Summary
Accompanying recent advances in determining RNA secondary structure is the growing appreciation for the importance of relatively simple topological constraints, encoded at the secondary structure level, in defining the overall architecture, folding pathways, and dynamic adaptability of RNA. A new view is emerging in which tertiary interactions do not define RNA 3D structure, but rather, help select specific conformers from an already narrow, topologically pre-defined conformational distribution. Studies are providing fundamental insights into the nature of these topological constraints, how they are encoded by the RNA secondary structure, and how they interplay with other interactions, breathing new meaning to RNA secondary structure. New approaches have been developed that take advantage of topological constraints in determining RNA backbone conformation based on secondary structure, and a limited set of other, easily accessible constraints. Topological constraints are also providing a much-needed framework for rationalizing and describing RNA dynamics and structural adaptation. Finally, studies suggest that topological constraints may play important roles steering RNA folding pathways. Here, we review recent advances in our understanding of topological constraints encoded by the RNA secondary structure.
Introduction
The strongest forces that define the structures of macromolecules are also the simplest ones; atoms must satisfy stereochemical bonding constraints and avoid steric collisions. For example, these constraints alone restrict torsion angles in nucleic acids to <5% of all possible conformations[1]. At longer length scales, RNA secondary structure consists of A-form helical domains that carry various receptors involved in tertiary interactions and that are linked together by a variety of junctions, including bulges, internal loops, three-way and other higher order junctions, and single strands. If we think of A-form helices as objects with variable shape and size, and junctions as linkers with variable stereochemical properties, higher order topological constraints come to existence that severely restrict, and in some cases define, the allowed range of RNA conformations. Recent biophysical studies are providing insights into these constraints while others are exploiting them to make predictions about RNA structure and dynamic-adaptation.
Overview of Topological Constraints
We begin discussion by defining what we mean by topological constraints. For this purpose, we consider the secondary structure of an RNA in its native folded state, which can be determined with ever-increasing accuracy using experimental-computational approaches (Figure 1a). Even in the absence of tertiary interactions, and ignoring electrostatic interactions, the RNA secondary structure alone imposes significant topological constraints on the allowed range of conformations. In particular, stable A-form helices exist in great abundance and their backbone conformation can to a very good approximation be modeled assuming an A-form helix geometry[2]. These helical elements are in turn tethered together at two positions on either side of the helix perimeter by single strands that are short (~5 Å per nt) in comparison to the local diameter of the A-form helix (~17 Å across a base pair). This gives rise to connectivity constraints i.e. reorientation and translation of one helix relative to another, is severely restricted by the short length of the two tethers on either side of the helix (Figure 1b). The specific nature of these connectivity constraints will vary depending on the junction topology, including junction order (two-way, three-way, etc.), relative lengths of individual strands, and whether bases in junctions adopt a looped in or looped out conformation. In addition, steric constraints between RNA helices and other elements, further restrict the allowed translation and reorientation of helices with respect to one another (Figure 1b). These steric collisions can be long-range in nature, occurring between helices that are far apart in sequence. Together, the connectivity and steric constraints define the topological constraints encoded by an RNA secondary structure.
Figure 1.
Topological constraints encoded by the RNA secondary structure. (a) Secondary and tertiary structure of group IIC intron from O. iheyensis (PDB 3IGI) [57]. (b) Connectivity and steric constraints in a two-way junction.
Inter-helical Junctions
Using electropheretic gel mobility measurements, fluorescence resonance energy transfer, transient electric birefringence, and electron microscopy, Lilley[3–5], Griffith[6], and Hagerman[7,8] showed more than two decades ago that bulges and internal loops induce directional bends in DNA and RNA duplexes by amounts dependent on the length, asymmetry, and sequence of the junction as well as on ionic strength. Longer bulges induced greater bends but only up to a certain length of ~5–7 nucleotides. U-rich bulges induced smaller bends compared to A-rich bulges and were more affected by addition of Mg2+ ions. Subsequent studies employing single molecule techniques[9] and NMR spectroscopy[10] confirmed that junctions do not induce rigid bends, but rather, a dynamic range of conformations and that this flexibility is important for allowing adaptive conformational changes to take place during recognition[11] and catalysis[12].
Recent studies on simple two-way junction model systems are helping explain these early observations, resulting in the establishment of a framework for quantitatively predicting certain aspects of inter-helical junction behavior based on secondary structure alone. Chu and Herschlag[13••] used stochastic dynamics simulations to understand how junction topologies define the allowed orientation sampled by helices. They used a duplex model system consisting of two DNA helices adjoined by either one or two polyethylene glycol (PEG) tethers as a mimic for single and double stranded junctions, respectively. The PEG junctions allowed the authors to focus on topological constraints and avoid contributions from sequence-specific interactions. In these simulations, the helices were coarse grained thus significantly increasing the efficiency with which the inter-helical space could be sampled. The simulations revealed that the double stranded PEG junction significantly confines the allowed inter-helical orientations along a hinge-like pathway and that the confinement was less severe and directional for the single stranded junction (Figure 2a). Thus, the anisotropic kinking of helices is simply the result of the junction topology and does not require the invocation of any sequence-specific interactions.
Figure 2.
Topological constraints confine the allowed inter-helical orientations across bulges. (a) Three inter-helical Euler angles (αhβhγh) used to define the relative orientation of 5' and 3' helices across two-way junctions containing X and Y (X ≥ Y) single stranded (S) residues. (b) 3D inter-helical orientation maps showing individual 2D projections along each plane of a 3D cube with associated correlation coefficients (R) between the inter-helical twist angles for three-nucleotide bulged RNA TAR (S3S0, HIV-1 TAR). The ligand-bound, NMR-RDC, NMR-MD, PDB-derived, and topologically computed inter-helical distributions are shown in black, yellow, blue, red, and grey, respectively. (c) The PDB-derived inter-helical distributions for various families of Y-strand length junctions.
