Abstract
Many regulatory RNAs undergo large changes in structure upon recognition of proteins and ligands but the mechanism by which this occur remains poorly understood. Using NMR residual dipolar coupling (RDCs), we characterized Na+ induced changes in the structure and dynamics of the bulge-containing HIV-1 transactivation response element (TAR) RNA that mirror changes induced by small molecules bearing a different number of cationic groups. Increasing the Na+ concentration from 25 mM to 320 mM led to a continuous reduction in the average inter-helical bend angle (from 46° to 22°), inter-helical twist angle (from 66° to −18°) and inter-helix flexibility (as measured by an increase in the internal generalized degree of order from 0.56 to 0.74). Similar conformational changes were observed with Mg2+, indicating that non-specific electrostatic interactions drive the conformational transition, although results also suggest that Na+ and Mg2+ may associate with TAR in distinct modes. The transition can be rationalized based on a population-weighted average of two ensembles comprising an electrostatically relaxed bent and flexible TAR conformation that is weakly associated with counterions, and a globally rigid coaxial conformation which has stronger electrostatic potential and association with counterions. The TAR inter-helical orientations that are stabilized by small molecules fall around the metal-induced conformational pathway, indicating that counterions may help predispose the TAR conformation for target recognition. Our results underscore the intricate sensitivity of RNA conformational dynamics to environmental conditions and demonstrate the ability to detect subtle conformational changes using NMR RDCs.
Keywords: RNA folding, metal binding, electrostatic interactions, induced-fit, tertiary capture, adaptive recognition
Many regulatory ribonucleic acids (RNAs) undergo large changes in conformation upon binding to proteins and small ligand molecules but the mechanism by which this occurs remains poorly understood (1–4). Numerous studies have examined the extent to which molecules induce new RNA conformations versus capture conformations that are dynamically accessible in the unbound state (1, 5–11). In contrast, fewer studies have examined the potential role of monovalent and divalent cations in conformational adaptation and target recognition (12).
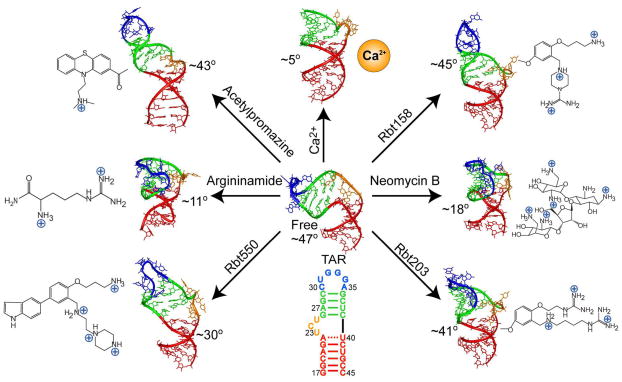
The interaction between the transactivation response element (TAR) RNA (13) from the human immunodeficiency type I virus (HIV-1) and the viral transactivator protein (Tat) is a paradigm for understanding the rules of RNA adaptive recognition (1, 5, 8, 14) and a primary target for developing anti-HIV therapeutics (15–18). Several high-resolution structures have been reported for HIV-I TAR (Figure 1), including the free form (19) and bound to divalent cations (20), peptide mimics of Tat (21, 22), and six distinct small molecules containing a different number of cationic groups designed to inhibit its interaction with Tat (23–26). These TAR conformations differ dramatically both in the global orientation of helices (inter-helical bend angle ranging between ~5° and ~47°) as well as the local conformation of bulge and neighboring residues where molecules bind. Thus, by adopting different conformations, TAR is capable of binding chemically diverse targets (Figure 1).
Figure 1.
Conformation of HIV-1 TAR in free form and bound to distinct molecular targets (19, 20, 22–26). The inter-helical bend angle is indicated next to each conformation.
The molecular basis for TAR conformational adaptation is key for understanding its role in Tat-mediated transcription elongation of HIV-1 genes and for rationally designing inhibitors of its interaction with Tat (17, 18). Electrostatic interactions play important roles in both RNA folding and recognition (27–32) and thus can provide a basis for driving conformational changes accompanying complex formation. Previous studies have emphasized the importance of electrostatic interactions in TAR recognition. The spatial arrangements of basic groups in Tat relative to the negative TAR surface is an important determinant of Tat binding and its induced TAR conformational changes (33). The Tat bound TAR conformation (21, 22) can be stabilized by distinct small molecules that can satisfy two key electrostatic interactions (26). Aminoglycosides bind TAR with affinities that correlate with their total number of positive amines (34). Our NMR studies suggest that the changes in the TAR conformational dynamics induced by small molecules are also correlated to their total number of positive groups (35).
RNA electrostatic hot-spots, which would normally be stabilized by cationic groups on proteins and small molecules, are expected to be energetically unfavorable in the unbound RNA. One can therefore expect that these structural elements only become significantly stabilized during or following complex formation. However, metal cations can also stabilize such electrostatic hot-spots and thus pre-adapt the RNA conformation for target recognition. Several studies have shown that a variety of monovalent and divalent cations can associate with the TAR bulge (20, 36–38). Transient electric birefringence (39) and NMR (40) studies have shown that the addition of Mg2+ results in a significant reduction in the TAR inter-helical bend angle and dynamics (20). However, as is often the case, it remains unclear whether the Mg2+ induced transition is driven by non-specific electrostatic interactions with diffusive counterions, which could be recapitulated by cationic groups on small molecules, or by inner-sphere contacts with specifically bound metals. Another unresolved question which has also proven generally difficult to address is whether the TAR conformations observed at various metal concentrations represent distinct conformations or a dynamical average of two or more states.
A number of studies suggest that the metal-induced TAR structural transition is driven by specific inner sphere contacts. An X-ray structure of TAR shows four specifically bound Ca2+ ions that stabilize a unique UCU bulge conformation through several inner-sphere contacts with TAR ligands (20). Furthermore, transient electric birefringence studies have shown that Mg2+ induced transitions in bulge containing RNAs are not recapitulated by Na+ even when using up to 50-fold higher concentrations (41). On the other hand, EPR studies show that Na+ and Ca2+ induce similar TAR conformational changes (38) but that other metals can induce distinct effects (39).
Metal induced RNA conformational transitions have been studied using a variety of experimental biophysical techniques including EPR (37,38), fluorescence spectroscopy (42), hydroxyl radical footprinting (43), analytical ultracentrifugation(44), small angle x-ray scattering (45), and small angle neutron scattering (46). Although it can uniquely provide information about both structure and dynamics at site-specific resolution, few studies have employed NMR to structurally characterize metal-induced RNA transitions. This is in part because conventional NOE derived distance restraints can be insensitive to global conformational changes (47) and because high-resolution structure determination remains time consuming which makes characterizing structures at multiple metal concentrations impractical (48, 49).
