Abstract
The application of residual dipolar couplings (RDCs) in studies of RNA structure and dynamics can be complicated by the presence of couplings between collective helix motions and overall alignment and by the inability to modulate overall alignment of the molecule by changing the ordering medium. Here, we show for a 27-nt TAR RNA construct that variable levels of helix elongation can be used to alter both overall alignment and couplings to collective helix motions in a semi-predictable manner. In the absence of elongation, a four base-pair helix II capped by a UUCG apical loop exhibits a higher degree of order compared to a six base-pair helix I (ϑI/ϑII = 0.56±0.1). The principal Szz direction is nearly parallel to the axis of helix II but deviates by ∼40° relative to the axis of helix I. Elongating helix I by three base-pairs equalizes the alignment of the two helices and pushes the RNA into the motional coupling limit such that the two helices have comparable degrees of order (ϑI/ϑII = 0.92±0.04) and orientations relative to Szz (∼17°). Increasing the length of elongation further to twenty-two base-pairs pushes the RNA into the motional decoupling limit in which helix I dominates alignment (ϑII/ϑI = 0.45±0.05), with Szz orientated nearly parallel to its helix axis. Many of these trends can be rationalized using PALES simulations that employ a previously proposed three-state dynamic ensemble of TAR. Our results provide new insights into motional couplings, offer guidelines for assessing their extent, and suggest that variable degrees of helix elongation can allow access to independent sets of RDCs for characterizing RNA structural dynamics.
Keywords: Residual dipolar couplings, collective helix motions, transactivation response element, HIV
1. Introduction
There is great interest in utilizing NMR residual dipolar couplings (RDCs) [1,2] to characterize the structure and dynamics of biomolecules [3-5]. RDCs can be measured in molecules that are partially aligned, either spontaneously when they have a significant magnetic susceptibility anisotropy [1,6] or, more commonly, by dissolution in an appropriate ordering medium [2,7-10]. Several studies have established the utility of measuring RDCs under multiple linearly independent alignment conditions [11] to increase the spatial resolution with which structure and dynamics can be simultaneously characterized [12-18].
Two challenges can arise when using RDCs to characterize the conformational dynamics of nucleic acids, particularly globally flexible RNAs. First, it cannot be generally assumed, as is done in most formalisms, that internal motions do not lead to coupled changes in overall alignment [12,13,19]. Rather, rigid-body collective movements of helical domains about flexible junctions can lead to significant changes in the RNA overall molecular shape and thus global alignment [20,21]. Second, though the alignment of proteins can be modulated by changing the alignment medium and altering the balance between electrostatic and steric forces governing alignment [11,22], this has proven difficult if not impossible for nucleic acids because their uniform charge distribution closely follows that of their molecular shape [23-28]. Although spontaneous magnetic field alignment can yield one additional independent alignment [26,28], the degree of order generally remains unfavorably small under current magnetic field strengths.
Recently, we showed that correlations between internal motions and overall alignment could be reduced and overall alignment altered by chemically perturbing the overall RNA molecular shape [20]. In particular, the alignment of the transactivation response element (TAR) RNA from HIV-1 dissolved in the Pf1 phage ordering medium [8,9] was modulated by independently elongating each of its two helices by twenty-two Watson-Crick base-pairs [20,29]. The elongation renders the overall molecular shape, and consequently overall alignment, far less sensitive to collective motions of helices. It also serves to predefine overall alignment by fixing the principal axis of order (Szz) to be nearly parallel to the elongated helix axis. A similar mutagenesis strategy that serves to alter the surface electrostatic properties has successfully been used by Bax and co-workers to modulate the alignment of proteins [30].
In RNA, extensive helix elongation (>20 base-pairs) is generally required to adequately decouple internal and overall motions. However, this results in an unfavorable increase in the size of the RNA under study and also necessitates preparation of two specifically labeled samples to minimize spectral overcrowding from elongation residues [29]. Many RNAs, including TAR bound to ligands [31] or at high monovalent and divalent ion concentrations [32], are globally rigid and may not require extensive helix elongation to decouple motions. In these cases, moderate degrees of elongation may offer an approach for modulating alignment. Even for globally flexible RNAs, moderate elongation may help expose collective helix motions and provide physical insights into the motional couplings themselves [33]. Here, we examine the utility of moderate degrees of helix elongation in modulating both the alignment and degree of motional couplings in a TAR mutant, EI(3)-TARm, in which the apical wild-type loop has been replaced with a UUCG loop and in which the terminal helix is elongated by three base-pairs (Figure 1a).
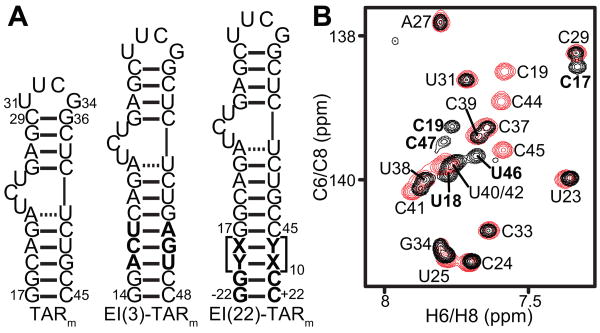
Figure 1.