Concurrently, Bailor et al[14••] showed that topological constraints severely confine the global orientation of helices across two-way junctions. The authors developed and standardized three inter-helical Euler angles (αhβhγh) for describing the orientation of helices across junctions[15]; αh and γh specify a twist around the axis of each of the two helices and βh specifies an inter-helical bend angle (Figure 2b). The authors measured the inter-helical Euler angles for all two-way junctions in the protein databank (PDB) and found that the range of orientations sampled was restricted to ~1%–4% of the total space (Figure 2a). Increasing the length of the internal loop resulted in an increase in the range of orientations sampled, whereas increasing the asymmetry resulted in a systematic shift in the inter-helical twist angle (αh + γh) by an amount comparable to the helical periodicity (~33°) (Figure 2a). The authors rationalized these observations by computing the allowed inter-helical orientations subject to two trivial topological constraints: helices cannot collide with one another and cannot assume conformations that cannot be satisfactorily linked based on the length of the junction. The computed space sampled only 4%–20% and on average 7% (for internal loops <4 nucleotide long) of the total space, yet it accommodated ~85% of all orientations observed in the PDB (Figure 2a). The systematic ~33° shift in αh + γh was rationalized by the tendency of junction residues to maximize non-canonical base-pairing, which in turn leads to increased inter-helical over twisting.
Recent follow up studies by Mustoe et al[16] showed that nucleotides within two-way junctions have a strong bias towards looping into the junction to maximize stacking interactions. Manual annotation of several hundred RNA structures revealed that the majority of these junctional residues form non-canonical base-pairs. This makes it possible to assign a ‘default’ bulge topology for any two-way junction, and to thereby compute the topologically allowed inter-helical space for any two-way junction based on secondary structure alone. Notable exceptions to this ‘default’ behavior are cases in which tertiary interactions stabilize looped out junctional nucleotides[16]. Residues in symmetric internal loops tend to form non-canonical base pairs and give rise to a conformational behavior similar to that observed for a continuous A-form helix.
One can expect topological constraints similar to those mentioned above to manifest themselves in higher-order junctions. Early studies on a small number of model systems showed that helices in such junctions tend to adopt a limited set of conformations often involving coaxial alignment of one or two pairs of helices[5]. The ever-growing number of X-ray structures of higher order junctions provide support for these early observations. For example, Lescoute and Westhof[17•] showed that three-way junctions are dominated by only three classes of global structure and suggested that global orientation may be predicted based on secondary structure alone. Laing and Schlick[18] analyzed four-way junctions and showed correlations between global structure and secondary structure, where helices with short single-stranded connectors are more likely to coaxially stack. Interestingly, if one defines the helices as H1, H2, H3, and H4 according to the order they are encountered when following the RNA strand from the 5'→3' direction, one finds a strong tendency for H1 to stack on H4 and H2 to stack on H3. The authors speculate that this is due to the right-handedness of A-form helices. Laing et al’s follow up studies on higher order junctions also revealed similar helical motifs[19].
A deeper appreciation of these topological constraints is making it possible to better interpret mutagenesis data. A salient example is a recent study by Zhang et al[20•] on a long five nucleotide pyrimidine rich J2ab bulge in human telomerase. Using RDC NMR analysis, the authors found that the bulge induces a sharp and flexible bend of ~90° in a manner that appears to be relatively independent of sequence. Decreasing the bulge length resulted in a marked decrease in telomerase activity. The authors conclude that the bulge is necessary for both inducing a highly bent structure and allowing for dynamic adaptation needed to guide telomerase activity.
Interplay Between Junction-included Topological Constraints and Other Interactions
Recent studies are providing insights into the interplay between topological constraints encoded by RNA inter-helical junctions and other interactions. Most notably, it is now becoming apparent that secondary structure encoded topological constraints play a prominent role specifying the geometric positioning of groups involved in key tertiary contacts. For example, simulations by Reymond et al[21] show that constraining the HCV ribozyme to its secondary structure results in a global fold that places distant loops, which participate in a tertiary pseudoknot motif, in close proximity to one another. This suggests that the topology of the RNA secondary structure is optimized to stabilize specific tertiary interactions. Indeed, prior studies by the Herschlag group on model PEG tethered duplexes[13••] showed that junction-encoded topological constraints can geometrically influence the ability of an RNA to form tertiary contacts. Specifically, the authors showed that the probability of forming hypothetical tertiary contacts between two helices depends on the specific geometrical position of the contact relative to the inter-helical junction and that this dependence weakens when linking helices by a single rather that double-stranded PEG tether (Figure 3). The study by Mustoe et al[16] mentioned above showed that the reverse scenario can also occur. Tertiary contacts can modify junction topology by inducing the looping out of residues that would otherwise participate in non-canonical base-pairing. The modified junction topology can in turn allow access to inter-helical conformations that would be inaccessible to the ‘default’ junction topology.
Figure 3.
Determining backbone global conformation of (a) tRNAAsp and (b) Tetrahymena ribozyme P4–P6 domain based on secondary structure constraints and limited tertiary contacts. Predicted structures (in red) are superimposed onto the native X-ray structure (in blue). Adapted from [34•] and [29•] and (Flores, Sherman, Bruns, Eastman, and Altman, Transactions on Computational Biology and Bioinformatics, 2010, in press). Secondary structure motifs within green boxes correspond to motif environments that were subjected to constraints derived from comparison to independent crystal and NMR structures. Blue bars between bases correspond to imposed stacking interactions and red connectors denote imposed tertiary contact constraints.
