represents the observation noise, Sr is the subsampling matrix, Br a blur matrix, Ws and Wr are geometric transformations of sth slice of the yr,s and of the rth low resolution image respectively. The purpose of super-resolution is to remove the effects of the blurring convolution and to increase the voxel grid density.
For simplicity, the four operators can be combined into a single matrix Hr,s: Hr,s = SrBrWsWr. The matrix Hr,s thus incorporates motion compensation, degradation effects, and sub-sampling for each slice yr,s. In this paper, we assume that the operators Dr and Br, and the noise characteristics are known and spatially invariant. The operators Ws and Wr are estimated using a similar method to the one proposed by Rousseau et al. in [4] using the pipeline detailed in Figure 2. Using this observation model, a straightforward definition of the data discrepancy functional is:
| (2) |
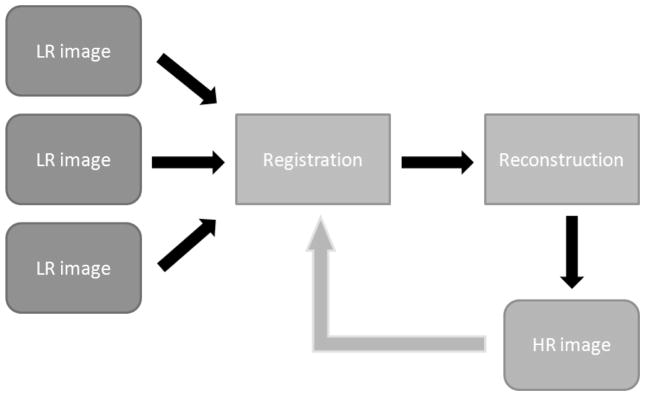
Fig. 2.
Pipeline used to estimate the matrix W and the HR image x. This pipeline is performed in an iterative fashion (this is represented by the gray arrow).
ψ is usually set as a quadratic functional by assuming that the image noise nr follows a Gaussian distribution. However, x̂ cannot be uniquely determined by minimizing D(x) since the image reconstruction process is an ill-posed problem. For such inverse problems, some form of regularization plays a crucial role and must be included in the cost function to stabilize the problem or constrain the space of solutions.
2.2 Variational regularization
One of the most important classes of linear deterministic regularization methods is the one introduced by Tikhonov in [14] for which the regularization term can be defined as follows:
| (3) |
where weights cp are strictly positive and x(p) is the pth order derivative of x. Much work has been carried out on the derivation of regularization terms of the form φ(|∇x|) or φ(|x|) where φ is a convex even function, increasing on R+ (see for instance [15]). If φ(t) = t2, one obtain L2 norm which smooths strong variations in the image. If φ(t) = t, one obtain L1 norm. One popular variational regularization functional relies on the total variation:
| (4) |
Charbonnier et al. in [15]) have proposed several edge-preserving regularization functionals whose one we use in our experiments is:
| (5) |
with .
3 Sufficient Number of LR Images
The principle of SR techniques is to make use of multiple LR images to estimate a single HR image. As it was pointed out in [11,12], two key points of SR algorithms are the magnification factor and the number of input images. Lin and Shum have shown in [12] that considering 2D images, the maximum magnification factor is 1.6 if the denoising and registration is not good enough and 5.7 under synthetic conditions. Moreover, when the magnification factor
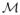 is an integer, the sufficient number of LR images is
is an integer, the sufficient number of LR images is
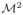 .
.
In the context of fetal MR data, we consider anisotropic data where the slice thickness is approximately three times bigger than the in-plane resolution. In this work, the sufficient number of LR fetal MR images is estimated by using simulated data. To explore the ability to reconstruct a high resolution volume for typical anatomical brain structures, we applied the SR reconstruction algorithm on a T1-weighted MR image from the Brainweb dataset [16]. Brainweb is a simulated brain database which is often used as a gold standard for the analysis of in vivo acquired data. In order to mimic the clinical fetal imaging protocol used in routine, orthogonal low resolution images (1 × 1 × 3 mm) have been created using the observation model described by Equation 1. When using more than 3 images, the LR orthogonal images are simulated using 1mm shift. In this noise free case, no noise except the rounding error is introduced to the LR images and the registration is exact. We also have evaluated the performance of the proposed method using noisy low resolution images (Gaussian noise with standard deviation set to 10). Reconstruction algorithms have been applied to these synthetic images in order to reconstruct an isotropic (1 × 1 × 1 mm) image.