In this study, we exploit the exquisite conformational sensitivity of NMR residual dipolar couplings (RDCs) (50–53) to characterize how the structure and dynamics of TAR change as a function of increasing Na+ concentration and compare findings with changes induced by Mg2+ and small molecules. Aiding our study is an order tensor based analysis of RDCs which permits studies of global conformational dynamics with high efficiency (14, 48, 49, 54, 55). The RDCs allowed us to detect Na+ induced changes in the TAR conformation which are indistinguishable from those induced by Mg2+ even though chemical shift mapping data suggest that the two metals may bind TAR using distinct modes. These results together with electrostatic calculations offer new insights into the possible role of electrostatic interactions and counterion condensation in TAR adaptive recognition.
MATERIALS AND METHODS
NMR sample preparation
Samples of uniformly 13C/15N labeled TAR were prepared by in vitro transcription using synthetic double stranded DNA templates containing the T7 promoter and sequence of interest (Integrated DNA Technologies, Inc.), T7 RNA polymerase (Takara Mirus Bio, Inc.), and 13C/15N labeled nucleotide triphosphates (ISOTEC, Inc.). The RNA was purified by 20% (w/v) denaturing polyacrylamide gel electrophoresis containing 8M urea and 1x TBE followed by electroelution in 20 mM Tris pH 8 buffer and ethanol precipitation. The RNA pellet was dissolved and exchanged into NMR buffer (15 mM sodium phosphate, 0.1 mM EDTA, and 25 mM NaCl at pH ~6.4,) using a centricon ultracel YM-3 concentrator (Millipore Corp.). The final RNA concentration in the aligned NMR samples was 0.3/0.5 mM for Na+/Mg2+ respectively. Three separate samples were used to measure RDCs at 160, 320 mM Na+, and 25 mM Na+/4 mM Mg2+. The aligned samples were prepared by adding Pf1 phage (50 mg/ml) (56, 57) in NMR buffer to a pre-concentrated TAR RNA sample to yield a final Pf1 phage concentration of 17–20 mg/ml. The addition of phage did not affect the ‘effective’ Na+ or Mg2+ concentration as judged from careful comparison of the chemical shifts in the absence and presence of phage (see Supporting Information).
Measurement of RDCs
All NMR experiments were performed at 298 K on an Avance Bruker 600 MHz spectrometer equipped with a triple-resonance cryogenic (5mm) probe with the exception of the 320 mM Na+ RDCs which were measured on a Varian Inova 800 MHz spectrometer equipped with a triple resonance Z-gradient probe. NMR spectra were analyzed using NMR Draw (58) or Felix (Accelrys Inc, 2002) and overlaid in Sparky 3 (59). The measurement of RDCs in 25 mM NaCl (14) and in the presence of Mg2+ (40) have been reported previously. The measurements of RDCs in the presence of Mg2+ (0.5 mM RNA and 25 mM Na+/4 mM Mg2+) were repeated using the same experiments and conditions used for the Na+ RDC measurements. Results were consistent with previous values (40). One bond 1DC6H6, 1DC8H8, 1DC5H5, 1DC2H2, 1DC1′H1′, and 1DN1/3H1/3 RDCs were measured using 2D 13C–1H (or 15N-1H) S3E HSQC experiments (35, 60) from the difference in splittings measured along the 13C (or 15N) dimension observed in the presence and absence of Pf1 phage (56, 57). The RDC measurement error was estimated from duplicate measurements using experiments that yield splittings along the 1H and 13C/15N dimension (standard deviation between two RDC sets was 0.9 Hz). The measured RDCs are listed in Supporting Information.
Chemical shift mapping
2D HSQC spectra were recorded following incremental increases in [Na+] (25, 40, 80, 160 and 320mM against 0.2 mM TAR) or [Mg2+] (0, 0.2, 0.4, 0.8, 1.6, 3.2 and 6.4 mM against 0.2mM TAR). Apparent dissociation constants (Kd) were obtained by fitting the observed changes in chemical shift to the equation (65):
| (1) |
where [M]T is the total concentration of metal (Na+ or Mg2+), [RNA]T is the TAR concentration based on UV absorbance at 260 nm, ΔδT is the difference in chemical shifts between the “free” and “metal associated” states (in ppm), δobs is the observed chemical shift (in ppm), and δFree is the chemical shift in the “free” state (in ppm). The data was fitted using the Origin software (OriginLab Corporation) in which ΔδT and Kd (and δFree for Na+) were allowed to vary during the fit. The Kd errors obtained from the fit were in excellent agreement with values obtained independently using a Monte Carlo approach. For the latter, “residuals” corresponding to the difference between the measured and best-fitted chemical shifts in the titration curves were computed for all fitted data. The resulting distribution of residuals was fitted to a Gaussian distribution. Next, simulations were performed in which perfect chemical shift titration data points corresponding to experimental values were generated and each point perturbed by a value randomly chosen from the Gaussian distribution. The perturbed points were then fitted to Equation 1 and the calculations repeated a hundred times. The difference between the “true” Kds used in the simulation and values obtained from curve-fitting were then fitted to a Gaussian distribution and the resulting standard deviation assumed to be the Kd error.
Order tensor analysis of RDCs
The RDCs measured in the TAR helices were subjected to an order tensor analysis using idealized A-form helices as input coordinates (14) as implemented in the program AFORM-RDC (55). The helices were constructed using the Biopolymer module in Insight II (Molecular Simulations, Inc.) followed by correction of propeller twist angles from +15° to −15° (61). The measured RDCs were fitted to idealized A-form helices using singular value decomposition (62) implemented in the in-house written program RAMAH (63). RDCs from terminal residues (17, 45, 22 and 40), bulge residues (23–25), and hairpin loop residues (31–34) were excluded from the analysis. The order tensor errors due to parameterized “A-form structural noise” and RDC uncertainty were estimated using the program AFORM-RDC (55). Key statistics for the order tensor analysis are summarized in Table 1.
Table 1.