While topological constraints can help define a range of inter-helical orientations that can be sampled by a given junction, other interactions can selectively stabilize specific conformers from this allowed range. One example is sequence-specific stacking interactions. By replacing a flexible AU base pair flanking a trinucleotide bulge in HIV-1 TAR with a stronger GC base-pair, Stelzer et al[22•] engineered a TAR mutant in which the helices coaxially stack in a rigid conformation, rather than adopt the bent and flexible conformation of wild-type TAR. Another example is interactions with the ion atmosphere [5,7,23]. In their same study on PEG linked-DNA duplexes, Chu and Herschlag[13••] showed that at low ionic strength, electrostatic repulsion between the helices limits the range of accessible conformations, but that increasing the ionic strength allows access to greater regions of the topologically allowed space. Other examples from recent work completed independently by the Woodson and Draper groups reveal that molecular crowding and some osmolytes lead to stabilization of more compact RNA conformations[24,58], presumably by selecting more compact conformations from the topologically allowed distribution.
From RNA Secondary Structure to 3D Conformation
A growing number of computational modeling studies are taking advantage of topological constraints in making useful inferences about RNA backbone conformation based primarily on secondary structure information. These are exciting developments considering recent advances that make it possible to determine secondary structures of very large, complex, and transient RNAs that do not lend themselves to high-resolution structure determination by NMR spectroscopy and X-ray crystallography[25]. In what follows, we review such applications; not reviewed are other approaches for atomic-level RNA structure prediction, which so far have primarily been applied to smaller (<50 nts) systems [26,27].
Weeks and Dokholyan[28••] explored the extent to which secondary structure constraints can help define RNA backbone conformation. They performed a series of computational simulations using a coarse-grained discrete molecular dynamics model to generate structural ensembles for RNAs ranging in size between 27 and 161 nucleotides. Inclusion of secondary structure constraints alone was enough to reduce the ensemble’s mean pairwise root-mean-square-deviation (RMSD) from ~30 to ~15 Å for a 100 nt system. Without tertiary restraints the structure of the P4–P6 domain (160 nt) from the Tetrahymena ribozyme (TR) is ill-defined and Flores et al’s RNABuilder software [29•] returns a ‘predicted’ structure with 45 Å all P RMSD from the native structure [29•]. Similarly, structures of TR P4–P6 predicted using FARNA [30] and secondary structure constraints superimpose upon the X-ray structure with all-C4' RMSDs ranging between 12 Å and 55 Å, and on average ~30 Å [31••]. Even for the smaller example of tRNAAsp (75 nt), 3D structure predictions using the DMD software [32] with secondary structure alone produce an 11.0 Å RMSD from the X-ray structure when considering phosphorus atoms of base paired nucleotides only[33].
Interestingly, much of the remaining uncertainty in defining RNA backbone conformation can be lifted using a limited number of strategically well chosen ‘other’ readily accessible constraints. Examples of these include tertiary contacts deduced by footprinting experiments, thermodynamic calculations, covariation/mutational analysis[29•,34•] as well as SAXS [35], Cryo-EM and NMR RDC data[36]. The addition of such constraints has been shown to improve the precision of candidate models for all phosphorous RMSDs to <10 Å. For example, by including constraints from ~5 tertiary contacts, Lavender et al[37•] built models for tRNAAsp (75 nt) and Thermus thermophilia group I intron P4–P6 domain (158 nt) with all phosphorus atom RMSDs relative to the X-ray structure of 6.4 Å and 11.3 Å, respectively. The agreement improves to 3.6 Å and 5.4 Å for the smaller HHR (67 nt) and CrPV (49 nt) RNA, respectively. Likewise, using constraints inferred from a limited number of tertiary contacts, stacking interactions, and NMR data, Flores and Altman [29•] generated models for tRNAPhe (76 nt) and the Tetrahymena group I intron P4–P6 domain (160 nt) with all phosphorus atom RMSDs of 9.6 Å and 10.0 Å respectively. As shown in Figure 4, these predictions accurately capture the global conformation of the molecules and represent the best predictions to date that have been achieved for these systems with less than ~10 tertiary constraints. Additional results obtained from Yang et al [35] and Jonikas et al [38], while achieving lower accuracy, demonstrate the robustness of this approach across varied sets of prediction algorithms and tertiary constraints. As might be expected, increasing the density of other constraints can improve the precision of models of all phosphorous RMSDs to <5 Å. For example, Wang et al[39] used a combination of secondary structure, SAXS data, and NMR RDCs to construct a 3D model of the riboA RNA (71 nt) that superimposed with 3.0 Å backbone-RMSD of the X-ray structure.
Figure 4.
Folding simulations of model HJH motifs demonstrate the formation of tertiary contacts is selected based on contact location linker – single PEG linker (sPEG) and double PEG linkers (dPEG) – properties. (a) Schematic representation of the sPEG HJH motif in a side-by-side helical configuration. Tertiary contacts, highlighted by diamond, circle, and triangle, were placed in three separate locations and investigated computationally. (b) Comparison of the fraction folded sPEG (grey) and dPEG (black) in 1 M monovalent salt concentration for three tertiary contact. The two junction topologies have different conformational preferences (sPEG prefers the diamond position, dPEG prefers circle) and folding specificities (sPEG is more receptive to tertiary contact location) which results in differences of folded fractions. Adapted from [13••].
As such, new biochemical approaches have been and are being developed to rapidly obtain distance constraints. In the so-called MOCHA method developed by Das et al [31••], modified nucleotides are randomly incorporated into an RNA molecule during transcription, tethered to Fe(II)-EDTA moieties, and then allowed to generate random cleavages that occur within 25 Å radius of the modified nucleotide. Two dimensional gel electrophoresis separation is used to locate the site of cleavage of the modified nucleotide, providing pairwise distance constraints of ~25 Å. Using this approach, the authors were able to measure 77 distance constraints in Tetrahymena group I intron P4–P6 domain and thereby generate a structure model with an all-C4' atom RMSD accuracy of 13.1 Å. Gherghe et al [40] used a sequence specific intercalating agent, MPE, bound to Fe(II)-EDTA moieties to perform cleavage experiments that provide distance constraints between the engineered intercalation site and nucleotides within ~30 Å. Application of this method to tRNAAsp yielded a total of ~200 distance constraints. These were combined with secondary structure as inputs to their DMD model, giving a prediction accurate to 4.0 Å all-P RMSD relative to the native structure.