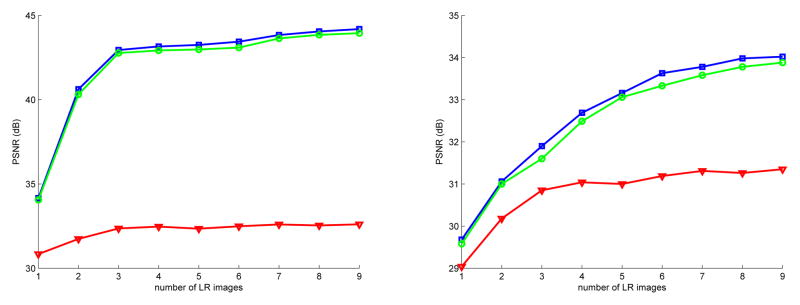
For quantitative comparison, the peak signal to noise ratio (PSNR) is also reported in decibels (dB): where d is the reference image dynamic. PSNR performance with respect to the number of considered LR images are reported in Figure. Two SR regularization functionals (JTV, JCh) are compared with the local scattered data interpolation approach proposed by Rousseau et al. in [4] (we found, in experiments not presented in this paper, that this interpolation method has similar performance to other scattered data interpolation techniques such as those using Radial Basis Functions).
Results are reported in Figure 3. When considering the noise free experiment, it clearly appears that using more than 3 LR images does not lead to significant improvement. This means that under optimal conditions, only 3 LR images are required to accurately estimate a HR image using SR techniques. When adding non negligible noise in LR images, the number of required LR images greatly increase (except for interpolation technique for which 3 remains a sufficient number of LR inputs) and is close to the estimated number obtained by Lin and Shum in [12]. Such experiment tends to show that efficient denoising method may have a substantial impact on SR results. In both cases, SR techniques lead to similar results and outperform the sparse interpolation approach.
Fig. 3.
PSNR in function of the number of input Brainweb images. Left: noise free experiment, Right: Gaussian noise experiment. □: SR technique with JCh regularisation, ○: SR technique with JTV regularisation, ▽: sparse interpolation technique [4].
4 Experiments on Fetal Brain MR Data
We applied the algorithm to fetal MR scans: T2 weighted HASTE sequence (TE/TR = 147/3190 ms) on a 1.5 T Siemens Avanto MRI Scanner (SIEMENS, Erlangen, Germany), resolution: 0.74 × 0.74 × 3.45mm. Pregnant women were briefed before the exam and signed informed consent. To reduce motion artefacts, fetal sedation was obtained with 1 mg of flunitrazepam given orally to the mother 30 minutes before the exam. The study was approved by the local ethics committee.