Order tensor analysis of TAR RDCs measured under different metal conditions.
| Metal Condition | Stem | N | CN | RMSD (Hz) | R | η | ϑ × 10−3 | ϑint | θ (°) | ξ (°) |
|---|---|---|---|---|---|---|---|---|---|---|
| 25 mM | I | 13 | 2.9 | 1.1 | 0.99 | 0.27±0.04 | 0.48±0.04 | 0.56±0.05 | 46±4 | 61±50 |
| NaCl | II | 11 | 3.1 | 1.1 | 0.99 | 0.11±0.07 | 0.88±0.04 | |||
| 160 mM | I | 15 | 2.9 | 1.3 | 0.99 | 0.15±0.03 | 0.57±0.03 | 0.67±0.05 | 37±7 | −1±50 |
| NaCl | II | 14 | 4.2 | 1.2 | 0.99 | 0.15±0.04 | 0.85±0.04 | |||
| 320 mM | I | 12 | 2.8 | 1.5 | 0.98 | 0.23±0.04 | 0.67±0.04 | 0.74±0.08 | 22±7 | −18±50 |
| NaCl | II | 12 | 4.0 | 1.1 | 0.99 | 0.20±0.04 | 0.90±0.08 | |||
| 4 mM | I | 13 | 2.9 | 1.0 | 0.99 | 0.26±0.04 | 0.80±0.03 | 0.85±0.04 | 17±7 | −58±50 |
| MgCl2 | II | 14 | 4.1 | 1.0 | 0.99 | 0.17±0.08 | 0.94±0.03 |
Shown for each helical stem are the number of RDCs (N), the condition number (CN) (69) describing the orientational spread of the RDC targeted bond vectors, the root-mean-square deviation (RMSD) and correlation coefficient (R) between measured and back-predicted RDCs, the order tensor asymmetry (η = |Syy−Sxx|/Szz), generalized degree of order (ϑ), internal generalized degree of order (ϑint), inter-helical bend (θ) and twist (ξ) angles. Errors are estimated using the program AFORM-RDC.
For each metal condition, helices were rotated into the principal axis system (PAS) of their best-fitted order tensor and assembled using the program Insight II (Molecular Simulations, Inc) by linking U40 (P) to C39 (O3′) (~1.59 Å). Due to the order tensor degeneracy (64), this yielded four possible inter-helical orientations, three of which could be discarded; two because they lead to anti-parallel helix alignments and one because it resulted in a distance between A22 (O3′) and G26 (P) (>30 Å) that cannot be satisfactorily linked using the trinucleotide bulge. Inter-helical angles for all TAR structures were calculated using an in-house program.
Electrostatic calculations
HIV-1 TAR RNA structures in free form (PDB ID#1ANR) and bound to argininamide (PDB ID#1ARJ), Ca2+ (PDB ID#397D), acetylpromazine (PDB ID#1LVJ), neomycin B (PDB ID#1QD3), Rbt158 (PDB ID#1UUI), Rbt203 (PDB ID#1UUD) and Rbt550 (PDB ID#UTS1) were obtained from the Protein Data Bank and used in the analysis. Electrostatic calculations were performed by solving the non-linear Poisson Boltzmann equation using the Delphi (66) module of Insight II (Molecular Simulations, Inc). No bound waters, ions or ligands were included in the calculation. The DNA-RNA AMBER force field (67) was used for partial charges of atoms. An interior dielectric constant of 2 was used for the RNA molecule. The continuum dielectric constant for the solvent (water) was set to 80 with a 1:1 electrolyte distribution according to the Boltzmann weighted average of the mean potential. A 2.0 Å exclusion radius was added to the surface of the RNA to account for ion size and a 1.4 Å probe used to determine the RNA molecular surface. A monovalent salt concentration of 0.025 M was used in the calculations to mimic the low ionic strength NMR conditions. 3D structures were mapped onto a grid (65 × 65 × 65 grid points/side) and the boundary potential at each lattice calculated using the Debye-Huckel and full Columbic equations implemented in Delphi (66). The calculated electrostatic potential maps were displayed using the program GRASP provided by the Honig lab (66).
RESULTS AND DISCUSSION
Chemical shift mapping of Na+ and Mg2+ association with TAR RNA
As shown in Figure 2A, large and specific changes in chemical shift were observed in 2D HSQC spectra of TAR (0.3 mM) upon incrementally increasing the NaCl concentration from 25 mM to 320 mM in a background buffer containing 15 mM sodium phosphate and 0.1 mM EDTA at pH ~6.4. The largest chemical shift changes were observed in and around the bulge. In contrast, little to no changes were observed for residues in the UUCG loop. For the majority of resonances, the directions of the chemical shift perturbations were similar to those induced by Mg2+ although the magnitude of the latter was uniformly larger (Figure 2A). As shown in Figure 2B, fitting of the chemical shift titration data to a two-state model yields apparent Kds that are more than two to orders of magnitude smaller for Mg2+ (Kd ~0.074 ± 0.002 – 0.10 ± 0.01 mM) compared to Na+ (Kd ~0.11 ± 0.01 −0.28 ± 0.09 M) as has been reported for other RNAs. No further changes in the TAR chemical shifts were apparent beyond 6.4 mM Mg2+ which is consistent with saturation kinetics and specific metal-induced transition. In the case of Na+, chemical shift data could not be recorded near saturation point as this would require salt concentrations (~ 1M) outside the tuning capacity of our probe.
Figure 2.
Chemical shift mapping of Na+ and Mg2+ association with TAR RNA. On the left, the secondary structure of the TAR construct used in this study. The wild-type loop is replaced with a UUCG loop. (A) 2D HSQC spectra of TAR recorded in the presence of Na+ (25 mM, 160 mM and 320 mM) and Mg2+ (25 mM Na+/4 mM Mg2+). Residues undergoing the largest chemical shift perturbations (top 20% for a given type of resonance) upon Na+ and Mg2+ binding are highlighted in blue and purple square boxes respectively on the TAR secondary structure. (B) Representative Na+ and Mg2+ titration curves with Kd values ( in M for Na+ and mM for Mg2+) shown at the end of each curve.
Despite many similarities, subtle but detectable differences between the Mg2+ and Na+ chemical shift perturbations were observed for residues G26, U23, and A27 (highlighted with a box, Figure 2A). Significantly, these are precisely the residues that are involved in inner sphere contacts with Ca2+ cations in the X-ray structure of TAR (20). Thus, Na+ and Mg2+ either stabilize a different conformation for these residues and/or associate with them in a different manner.
Structure and dynamics of TAR as a function of Na+ and Mg2+ using RDCs
The interpretation of chemical shift perturbations is complicated by the fact that they are sensitive to changes in structure, dynamics, and metal localization. To specifically characterize the metal induced TAR conformational changes, we measured RDCs in TAR (0.3 mM) at Na+ concentration of 25 mM, 160 mM, and 320 mM. Based on the apparent Kds (Figure 2B), these correspond to 10%, 43%, and 60% of TAR being in the Na+ “bound” state. For comparison, RDCs in the presence of 25 mM Na+/4 mM Mg2+ (83% of TAR bound) were measured using the same NMR experiments and sample conditions used to measure the Na+ RDCs. The Mg2+ RDCs were measured slightly below saturation, allowing insight into the dynamical nature of the metal-induced TAR transition under physiological Mg2+ concentrations. The Mg2+ RDCs were in very good agreement with values reported previously at slightly different Mg2+ concentrations (40).