Studies have also been recently reported of models of larger RNAs for which high resolution X-ray or NMR structures are currently not available. As an example, Lavender et al[34•] used a secondary structure along with six tertiary constraints to build a model for the 120 nucleotide HCV IRES pseudoknot domain that is consistent with hydroxyl radical cleavage patterns and cryo-EM electron density maps. Lipfert et al[41] were able to construct a model of the 192 nt VS ribozyme using manual fitting of secondary structure into a SAXS electron density profile with incorporation of tertiary contacts known to exist from biochemical experiments. Zhang et al used a combination of secondary structure and RDCs with the MC-Sym software to build a model for the human telomerase core domain [20•]. At an even larger scale, the Harvey group has been utilizing the YAMMP software along with hypothetical secondary structures to predict the structures viral genomes [42].
Another particularly interesting application of topological constraints, which involves the use of secondary and tertiary structure constraints to produce 3D RNA structures was demonstrated by the Perreault group [21] and the Herschlag group [31••]. Using prior experimental data on the tertiary contacts present in different intermediates along the folding pathway of the HDV ribozyme Reymond et al were able to use the MC-Sym software to make predictions about the 3D conformation of these intermediates and infer the 3D structural changes that occur during folding. Similarly, Das et al[31••] used MOCHA pairwise distance constraints obtained at different salt concentrations to build NAST models of the unfolded and non-native, compact states of the P4–P6 domain of the Tetrahymena ribozyme. While molecular dynamics simulations could theoretically provide information on structures of unfolded states (and to higher resolution), the computation at this time is infeasible. These studies highlight the power of using secondary structure along with tertiary constraints to address questions concerning RNA structure that would be otherwise intractable. With accurate secondary structures now obtainable for molecules as large as viral genomes [43,44], we anticipate the further development and application of these methods towards addressing 3D structural characteristics of truly large genomic RNA structures.
Folding on Frustrated Energy Landscapes: Role of Topology
The formation of competing helices during RNA co-transcriptional folding is expected to give rise to topological constraints that may in turn steer folding pathways [45]. So-called Gō models are proving to be a useful tool to explore these aspects of RNA folding. Gō models have been utilized in protein folding studies to delineate dominant folding pathways in cases where the formation of elements of structure, e.g., domains, secondary structural components, etc, compete to give rise to complex folding kinetics[46–48]. These models are parameterized to only include attractive non-bonded forces that are present in the native structure; all other non-bonded interactions are treated as repulsive Lennard-Jones potentials. Simulations with these models are especially well suited to studying how formation of secondary structure elements, and the topological constraints they induce, in differential orders impacts overall folding of the RNA.
For instance, Thirumalai and coworkers used coarse-grained Gō models to explore the folding of three RNA pseudo-knots from the viral genomes of MMTV and SRV-1 and from human telomerase [49]. By combining thermodynamic based studies with kinetic folding studies Cho et al[49], observe that the relative stabilities of the isolated helices control the folding mechanisms. The kinetic simulations they performed corroborate inferences drawn from the free energy profiles, suggesting that MMTV folds by a hierarchical mechanism with parallel pathways wherein the formation of one helix nucleates the assembly of the remainder of the structure. In the SRV-1 pseudo-knot they observe a more cooperative folding process in which preformed helices simultaneously consolidate the tertiary structure. Folding occurs by multiple pathways in the hTR pseudo-knot, and the potential for topological frustration is evident in the observation of a competing interaction pathway involving differing parts of the tertiary structure.
Moreover, Gō models have also been successfully applied to studies of riboswitches, a class of RNA molecules found on the 5' end of mRNAs that have several competing secondary structures where the choice of one structure or the other governs gene expression. Studies by Lin et al [50••] indicate that folding of the three-helical Ariboswitch aptamer proceeds hierarchically as determined by the stability of the helical elements. Furthermore, ligand binding can act to stabilize a meta-stable state on a time frame long enough to commit the RNA to a given secondary structure thereby preventing it from forming alternative, competing structures.