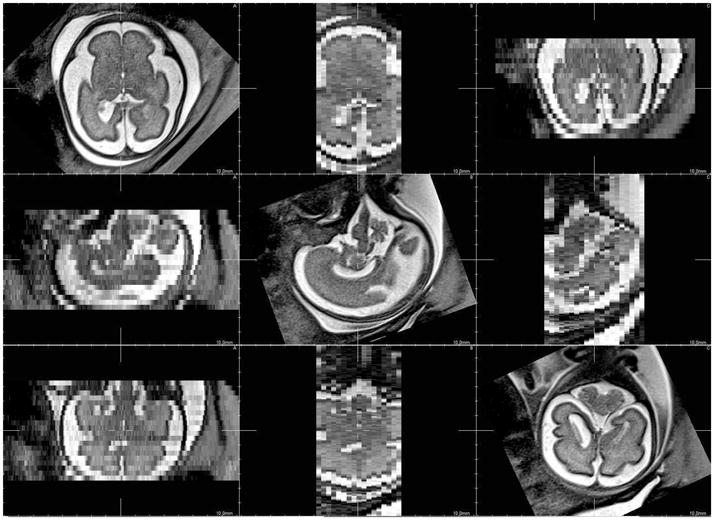
Figure 4 shows one original low resolution image compared to the high resolution reconstructed images for axial, coronal, sagittal views. Results obtained with the SR approach with JCh regularisation compare favorably with the local sparse interpolation approach proposed by Rousseau et al. in [4]. It can be especially noticed that the boundaries of brain structures (like the cortex) are better recovered. Moreover, the noise present in the LR image has no major impact on the reconstructed images. However, reconstruction artefacts appear outside the fetal head (see for instance the surrounding structures). In this case, the assumption concerning the “rigidity” of the scene is violated and registration errors disturb the reconstruction process. The result shown in Figure 4 has been obtained using 3 LR images. Using up to 6 images gives only marginal qualitative improvements. This experiment tends to show that the pipeline described in Figure 2 may be efficient enough to reduce, in our context, fetal imaging time by avoiding unnecessary acquisitions.
Fig. 4.
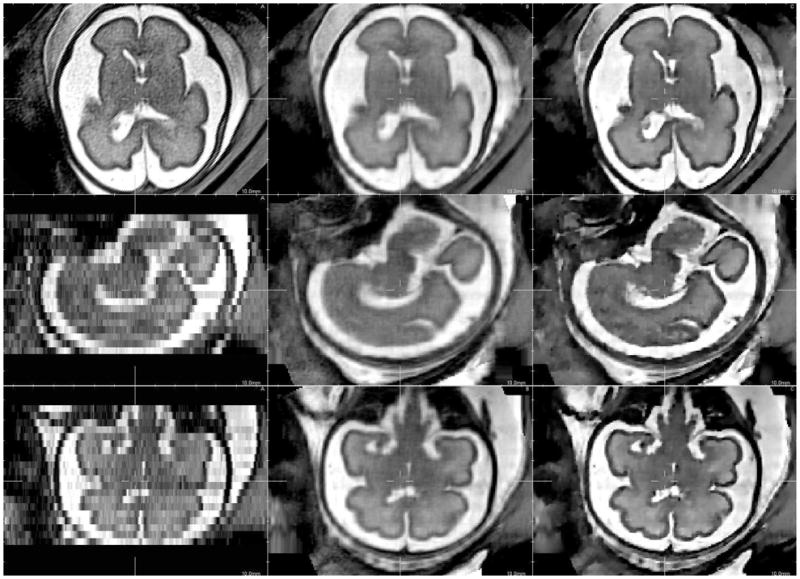
Details of a reconstructed fetal brain MR image using 3 orthogonal LR images. From left to right: A) original low resolution image, B) reconstructed image using local sparse interpolation [4], C) SR reconstruction using JCh regularisation.
5 Discussion
The first contribution of this work concerns the use of a SR framework for HR 3D isotropic fetal brain MR image reconstruction. Contrary to previous works [4,5,6] where sparse interpolation frameworks were used, an observation model is introduced in this paper in order to truly take into account physics of MR image acquisition (point spread function, sub-sampling and noise). This model allows potentially to recover fine image details compared to interpolation based approaches. Visual analysis of the obtained results on real data are very encouraging. Such high resolution image reconstruction algorithm represents an important step towards analysis of fine scale anatomical details.
The second contribution of this paper is the study of the sufficient number of LR images based on simulations using the Brainweb images. Considering a set of orthogonal stack of 2D slices with an anisotropic resolution of 1 × 1 × 3mm, the sufficient number of LR images is three in the case of noise free images. When adding Gaussian noise, the sufficient number of LR inputs is less than 9. Including an efficient denoising step in the overall image processing pipeline should lead to a significant decrease of the number of required LR images. As a consequence, further work is to investigate the building of such pipeline in order to reduce the fetal imaging time.