As shown in Figure 3A, continuous and significant changes in RDCs (normalized for differences in total degree of order, see Figure legend) were observed upon increasing the Na+ concentration, providing strong evidence for Na+ induced changes in the TAR conformation. The 320 mM Na+ RDCs are in better agreement with those measured at 25 mM Na+ in the presence rather than in the absence of 4 mM Mg2+ (Figure 3A), indicating that Na+ and Mg2+ induce similar TAR conformational changes.
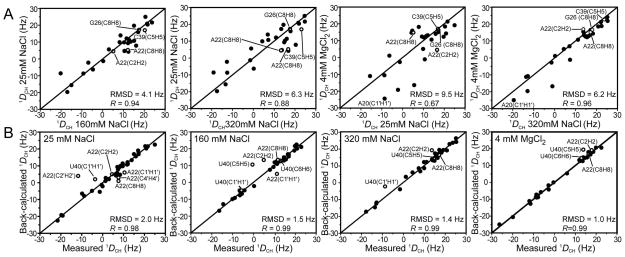
Figure 3.
(A) Comparison of RDCs measured at 25 mM, 160 mM, and 320 mM Na+ and Mg2+ (25 mM Na+/4 mM Mg2+). The RDCs are normalized (relative to 25 mM Na+) to account for differences in the degree of order. This was done by scaling the RDCs measured under salt condition “X” by the ratio of stem II degree of order measured at X and 25 mM (i.e. through multiplication by ϑNa-25mM/ϑX). Stem II was chosen since it dominates the total degree of alignment in all cases (Table 1). (B) Order tensor fits against an idealized A-form geometry carried out independently for stems I and II using RDCs measured at 25 mM, 160 mM, and 320 mM Na+ and 25 mM Na+/4 mM Mg2+. Shown in each case is the root-mean square deviation (rmsd) between measured and back-predicted RDCs as well as the correlation coefficient (R).
To further characterize the metal induced transition, the RDCs were subjected to an order tensor analysis (62, 68, 69). Here, non-terminal Watson-Crick (WC) base-pairs in individual helical stems were modeled assuming an idealized A-form geometry (14, 54, 55). The RDCs and idealized A-form geometry were then used to determine order tensors for each helical stem (statistics summarized in Table 1). As shown in Figure 3B, an excellent order tensor fit was obtained for RDCs measured under all metal conditions examined. In all cases, the root-mean-square-deviation (RMSD) between measured and back-predicted RDCs compared favorably with the RDC measurement uncertainty (0.9 Hz SD from comparison of 1H and 13C dimension RDCs). Thus, the local conformation of the WC pairs does not vary significantly from the ideal A-form geometry. The excellent RDC fits observed for residues G26 and A27 argue that the observed chemical shift differences with Na+ and Mg2+ (Figure 2A) are unlikely due to different metal-induced conformational changes at these sites but rather to differences in the modes of metal association with TAR. Likewise, both Na+ and Mg2+ yield slightly reduced values in the already attenuated bulge RDCs particularly for U23 (see Supporting Information). As mentioned previously (40), this is consistent with an increase in the local flexibility accompanying looped out bulge residues (20). The agreement for the flexible terminal A22-U40 base-pair RDCs (14, 40) (shown as open symbols, Figure 3) which were not included in the order tensor fit, improves with increasing metal concentration (rmsd = 6.9 Hz, 5.1 Hz, and 3.4 Hz at 160 mM, 320 mM Na+ and 25 mM Na+/4 mM Mg2+ respectively). This stabilization is consistent with the observed increase in coaxial stacking between the two stems with increasing metal concentration (see below).
Given the excellent fit to the A-form geometry, the observed changes in RDCs with increasing Na+ concentration must be attributed to changes in the global orientation and/or dynamics of the two helices. To this end, we used the order tensors computed for each helix to determine their relative orientation and dynamics (62, 68) with estimated errors calculated using the program AFORM-RDC, which yields order tensor errors due to a combination of A-form structural noise and RDC uncertainty (55). In this analysis, the relative orientation of helices is obtained by superimposing their order tensor frames describing helix alignment relative to the applied magnetic field. The amplitude of inter-helical motions is computed from the ratio of the generalized degree of order (ϑint = ϑi/ϑj; ϑi < ϑj) describing the degree of helix alignment relative to the applied magnetic field (14, 69). The ϑint value ranges between 1 for inter-helical rigidity and 0 for maximum inter-helical motions. Owing to possible correlations between helix motions and overall alignment, the ϑint value will generally underestimate the real motional amplitudes (14, 70).
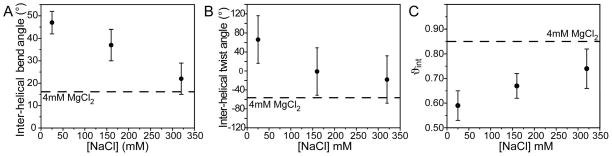
As shown in Figure 4, increasing the Na+ concentration from 25 to 320 mM led to a gradual reduction in the inter-helical bend from θ = 46° ± 4° to 22° ± 7° (Figure 4A). This was accompanied by a reduction in the inter-helical twist angle from ξ = 66° ± 50 to −18° ± 50 (Figure 4B) and amplitude of inter-helical motions from ϑint = 0.59 ± 0.06 to 0.74 ± 0.08 (Figure 4C). Assuming an isotropic cone motional model (13), the ϑint values correspond to inter-helical motional amplitudes of 48°, 41°, and 35° at Na+ concentrations of 25 mM, 160 mM, and 320 mM respectively. The comparatively large error in the inter-helical twist angle reflects the larger uncertainty in the principal Sxx−Syy directions arising due to near axial symmetry (η ~ 0) of the helix order tensors (Table 1). Similar but significantly larger conformational changes were induced by 25 mM Na+/4 mM Mg2+ (θ = 17° ± 7°, ξ = −58 ± 50, and ϑint = 0.85 ± 0.04, shown as horizontal lines in Figure 4). This suggests that the similar Na+ and Mg2+ induced TAR chemical shift perturbations (Figure 2A) likely reflect a similar TAR conformational change that is driven by non-specific electrostatic interactions with counterions.
Figure 4.
Probing the metal induced TAR structure-dynamical transition using an order tensor analysis of RDCs. Shown are the (A) inter-helical bend angle (θ), (B) inter-helical twist angle (ξ) (positive/negative values correspond to over/under twisting respectively), and (C) inter-helical mobility (ϑint) as a function of Na+ concentration. Values in the presence of 25 mM Na+/4 mM Mg2+ are shown using a horizontal line.