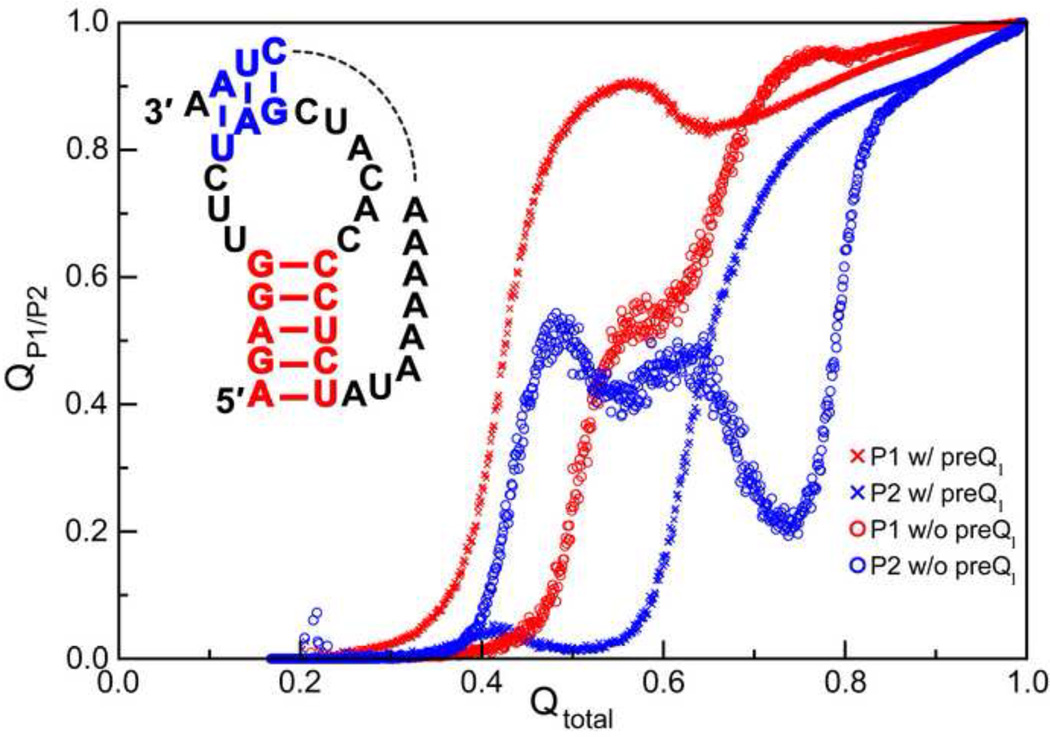
Recently, the preQ1 riboswitch has also been examined using an all-atom Gō model by Feng et al [51]. The authors found that folding was highly cooperative in the presence of metabolite and involved the sequential formation of helix P1 followed by the docking of helix P2 (Figure 5) [51]. These observations are consistent with the idea of local stability determining folding order as observed by Cho et al. [49] and Lin and Thirumalai [50••]. However, in the absence of preQ1, formation of P1 is less facile and competition for formation of helical interactions in P2 creates a topological trap for structure formation, where premature formation of P2 requires prior unfolding of P2 for assembly of the P1 helix (Figure 5). Clearly, preQ1 interactions with the P1 helix further stabilize this helix and nucleate the formation of P1 prior to the establishment of interactions involving the P2 helix.
Figure 5.
Formation of helical elements in the preQ1 riboswitch. All-atom Go model folding simulations of the preQ1 riboswitch showing formation of the P1 (red) and P2 (blue) helix contacts as a function of the total native contacts in the presence and absence of preQ1 ligand. Results are averaged over 78 kinetic folding simulations. Adapted from Feng et al [51].
Similar to the preQ1 system, all-atom Gō models studies of the S-adenosylmethionine-1 (SAM-1) riboswitch found that formation of the nonlocal helix (P1) is rate-limiting in aptamer domain formation [52•]. In the absence of metabolite the 3' end of the P1 helix is sequestered by involvement in an alternative helix. Binding of SAM-1 induced the switching of the conformational states and provides a mechanistic explanation of expression platform regulation. The authors also observe an interesting interplay between the formation of helix P1 and a neighboring pseudo-knot. In a trend that is particularly apparent in their kinetic simulations they find that partial unfolding of the pseudo-knot domain is correlated with formation of helix P1, which then itself must partially melt to allow cooperative refolding of the two elements. This surprising result suggests that the prior formation of the pseudo-knot results in a topological trap or knot that must be undone in order to form the final folded structure.
Dynamics and Conformational Adaptation
Most regulatory RNAs not only have to fold into specific 3D structures, they also have to retain a degree of flexibility to allow their structure to change adaptively during ribonucleoprotein assembly, recognition, catalysis, and signaling. Recent approaches that employ NMR[53] and SAXS[54] are providing insights into the nature of RNA dynamic ensembles and topological constraints and providing a framework for rationalizing these observation.
For example, domain-elongation RDC NMR studies by Zhang et al[55] and subsequent studies combining NMR and MD simulations by Stelzer et al[56] showed that two helices in HIV-1 TAR RNA dynamically sample a limited range of the total inter-helical space forming a distribution in which the helices twist in a correlated manner. Decreasing the bulge length resulted in a more confined distribution and greater motional correlations. These aspects of the inter-helical distributions could readily be rationalized based on the topologically allowed inter-helical space [14••]. The computed space closely coincided with that of the dynamic distribution, implying that the two helices efficiently explore all topologically allowed inter-helical orientations (see Figure 2b).
A number of other studies are also revealing a link between topological constraints and conformational adaptation. By changing the sequence-identity of Watson-Crick base-pairs neighboring the HIV-1 TAR bulge, Stelzer et al[22•] engineered an HIV-1 TAR mutant that favors a coaxial conformation similar to that observed for TAR when bound to ligand mimics of its cognate target Tat. By biasing the inter-helical distribution to the bound conformation, the authors showed that the TAR mutant bound to a Tat ligand mimic with slightly increased affinity without affecting the resulting ligand bound TAR conformation.