Acknowledgments
The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013 Grant Agreement no. 207667). This work is also funded by NIH/NINDS Grant R01 NS 055064 and a CNRS grant for collaboration between LSIIT and BICG.
Footnotes
References
- 1.Prayer D, Brugger PC, Prayer L. Fetal MRI: techniques and protocols. Pediatric Radiology. 2004 September;34(9):685–693. doi: 10.1007/s00247-004-1246-0. [DOI] [PubMed] [Google Scholar]
- 2.Moore J, Drangova M, Wierzbicki M, Barron J, Peters T. A high resolution dynamic heart model based on averaged MRI data. Medical Image Computing and Computer-Assisted Intervention - MICCAI; 2003; 2003. pp. 549–555. [Google Scholar]
- 3.Prager RW, Gee A, Berman L. Stradx: real-time acquisition and visualization of freehand three-dimensional ultrasound. Medical Image Analysis. 1999;3(2):129–140. doi: 10.1016/s1361-8415(99)80003-6. [DOI] [PubMed] [Google Scholar]
- 4.Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza C, Vigneron DB, Barkovich JA, Studholme C. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Academic Radiology. 2006 September;13(9):1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. IEEE Transactions on Medical Imaging. 2007 July;26(7):967–980. doi: 10.1109/TMI.2007.895456. [DOI] [PubMed] [Google Scholar]
- 6.Kim K, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Intersection based motion correction of multislice MRI for 3-D in utero fetal brain image formation. IEEE Transactions on Medical Imaging. 2010;29(1):146–158. doi: 10.1109/TMI.2009.2030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmi E, Liu S, Alon N, Fiat A, Fiat D. Resolution enhancement in MRI. Magnetic Resonance Imaging. 2006 February;24(2):133–154. doi: 10.1016/j.mri.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Peeters RR, Kornprobst P, Nikolova M, Sunaert S, Vieville T, Malandain G, Deriche R, Faugeras O, Ng M, Hecke PV. The use of super-resolution techniques to reduce slice thickness in functional MRI. International Journal of Imaging Systems and Technology. 2004;14(3):131–138. [Google Scholar]
- 9.Greenspan H, Peled S, Oz G, Kiryati N. MRI inter-slice reconstruction using Super-Resolution. Medical Image Computing and Computer-Assisted Intervention MICCAI; 2001; 2001. pp. 1204–1206. [Google Scholar]
- 10.Shilling RZ, Robbie TQ, Bailloeul T, Mewes K, Mersereau RM, Brummer ME. A super-resolution framework for 3-D high-resolution and high-contrast imaging using 2-D multislice MRI. IEEE Transactions on Medical Imaging. 2009 May;28(5):633–644. doi: 10.1109/TMI.2008.2007348. [DOI] [PubMed] [Google Scholar]
- 11.Baker S, Kanade T. Limits on Super-Resolution and how to break them. IEEE Trans Pattern Anal Mach Intell. 2002;24(9):1167–1183. [Google Scholar]
- 12.Lin Z, Shum H. Fundamental limits of Reconstruction-Based superresolution algorithms under local translation. IEEE Trans Pattern Anal Mach Intell. 2004;26(1):83–97. doi: 10.1109/tpami.2004.1261081. [DOI] [PubMed] [Google Scholar]
- 13.Bose N, Chan R, Ng M. Special issue: High resolution image reconstruction. International Journal of Imaging Systems and Technology. 2004;14:2–3. [Google Scholar]
- 14.Tikhonov A. Regularization of incorrectly posed problems. Sov Math Dokl. 1963;4:1624–1627. [Google Scholar]
- 15.Charbonnier P, Blanc-Feraud L, Aubert G, Barlaud M. Deterministic edge-preserving regularization in computed imaging. IEEE Transactions on Image Processing. 1997 February;6(2):298–311. doi: 10.1109/83.551699. [DOI] [PubMed] [Google Scholar]
- 16.Cocosco CA, Kollokian V, Kwan RK, Evans AC. BrainWeb: online interface to a 3D MRI simulated brain database. 1997. [Google Scholar]