A two state non-specific electrostatic switch underlies the metal-induced TAR structure-dynamical transition
The TAR conformations observed at 25 mM, 160 mM and 320 mM Na+ and 25 mM Na+/4 mM Mg2+ may either represent distinct conformations, or a population weighted average of two or more conformational states. Although these scenarios can be difficult to resolve (46), the latter can be tested for a simple two-state phenomenological model that is consistent with apparent good fit of chemical shift perturbations to a two state model (Figure 2B). Here, it is assumed that TAR consists of a population weighted average of two states; a metal “free” ensemble, characterized by inter-helical bend angle θ(free), and inter-helical flexibility ϑint(free), that is weakly associated with metals and which is favored at low ionic strength conditions (Figure 5A). The metal “bound” state, characterized by inter-helical bend angle θ(bound) and inter-helical flexibility ϑint(bound), is more strongly associated with metals and within our metal concentration range is favored at high ionic strength conditions (Figure 5A). Assuming that the dynamical inter-conversion between the “free” and “bound” states occurs at timescales faster than the inverse of the measured RDCs (i.e. faster than milliseconds), the observed RDCs will be a population weighted average over the two states. Simulations using the TAR helices (data not shown) show that to a good approximation, the observed ϑint (ϑint(obs)) and inter-helical bend angle (θ(obs)) at a given Na+ (or Mg2+) will be given by a population weighted average of “free” and “bound” ensembles (40, 71):
| (2) |
where the values of pbound can be computed from the apparent Kd values determined for Na+ and Mg2+ based on the chemical shift titrations (Figure 2B). Assuming that Mg2+ stabilizes a similar bound structure as Na+, which is supported by RDC measurements (Figure 3), the ϑint(obs) and θ(obs) measured under all four metal conditions (25 mM, 160 mM, and 320 mM Na+ and 25 mM Na+/4 mM Mg2+) can be fitted to Equation 2 to solve for the two unknowns in each case (ϑint(free), ϑint(bound) and θ (free), θ (bound) respectively).
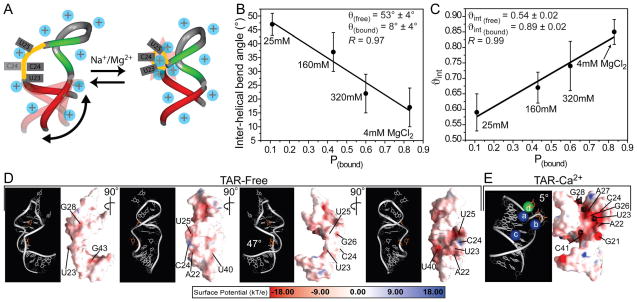
Figure 5.
(A) Two-state model for the metal-induced TAR structural transition. (B–C) Fitting of the observed (B) inter-helical bend angle (θ(obs)) and (C) amplitude of inter-helical motions (ϑint(obs) as a function of the fractional bound populations (pbound) using Equation 2. The “free” and “bound” θ and ϑint values obtained from the fit are shown together with the correlation coefficient (R). (D–E) Electrostatic surfaces for TAR (D) in “free” form (PDB ID#1ANR) (19) under moderate ionic strength conditions (50mM NaCl and 5mM phosphate buffer) with different views showing the weaker electrostatic potential and (E) bound to Ca2+ cations (PDB ID#397D) (20) with residues undergoing the largest metal-induced chemical shift perturbations highlighted.
As shown in Figure 5, a good fit could be obtained for both the inter-helical bend angle θ(obs) (Figure 5B) and flexibility ϑint(obs) (Figure 5C). The average Kd values obtained for the largest chemical perturbations (see methods) were used in computing pbound with minor differences observed when using various Kd values from the range observed (Figure 2B). No attempts were made to fit the inter-helical twist angles given their much larger uncertainty. The fit yields parameters for the metal “bound” state (ϑint(bound) = 0.89 ± 0.02 and θ(bound) = 8° ± 4°) that is in very good agreement with expectations based on the X-ray structure of TAR (θ(bound) = −11° and (ϑint(bound) = 1.0) which was determined in the presence of saturating amounts of divalent ions (100mM CaCl2) (20).
Model conformations for the “free” and “bound” TAR states have been reported, namely an NOE-based NMR structure of unbound TAR under low ionic strength conditions in the absence of divalent ions (50 mM NaCl and 5 mM phosphate buffer) (19) and an X-ray structure of TAR determined in the presence of saturating Ca2+ concentrations (100 mM CaCl2, 50 mM Na-cacodylate and 200 mM NH4Cl) (20). To gain insight into the molecular basis for the metal-induced structural transition, we performed nonlinear Poisson-Boltzmann calculations (27, 66) and compared the surface electrostatic potential for these two TAR conformations. The negative electrostatic potential was significantly weaker for the unbound TAR structure (Figure 5D). Here, inter-helical bending allows the bulge to adopt an extended conformation that minimizes negative charge repulsion while allowing U23 to adopt a looped in stacked conformation (19) (Figure 5A). The looping in of this bulge residue accounts for the over-twisting observed at low ionic strength (+ve ξ angles, Figure 4B) (19). In stark contrast, as shown in Figure 5E, a strong negative electrostatic potential was observed for the Ca2+ bound TAR structure in and around the bulge (G21-C24, and A27) precisely at residues that exhibit large metal-induced chemical shift perturbations (Figure 2A). This strong negative potential explains in part why coaxial TAR conformations are not favored at low ionic strength conditions despite the energetic benefits of helical stacking, metals are required to screen repulsive forces. The metal bound TAR conformation is also likely disfavored by the looping out of U23 and loss of stacking interactions with A22 and U24 (Figure 5A).
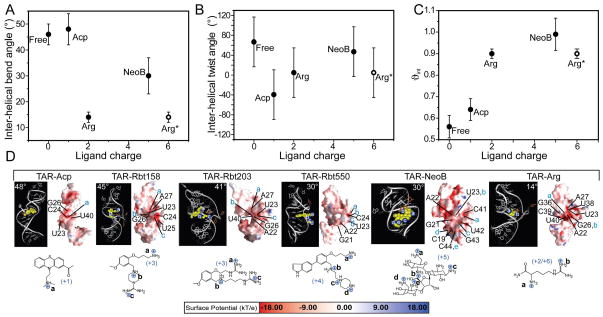
Comparison with small molecule binding
We previously used RDCs to characterize the conformational dynamics of TAR bound to the small molecules argininamide (72), acetylpromazine, and neomycin B (35). We noted qualitatively that the small molecules induce a reduction in the TAR inter-helical bend angle and dynamics by an amount that is apparently dependent on their total number of positive groups. These trends are shown more quantitatively in Figure 6 using an expanded set of TAR structures. An inverse correlation is apparent between the TAR inter-helical bend angle (θ) (Figure 6A) and dynamics (ϑint) (Figure 6C) and the net positive charge delivered by the small molecule. This is analogous to the trend observed with increasing metal concentration (Figure 4). In contrast, no significant correlation is observed with the inter-helical twist (ξ) angle (Figure 6B) which as mentioned previously is also influenced by the local bulge conformation. For the Tat mimic argininamide (ARG), a charge of both +2 and +6 is shown. The latter is based on surface plasmon resonance measurements indicating that up to three ARG molecules bind TAR under NMR conditions (26).