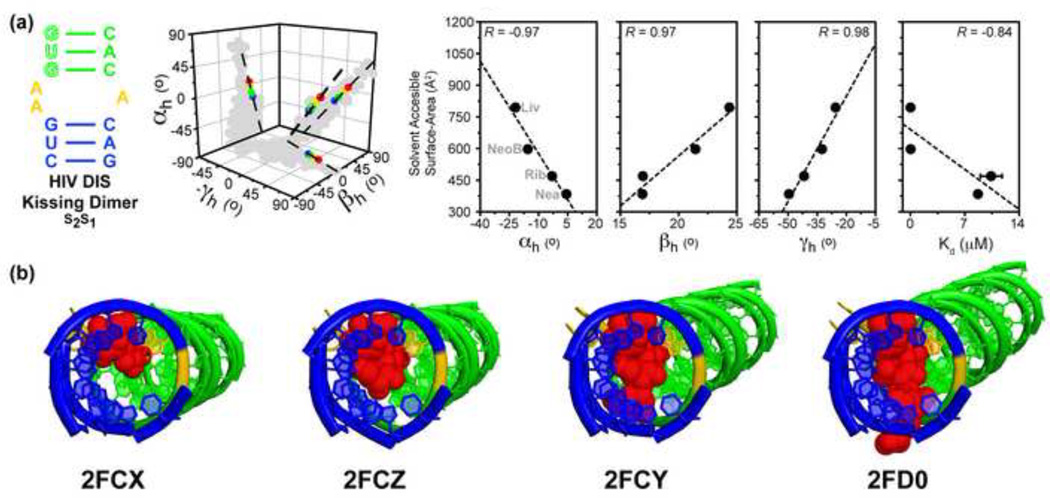
Finally, a study by Bailor et al[14••] suggests that topology not only helps define dynamic aspects of RNA global structures, but also, which structures are selected and stabilized upon binding to specific ligands. Comprehensive analysis of RNA structures bound to small molecules that differ in size, shape, and charge, revealed that small changes in the chemical structure of small molecules leads to stabilization of slightly different RNA structures within the allowed topological space. In particular, a linear correlation was observed between the small molecule size and the inter-helical angles describing the bound RNA inter-helical conformation, with larger small molecules stabilizing more bent and twisted conformations (Figure 6). Conversely, changing other aspects of the small molecule, such as its charge, without affecting size, did not lead to sizeable changes in the bound RNA conformation. Close inspection of X-ray structures of RNA complexes reveals that small molecules tend to act as a wedge between helices, thus inducing bending along the topologically allowed pathway by amounts dependent on their size (Figure 6). These results suggest that different RNA conformations from a topologically allowed distribution can be selected by the added topological constraints arising from the interactions with small molecule ligand.
Figure 6.
Size-encoded adaptation of RNA by conformational selection using small molecules. (a) Inter-helical orientation maps of small-molecule bound RNA crystal structures for HIV DIS kissing dimers (2FCX, 2FCY, 2FCZ, 2FD0) according to solvent accessible surface area (SAS) of the ligand. Predicted junction-allowed topological distributions (in grey) for corresponding two-way junctions of type 2–1 in the “free” state are shown in the cube plot. Shown to the right are correlation plots between each small molecule’s SAS and the corresponding bound RNA’s inter-helical angles (αhβhγh) and Kd. (b) X-ray structures of two-way RNA junction from a are shown bound to their small molecules in order of increasing size. Aminoglycosides are shown in red.
Future Perspective.
On one hand, studies are providing fundamental new insights into topological constraints and how they vary with RNA secondary structure. As a result, we are now in a much better position to appreciate why a given RNA secondary structure conserves aspects of topology such as length of helices and asymmetry of junctions. Likewise, we are in a better position to make and interpret mutations. Although we are far from it, it seems that a complete and quantitative understanding of junction behavior is within our reach. The major challenges are to better understand, and indeed predict, sequence specific interactions involving junction residues and flanking base-pairs, and of course, to properly take into account interactions involving the ion atmosphere and bound metals. Studies should pursue a deeper understanding of these basic interactions while also making interesting and bold predications about RNA conformation. Further studies are also needed to explore the potentially very significant role topological constraints can play in steering the co-transcriptional folding pathways of RNA.
On the other hand, studies are already taking advantage of topological constraints encoded by the RNA secondary structure to make predictions about 3D conformation and dynamic-adaptation. Recent results show unequivocally that this approach has merit. Looking ahead, we can anticipate that low-resolution models of RNA backbone conformation will routinely be depicted, alongside secondary structure, regardless of its size and complexity. Such models will provide a starting framework for interpreting data and for informing study designs. These approaches will likely be the first to unveil new principles of RNA architecture that manifest at much larger length scales and to provide the first insights into higher order genomic structure. A major challenge will be to bridge the scale back to the atomic level. Here, advances in RNA atomic-resolution structure prediction will undoubtedly play an important role.
But perhaps the most important and gratifying aspect of topological constraints is that we can look at a given RNA secondary structure and better understand, and indeed appreciate, how evolutionary pressures on folding, structure, and dynamic adaptation led to its selection.
Highlights.
Secondary structures can define RNA global architecture and dynamic adaptability.
Tertiary interactions select for specific conformers from structural ensembles.
New structure prediction approaches are utilizing topological derived constraints.
Topological constraints rationalize and describe RNA dynamics and adaptation.
Topological constraints may direct RNA down specific folding pathways.