Figure 6.
TAR conformational dynamics when bound to the small molecules argininamide (ARG), acetylpromazine (ACP), neomycin B (NeoB), Rbt 158, Rbt 203, and Rbt 550. Shown are the (A) inter-helical bend angle (θ), (B) inter-helical twist angle (ξ), and (C) amplitude of inter-helical motions (ϑint) as a function of total positive charge delivered by the small molecules. The inter-helical bend angles for ARG, ACP, and NeoB were obtained from order tensor analysis of RDCs, as reported previously (35, 72). For remaining structures, the angles were obtained from model 1 of the NOE-based NMR structure (Rbt 158, Rbt 203, and Rbt 550) (25, 26) or the X-ray structure (Ca2+) (20). For ARG, which has a charge of +2, a total charge of +6 is also shown based on surface plasmon resonance measurements that indicate that up to three ARG molecules bind TAR (26). For Ca2+, a charge of +8 is assumed based on observation of 4 × Ca2+ ions in the X-ray structure (20). (D) Electrostatic surfaces for the bound TAR structures following removal of ligands. Highlighted in blue letters are the positions of cationic groups on small molecules relative to the TAR electrostatic surface. The TAR orientation in each case was chosen to illustrate proximity of cationic groups near the strong negative TAR charge potential.
The above results suggest that electrostatic interactions also dominate the TAR global conformational changes that are induced by small molecule recognition. To explore this further, we performed nonlinear Poisson-Boltzmann calculations on the TAR complexes following removal of bound ligands. Despite variations due to the structural uncertainty of the NMR ensemble (see supporting), cationic groups from small molecules were frequently observed near TAR regions of strong electrostatic potential (Figure 6D), particularly for regions in and around the bulge, as previously noted for the Rbt family of ligands (26). Furthermore, as expected, linear TAR conformations (e.g. TAR-NeoB, TAR-Ca2+ and TAR-ARG) generally have a stronger electrostatic potential compared to the more bent conformations (e.g. TAR-ACP) (Figure 6D). The electrostatic hot-spots in the linear conformations are primarily composed of backbone phosphates (O1P, O2P) which explains why for example Mg2+, NeoB, and ARG are effective at stabilizing the TAR global backbone conformation. In contrast, for the highly bent conformations (TAR-ACP, TAR-Rbt158 and TAR-Rbt203), they are primarily composed of oxygen and nitrogen atoms in sugar (O2′, O3′, O5′) and base (U(O4, O2), G(N7, O6), A(N7)) moieties. The lack of involvement of backbone phosphates helps rationalize why ACP is ineffective at stabilizing the TAR overall conformation. No dynamical data has been obtained for the Rbt family of small molecules.
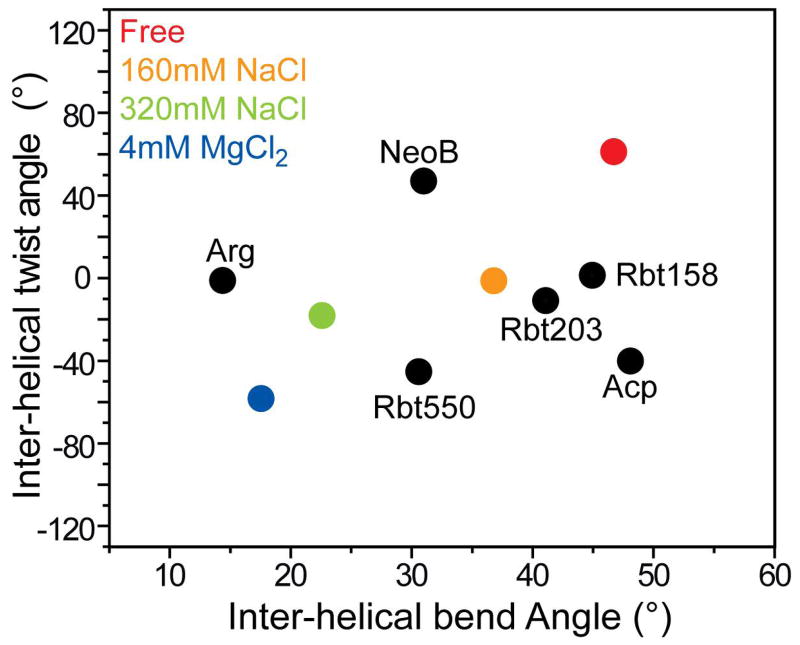
Possible role for counterions in TAR adaptive recognition
In Figure 7, we show the inter-helical bend (θ) and twist angles (ξ) for TAR in different ligand bound states along with the conformational changes that are induced by metals ions. Many of the ligand bound TAR conformations fall along or near the predicted two-state metal induced conformational transition. This highlights the similar changes in the TAR conformation that are induced by small molecules and metals. The largest deviation from the pathway is observed for ACP which among the bound TAR conformations has the weakest electrostatic potential composed of sugar and base moieties. Interestingly, at the extremity of our two state metal transition, we find the functionally relevant TAR conformation that is stabilized by the Tat mimic ARG (Figure 7). Thus, metals may act to increase the probability of sampling productive conformations that mimic the functional protein bound state by screening unfavorable backbone repulsive forces. By also reducing the population of non-productive conformations, counterions may decrease the likelihood for non-specific adaptation and promiscuous recognition. In this context, metals may act to bias the specificity of internal motions towards functionally active conformations.
Figure 7.
Comparison of metal and small molecule induced changes in the TAR inter-helical conformation.
CONCLUSION
Our results show that Na+ and Mg2+ ions induce a similar TAR structural and dynamical transition from a bent flexible to coaxial rigid state though the binding modes of the two metals may be different. This is in contrast to previous transient electric birefringence studies indicating that the Mg2+ induced transitions in model bulge containing RNAs are not reproduced by Na+ even when using up to 50-fold higher concentrations (41). It remains to be established if the observed effects of monovalent ions on the TAR conformation are a general feature of bulge containing RNAs.
The similar structural and dynamical changes induced by Na+ and Mg2+ strongly suggests that non-specific electrostatic interactions with diffusive counterions, and not specifically bound metals observed in the X-ray structure, drive the TAR global structure-dynamical transition. Our results do not however rule out the presence of inner sphere contact(s) particularly with Mg2+. Similar electrostatic interactions seem to dominate the TAR conformational transitions that are induced by Tat derived peptides and other small molecules. Many of these ligand bound TAR conformations fall near the pathway of the metal-induced conformational transition (Figure 7) suggesting that metals may be involved in adapting the TAR conformation for target recognition.
Finally, our study underscores the intricate sensitivity of RNA structural dynamics to environmental conditions. The range of ionic strength (25 mM – 320 mM) over which significant changes in the TAR structural and dynamical changes could be detected falls within a range of buffer conditions that are often used interchangeably. Our results also demonstrate the ability to use RDCs in quantitatively measuring subtle differences in RNA conformational dynamics.
Supplementary Material
Acknowledgments
We thank members of the Al-Hashimi lab, especially Qi Zhang, for insightful comments and help, and Dr. Alex Kurochkin for maintenance of the NMR instruments.
ABBREVIATIONS
- RDC
residual dipolar couplings
- HSQC
heteronuclear single quantum coherence spectroscopy
- TAR
transactivation response element
- HIV
human immunodeficiency virus
Footnotes
This work was supported by funding from the NIH (RO1 AI066975-01) and the Rackham Science Award (University of Michigan). The Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor supported and purchased the 600 MHz spectrometer.
SUPPORTING INFORMATION. Figures showing bulge RDCs as a function of ionic strength, RDC measurement error, simulations assessing two-state averaging of RDCs, comparison of spectra with and without phage, electrostatic surfaces for various models of the NMR ensemble. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Leulliot N, Varani G. Current topics in RNA-protein recognition: Control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 2.Micura R, Hobartner C. On secondary structure rearrangements and equilibria of small RNAs. Chembiochem. 2003;4:984–990. doi: 10.1002/cbic.200300664. [DOI] [PubMed] [Google Scholar]
- 3.Al-Hashimi HM. Dynamics-Based Amplification of RNA Function and Its Characterization by Using NMR Spectroscopy. Chembiochem. 2005;6:1506–1519. doi: 10.1002/cbic.200500002. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 5.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 6.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu Y, Stock G. Conformational dynamics of RNA-peptide binding: a molecular dynamics simulation study. Biophys J. 2006;90:391–399. doi: 10.1529/biophysj.105.069559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 9.Dayie KT, Brodsky AS, Williamson JR. Base Flexibility in HIV-2 TAR RNA Mapped by Solution (15)N, (13)C NMR Relaxation. J Mol Biol. 2002;317:263–278. doi: 10.1006/jmbi.2001.5424. [DOI] [PubMed] [Google Scholar]
- 10.Pitici F, Beveridge DL, Baranger AM. Molecular dynamics simulation studies of induced fit and conformational capture in U1A-RNA binding: do molecular substates code for specificity? Biopolymers. 2002;65:424–435. doi: 10.1002/bip.10251. [DOI] [PubMed] [Google Scholar]
- 11.Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermann T, Westhof E. Simulations of the dynamics at an RNA-protein interface. Nat Struct Biol. 1999;6:540–544. doi: 10.1038/9310. [DOI] [PubMed] [Google Scholar]
- 13.Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumilation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 14.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 15.Weeks KM, Ampe C, Schultz SC, Steitz TA, Crothers DM. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990;249:1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- 16.Bannwarth S, Gatignol A. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Curr HIV Res. 2005;3:61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- 17.Krebs A, Ludwig V, Boden O, Gobel MW. Targeting the HIV trans-activation responsive region--approaches towards RNA-binding drugs. Chembiochem. 2003;4:972–978. doi: 10.1002/cbic.200300652. [DOI] [PubMed] [Google Scholar]
- 18.Froeyen M, Herdewijn P. RNA as a target for drug design, the example of Tat-TAR interaction. Curr Top Med Chem. 2002;2:1123–1145. doi: 10.2174/1568026023393200. [DOI] [PubMed] [Google Scholar]
- 19.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ippolito JA, Steitz TA. A 1.3-angstrom resolution crystal structure of the HIV-1 trans- activation response region RNA stem reveals a metal ion- dependent bulge conformation. Proc Natl Acad Sci U S A. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 22.Aboul-ela F, Karn J, Varani G. The Structure of the Human-Immunodeficiency-Virus Type-1 Tar RNA Reveals Principles of RNA Recognition By Tat Protein. J Mol Biol. 1995;253:313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 23.Faber C, Sticht H, Schweimer K, Rosch P. Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin B. J Biol Chem. 2000;275:20660–20666. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 24.Du Z, Lind KE, James TL. Structure of TAR RNA complexed with a Tat-TAR interaction nanomolar inhibitor that was identified by computational screening. Chem Biol. 2002;9:707–712. doi: 10.1016/s1074-5521(02)00151-5. [DOI] [PubMed] [Google Scholar]
- 25.Murchie AI, Davis B, Isel C, Afshar M, Drysdale MJ, Bower J, Potter AJ, Starkey ID, Swarbrick TM, Mirza S, Prescott CD, Vaglio P, Aboul-ela F, Karn J. Structure-based drug design targeting an inactive RNA conformation: exploiting the flexibility of HIV-1 TAR RNA. J Mol Biol. 2004;336:625–638. doi: 10.1016/j.jmb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 26.Davis B, Afshar M, Varani G, Murchie AI, Karn J, Lentzen G, Drysdale M, Bower J, Potter AJ, Starkey ID, Swarbrick T, Aboul-ela F. Rational design of inhibitors of HIV-1 TAR RNA through the stabilisation of electrostatic “hot spots”. J Mol Biol. 2004;336:343–356. doi: 10.1016/j.jmb.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 27.Chin K, Sharp KA, Honig B, Pyle AM. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat Struct Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Garcia C, Draper DE. Electrostatic interactions in a peptide--RNA complex. J Mol Biol. 2003;331:75–88. doi: 10.1016/s0022-2836(03)00615-6. [DOI] [PubMed] [Google Scholar]
- 29.Woodson SA. Metal ions and RNA folding: a highly charged topic with a dynamic future. Curr Opin Chem Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 31.Draper DE. A guide to ions and RNA structure. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Law MJ, Linde ME, Chambers EJ, Oubridge C, Katsamba PS, Nilsson L, Haworth IS, Laird-Offringa IA. The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 2006;34:275–285. doi: 10.1093/nar/gkj436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao J, Frankel AD. Electrostatic interactions modulate the RNA-binding and transactivation specificities of the human immunodeficiency virus and simian immunodeficiency virus Tat proteins. Proc Natl Acad Sci U S A. 1993;90:1571–1575. doi: 10.1073/pnas.90.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blount KF, Tor Y. Using pyrene-labeled HIV-1 TAR to measure RNA-small molecule binding. Nucleic Acids Res. 2003;31:5490–5500. doi: 10.1093/nar/gkg755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pitt SW, Zhang Q, Patel DJ, Al-Hashimi HM. Evidence that electrostatic interactions dictate the ligand-induced arrest of RNA global flexibility. Angew Chem Int Ed Engl. 2005;44:3412–3415. doi: 10.1002/anie.200500075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olejniczak M, Gdaniec Z, Fischer A, Grabarkiewicz T, Bielecki L, Adamiak RW. The bulge region of HIV-1 TAR RNA binds metal ions in solution. Nucleic Acids Res. 2002;30:4241–4249. doi: 10.1093/nar/gkf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards TE, Sigurdsson ST. EPR spectroscopic analysis of TAR RNA-metal ion interactions. Biochem Biophys Res Commun. 2003;303:721–725. doi: 10.1016/s0006-291x(03)00411-x. [DOI] [PubMed] [Google Scholar]