Acknowledgements
AMM acknowledges support from a Graduate Research Fellowship from the National Science Foundation. CLB acknowledges support for the Center for Multi-scale Modeling Tools in Structural Biology (MMTSB) from the NIH through grant number RR012255. HMA acknowledges support NSF CAREER award (MCB 0644278) and an NIH grant (R01GM089846).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murthy VL, Srinivasan R, Draper DE, Rose GD. A complete conformational map for RNA. J Mol Biol. 1999;291:313–327. doi: 10.1006/jmbi.1999.2958. [DOI] [PubMed] [Google Scholar]
- 2.Musselman C, Pitt SW, Gulati K, Foster LL, Andricioaei I, Al-Hashimi HM. Impact of static and dynamic A-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J Biomol NMR. 2006;36:235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 3.Riordan FA, Bhattacharyya A, McAteer S, Lilley DM. Kinking of RNA helices by bulged bases, and the structure of the human immunodeficiency virus transactivator response element. J Mol Biol. 1992;226:305–310. doi: 10.1016/0022-2836(92)90947-i. [DOI] [PubMed] [Google Scholar]
- 4.Lilley DM, Clegg RM, Diekmann S, Seeman NC, Von Kitzing E, Hagerman PJ. A nomenclature of junctions and branchpoints in nucleic acids. Nucleic Acids Res. 1995;23:3363–3364. doi: 10.1093/nar/23.17.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lilley DM. Structures of helical junctions in nucleic acids. Q Rev Biophys. 2000;33:109–159. doi: 10.1017/s0033583500003590. [DOI] [PubMed] [Google Scholar]
- 6.Hsieh CH, Griffith JD. Deletions of bases in one strand of duplex DNA, in contrast to single-base mismatches, produce highly kinked molecules: possible relevance to the folding of single-stranded nucleic acids. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:4833–4837. doi: 10.1073/pnas.86.13.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zacharias M, Hagerman PJ. The influence of symmetric internal loops on the flexibility of RNA. J Mol Biol. 1996;257:276–289. doi: 10.1006/jmbi.1996.0162. [DOI] [PubMed] [Google Scholar]
- 8.Zacharias M, Hagerman PJ. Bulge-induced bends in RNA: quantification by transient electric birefringence. J Mol Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- 9.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 11.Williamson JR. Induced fit in RNA-protein recognition. Nature Structural Biology. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 12.Lilley DM. Structure, folding and mechanisms of ribozymes. Curr Opin Struct Biol. 2005;15:313–323. doi: 10.1016/j.sbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 13. Chu VB, Lipfert J, Bai Y, Pande VS, Doniach S, Herschlag D. Do conformational biases of simple helical junctions influence RNA folding stability and specificity? RNA. 2009;15:2195–2205. doi: 10.1261/rna.1747509. A study combining molecular dynamics simulations and SAXS measurements on B-DNA duplexes linked by PEG tethers provides fundemental insights into the nature of topological contraints, their interplay with ion atmosphere, and their role in directing the specificty of tertiary interactions.
- 14. Bailor MH, Sun X, Al-Hashimi HM. Topology links RNA secondary structure with global conformation, dynamics, and adaptation. Science. 2010;327:202–206. doi: 10.1126/science.1181085. A simple topological model is used to rationalize the anistropic confinement of inter-helical orientations across two-way junctions in the PDB as well as the differential adaptive changes in inter-helical orientations that follow binding of related ligands.
- 15.Bailor MH, Musselman C, Hansen AL, Gulati K, Patel DJ, Al-Hashimi HM. Characterizing the relative orientation and dynamics of RNA A-form helices using NMR residual dipolar couplings. Nat Protoc. 2007;2:1536–1546. doi: 10.1038/nprot.2007.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mustoe AM, Bailor M, Brooks CL, III, Al-Hashimi HM. On the Role of Topological Contraints in Defining RNA Backbone Conformation. 2011 In submission. [Google Scholar]
- 17. Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. Rna-a Publication of the Rna Society. 2006;12:83–93. doi: 10.1261/rna.2208106. A survey of thee-way junctions in the PDB shows that there is a limited set of global helix conformations adopted by these higher-order junctions.
- 18.Laing C, Schlick T. Analysis of four-way junctions in RNA structures. Journal of Molecular Biology. 2009;390:547–559. doi: 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laing C, Jung S, Iqbal A, Schlick T. Tertiary motifs revealed in analyses of higher-order RNA junctions. Journal of Molecular Biology. 2009;393:67–82. doi: 10.1016/j.jmb.2009.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Q, Kim NK, Peterson RD, Wang Z, Feigon J. Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA. Proc Natl Acad Sci U S A. 2010;107:18761–18768. doi: 10.1073/pnas.1013269107. Experimental characterization of the J2a/b five nucleotide bulge in human telomerase shows that the number nucleotides in the bulge defines global structure and dynamics and is intimately linked to determining telomerase activity.
- 21.Reymond C, Levesque D, Bisaillon M, Perreault JP. Developing three-dimensional models of putative-folding intermediates of the HDV ribozyme. Structure. 2010;18:1608–1616. doi: 10.1016/j.str.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM. RNA Dynamics by Design: Biasing Ensembles Towards the Ligand-Bound State. Angew Chem Int Ed Engl. 2010 doi: 10.1002/anie.201000814. Changing the identity of a base-pair flanking a bulge junction biases the average inter-helical structure and dynamics towards a ligand bound state, resulting in higher affinity ligand binding.
- 23.Casiano-Negroni A, Sun X, Al-Hashimi HM. Probing Na(+)-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: new insights into the role of counterions and electrostatic interactions in adaptive recognition. Biochemistry. 2007;46:6525–6535. doi: 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilburn D, Roh JH, Guo L, Briber RM, Woodson SA. Molecular crowding stabilizes folded RNA structure by the excluded volume effect. Journal of the American Chemical Society. 2010;132:8690–8696. doi: 10.1021/ja101500g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol. 20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 28. Hajdin CE, Ding F, Dokholyan NV, Weeks KM. On the significance of an RNA tertiary structure prediction. Rna-a Publication of the Rna Society. 2010;16:1340–1349. doi: 10.1261/rna.1837410. Simulations show that inclusion of secondary structure constraints greatly improves the quality of RNA tertiary structure predictions.
- 29. Flores SC, Altman RB. Turning limited experimental information into 3D models of RNA. Rna-a Publication of the Rna Society. 2010;16:1769–1778. doi: 10.1261/rna.2112110. These papers (see also Refs [34•,37•]) produce accurate global tertiary structure predictions for RNAs as large as 160 nts utilizing secondary structure and <10 inferred tertiary structure constraints.
- 30.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc Natl Acad Sci U S A. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Das R, Kudaravalli M, Jonikas M, Laederach A, Fong R, Schwans JP, Baker D, Piccirilli JA, Altman RB, Herschlag D. Structural inference of native and partially folded RNA by high-throughput contact mapping. Proc Natl Acad Sci U S A. 2008;105:4144–4149. doi: 10.1073/pnas.0709032105. The use of secondary structure and distance constraints obtained from cleaavage experiments is able to produce an accurate global tertiary structure of the P4–P6 domain of the Tetrahymena ribozyme. The authors are also able to produce models of different structural intermediates using the same procedure at different salt conditions.