- 38.Edwards TE, Okonogi TM, Sigurdsson ST. Investigation of RNA-protein and RNA-metal ion interactions by electron paramagnetic resonance spectroscopy. The HIV TAR-Tat motif. Chem Biol. 2002;9:699–706. doi: 10.1016/s1074-5521(02)00150-3. [DOI] [PubMed] [Google Scholar]
- 39.Zacharias M, Hagerman PJ. The Bend in RNA Created By the Transactivation Response Element Bulge of Human-Immunodeficiency-Virus Is Straightened By Arginine and By Tat-Derived Peptide. Proc Natl Acad Sci U S A. 1995;92:6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. Mg2+-induced variations in the conformation and dynamics of HIV-1 TAR RNA probed using NMR residual dipolar couplings. J Mol Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zacharias M, Hagerman PJ. Bulge-induced bends in RNA: quantification by transient electric birefringence. J Mol Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- 42.Bokinsky G, Zhuang X. Single-molecule RNA folding. Acc Chem Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- 43.Brenowitz M, Chance MR, Dhavan G, Takamoto K. Probing the structural dynamics of nucleic acids by quantitative time-resolved and equilibrium hydroxyl radical “footprinting”. Curr Opin Struct Biol. 2002;12:648–653. doi: 10.1016/s0959-440x(02)00366-4. [DOI] [PubMed] [Google Scholar]
- 44.Takamoto K, He Q, Morris S, Chance MR, Brenowitz M. Monovalent cations mediate formation of native tertiary structure of the Tetrahymena thermophila ribozyme. Nat Struct Biol. 2002;9:928–933. doi: 10.1038/nsb871. [DOI] [PubMed] [Google Scholar]
- 45.Caliskan G, Hyeon C, Perez-Salas U, Briber RM, Woodson SA, Thirumalai D. Persistence length changes dramatically as RNA folds. Phys Rev Lett. 2005;95:268303-1–268303-4. doi: 10.1103/PhysRevLett.95.268303. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Salas UA, Rangan P, Krueger S, Briber RM, Thirumalai D, Woodson SA. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry. 2004;43:1746–1753. doi: 10.1021/bi035642o. [DOI] [PubMed] [Google Scholar]
- 47.Mollova ET, Pardi A. NMR solution structure determination of RNAs. Curr Opin Struct Biol. 2000;10:298–302. doi: 10.1016/s0959-440x(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 48.Al-Hashimi HM, Gorin A, Majumdar A, Gosser Y, Patel DJ. Towards structural genomics of RNA: rapid NMR resonance assignment and simultaneous RNA tertiary structure determination using residual dipolar couplings. J Mol Biol. 2002;318:637–649. doi: 10.1016/S0022-2836(02)00160-2. [DOI] [PubMed] [Google Scholar]
- 49.Al-Hashimi HM, Patel DJ. Residual dipolar couplings: Synergy between NMR and structural genomics. J Biomol NMR. 2002;22:1–8. doi: 10.1023/a:1013801714041. [DOI] [PubMed] [Google Scholar]
- 50.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear Magnetic Dipole Interactions in Field-Oriented Proteins - Information For Structure Determination in Solution. Proc Natl Acad Sci U S A. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 52.Prestegard JH, Al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 53.Bax A, Grishaev A. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol. 2005;15:563–570. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 54.Mollova ET, Hansen MR, Pardi A. Global structure of RNA determined with residual dipolar couplings. J Am Chem Soc. 2000;122:11561–11562. [Google Scholar]
- 55.Musselman C, Pitt SW, Gulati K, Foster LL, Andricioaei I, Al-Hashimi HM. Impact of static and dynamic A-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J Biomol NMR. 2006;36:235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 56.Hansen MR, Hanson P, Pardi A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Methods in Enzymology. 2000;317:220–240. doi: 10.1016/s0076-6879(00)17017-x. [DOI] [PubMed] [Google Scholar]
- 57.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 58.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based On Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 59.Goddard TD, Kneller DG. SPARKY 3. University of California; San Franscisco: [Google Scholar]
- 60.Meissner A, Sorensen OW. The role of coherence transfer efficiency in design of TROSY- type multidimensional NMR experiments. J Magn Reson. 1999;139:439–442. doi: 10.1006/jmre.1999.1788. [DOI] [PubMed] [Google Scholar]
- 61.Neidle S. Oxford Handbook of Nucleic Acid Structure. Oxford University Press; New York: 1999. [Google Scholar]
- 62.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order matrix analysis of residual dipolar couplings using singular value decomposition. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 63.Hansen AL, Al-Hashimi HM. Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: towards new long-range orientational constraints. J Magn Reson. 2006;179:299–307. doi: 10.1016/j.jmr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 64.Al-Hashimi HM, Valafar H, Terrell M, Zartler ER, Eidsness MK, Prestegard JH. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J Magn Reson. 2000;143:402–406. doi: 10.1006/jmre.2000.2049. [DOI] [PubMed] [Google Scholar]
- 65.Gonzalez RL, Jr, Tinoco I., Jr Solution structure and thermodynamics of a divalent metal ion binding site in an RNA pseudoknot. J Mol Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- 66.Nicholls A, Sharp K, Honig B. Protein folding and association:insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 67.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A 2nd Generation Force-Field For The Simulation Of Proteins, Nucleic-Acids, And Organic-Molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 68.Saupe A. Recent results in the field of liquid crystals. Angew Chem, Int Ed Engl. 1968;7:97–112. [Google Scholar]
- 69.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. J Am Chem Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Q, Throolin R, Pitt SW, Serganov A, Al-Hashimi HM. Probing motions between equivalent RNA domains using magnetic field induced residual dipolar couplings: accounting for correlations between motions and alignment. J Am Chem Soc. 2003;125:10530–10531. doi: 10.1021/ja0363056. [DOI] [PubMed] [Google Scholar]
- 71.Bolon PJ, Al-Hashimi HM, Prestegard JH. Residual dipolar coupling derived orientational constraints on ligand geometry in a 53 kDa protein-ligand complex. J Mol Biol. 1999;293:107–115. doi: 10.1006/jmbi.1999.3133. [DOI] [PubMed] [Google Scholar]
- 72.Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. Argininamide binding arrests global motions in HIV-1 TAR RNA: comparison with Mg2+-induced conformational stabilization. J Mol Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.