- 32.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–1173. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J Am Chem Soc. 2009;131:2541–2546. doi: 10.1021/ja805460e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lavender CA, Ding F, Dokholyan NV, Weeks KM. Robust and Generic RNA Modeling Using Inferred Constraints: A Structure for the Hepatitis C Virus IRES Pseudoknot Domain (vol 49, pg 4931, 2010) Biochemistry. 2010;49:5968–5968. doi: 10.1021/bi100142y. See annotation for Ref. [29•].
- 35.Yang SC, Parisien M, Major F, Roux B. RNA Structure Determination Using SAXS Data. Journal of Physical Chemistry B. 2010;114:10039–10048. doi: 10.1021/jp1057308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Zuo X, Yu P, Xu H, Starich MR, Tiede DM, Shapiro BA, Schwieters CD, Wang YX. A method for helical RNA global structure determination in solution using small-angle x-ray scattering and NMR measurements. J Mol Biol. 2009;393:717–734. doi: 10.1016/j.jmb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lavender CA, Ding F, Dokholyan NV, Weeks KM. Robust and generic RNA modeling using inferred constraints: a structure for the hepatitis C virus IRES pseudoknot domain. Biochemistry. 2010;49:4931–4933. doi: 10.1021/bi100142y. See annotation for Ref. [29•].
- 38.Jonikas MA, Radmer RJ, Laederach A, Das R, Pearlman S, Herschlag D, Altman RB. Coarse-grained modeling of large RNA molecules with knowledge-based potentials and structural filters. Rna-A Publication of the Rna Society. 2009;15:189–199. doi: 10.1261/rna.1270809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JB, Zuo XB, Yu P, Xu H, Starich MR, Tiede DM, Shapiro BA, Schwieters CD, Wang YX. A Method for Helical RNA Global Structure Determination in Solution Using Small-Angle X-Ray Scattering and NMR Measurements. Journal of Molecular Biology. 2009;393:717–734. doi: 10.1016/j.jmb.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA Tertiary Structures Using a Sequence-Encoded Cleavage Agent and Refinement by Discrete Molecular Dynamics. Journal of the American Chemical Society. 2009;131:2541–2546. doi: 10.1021/ja805460e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipfert J, Ouellet J, Norman DG, Doniach S, Lilley DMJ. The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure. 2008;16:1357–1367. doi: 10.1016/j.str.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devkota B, Petrov AS, Lemieux S, Boz MB, Tang L, Schneemann A, Johnson JE, Harvey SC. Structural and electrostatic characterization of pariacoto virus: implications for viral assembly. Biopolymers. 2009;91:530–538. doi: 10.1002/bip.21168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thirumalai D, Lee N, Woodson SA, Klimov D. Early events in RNA folding. Annu Rev Phys Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 46.Karanicolas J, Brooks CL., III Integrating folding kinetics and protein function: Biphasic kinetics and dual binding specificity in a WW domain. Proc Natl Acad Sci U S A. 2004;101:3432–3437. doi: 10.1073/pnas.0304825101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hills RD, Jr, Brooks CL., III Subdomain competition, cooperativity, and topological frustration in the folding of CheY. J Mol Biol. 2008;382:485–495. doi: 10.1016/j.jmb.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hills RD, Jr, Kathuria SV, Wallace LA, Day IJ, Brooks CL, III, Matthews CR. Topological frustration in beta alpha-repeat proteins: sequence diversity modulates the conserved folding mechanisms of alpha/beta/alpha sandwich proteins. J Mol Biol. 2010;398:332–350. doi: 10.1016/j.jmb.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho SS, Pincus DL, Thirumalai D. Assembly mechanisms of RNA pseudoknots are determined by the stabilities of constituent secondary structures. Proc Natl Acad Sci U S A. 2009;106:17349–17354. doi: 10.1073/pnas.0906625106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lin JC, Thirumalai D. Relative stability of helices determines the folding landscape of adenine riboswitch aptamers. Journal of the American Chemical Society. 2008;130:14080–14081. doi: 10.1021/ja8063638. Application of coarse-grained Gō models to adenine riboswitches demonstrate that ligand binding can stabilize a meta-stable secondary structure long enough to commit it to a given folding pathway.
- 51.Feng J, Walter NG, Brooks CL., III Cooperative and Directional Folding of the preQ1 Riboswitch Aptamer Domain. J Am Chem Soc. 2011 doi: 10.1021/ja110411m. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY. Nonlocal helix formation is key to understanding S-adenosylmethionine-1 riboswitch function. Biophys J. 2009;96:L7–L9. doi: 10.1016/j.bpj.2008.10.033. All-atom Gō model simulations show that helix formation can act as a conformational switching mechanism. Kinetic simulations also reveal that formation of secondary structures can result in topological frustration where neighboring helical elements must first unfold to allow assembly of downstream helices.
- 53.Frank AT, Stelzer AC, Al-Hashimi HM, Andricioaei I. Constructing RNA Dynamical Ensembles by Combining MD and Motionally Decoupled NMR RDCs: New insights into RNA Dynamics and Adaptive Ligand Recognition. Nucleic Acids Res. 2009;37:3670–3679. doi: 10.1093/nar/gkp156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 56.Stelzer AC, Frank AT, Bailor MH, Andricioaei I, Al-Hashimi HM. Constructing atomic-resolution RNA structural ensembles using MD and motionally decoupled NMR RDCs. Methods. 2009;49:167–173. doi: 10.1016/j.ymeth.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lambert D, Leipply D, Draper DE. The osmolyte TMAO stabilizes native RNA tertiary structures in the absence of Mg2+: evidence for a large barrier to folding from phosphate dehydration. J Mol Biol. 2010;404:138–157. doi: 10.1016/j.jmb.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]