TARm constructs. (a) Secondary structures of TARm, EI(3)-TARm, and EI(22)-TARm. Differences between constructs are shown in bold. (b) 2D 1H-13C HSQC spectrum of EI(3)-TARm (black) overlaid on the corresponding spectrum of TARm (red). TARm resonances are labeled in black, while peaks that belong only to EI(3)-TARm are shown in bold. All EI(3)-TARm experiments were conducted in NMR buffer (15 mM sodium phosphate, 25 mM sodium chloride, 0.1 mM EDTA and pH 6.4) at 298 K on an Avance Bruker 600 MHz NMR spectrometer equipped with a triple-resonance 5 mm cryogenic probe. 1H, 13C, and 15N resonances were assigned by spectra overlay [29,43] and using standard homonuclear and heteronuclear 2D experiments, including an exchangeable NOESY and a 2D HCN experiment that correlates intranucleotide H8/H6 to N1/N9 to H1′.
2. Results and Discussion
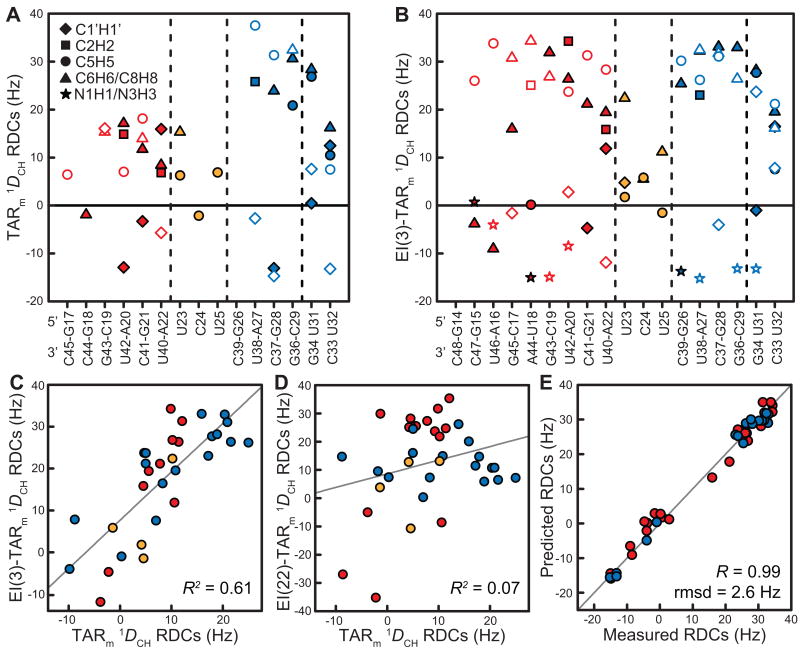
We prepared uniformly 13C/15N labeled EI(3)-TARm by in vitro transcription as previously described [34]. As shown in Figure 1b, spectra of EI(3)-TARm are in excellent agreement with those of non-elongated TARm, indicating that the elongation does not affect its structural and dynamical integrity, as reported previously for a twenty-two base-pair elongated TAR sample (EI(22)-TARm, Figure 1a) [20,29]. A total of 9 N-H and 53 C-H RDCs (Table S1) were measured in sugar (C1′H1′) and base (C2H2, C5H5, C6H6, C8H8, N1H1, and N3H3) moieties of EI(3)-TARm using ∼22 mg/ml of Pf1 phage as an ordering medium [8,35]. A plot of RDCs as a function of secondary structure is shown in Figure 2a and 2b for TARm and EI(3)-TARm, respectively. The RDCs measured in helix I of TARm are consistently attenuated relative to counterparts in helix II. This has previously been attributed to inter-helical motions and a smaller degree of order for helix I compared to helix II [36]. Differences between helix RDCs are far less pronounced in EI(3)-TARm, indicating that elongation of helix I by three base-pairs increases its degree of order relative to helix II. The poor agreement (R2=0.61) observed between the EI(3)-TARm and TARm RDCs (Figure 2c) qualitatively suggests that the three-base pair elongation modulates overall alignment and/or motional couplings. An even poorer correlation (R2=0.07) is observed upon elongating helix I by twenty-two base-pairs (Figure 2d), consistent with elongation-dependent modulation of alignment and/or motional couplings.
Figure 2.
Measurement of RDCs in EI(3)-TARm. One bond RDCs (1DCH and 1DNH) measured in (a) TARm and (b) EI(3)-TARm as a function of residue/secondary structure. Filled and open circles represent RDCs measured on the 5′and 3′ strand, respectively. RDCs have been normalized to those of EI(3)-TARm by the ratio of the ϑII values of EI(3)-TARm and TARm. See inset in Figure 2a for key. (c-d) Correlation plots between (c) TARm and EI(3)-TARm and (d) TARm and EI(22)-TARm RDCs. (e) Comparison of EI(3)-TARm RDCs measured in helix I (red) and helix II (blue) with values back-predicted using the best-fit order tensor and an idealized A-form helix and X-ray structure of the loop.
To characterize the alignment of EI(3)-TARm in phage, we used the RDCs measured in non-terminal Watson-Crick base-pairs and structurally stable loop residues to determine order tensors for each helix. The order tensors were determined by fitting measured RDCs to an idealized A-form helical geometry and an X-ray structure of the loop[37] as described previously [38-40]. An excellent fit was observed between the measured and back-predicted RDCs, indicating that both EI(3)-TARm helices are accurately modeled using an idealized A-form geometry as described previously for non-elongated [36] and elongated [20] TAR (Figure 2e).
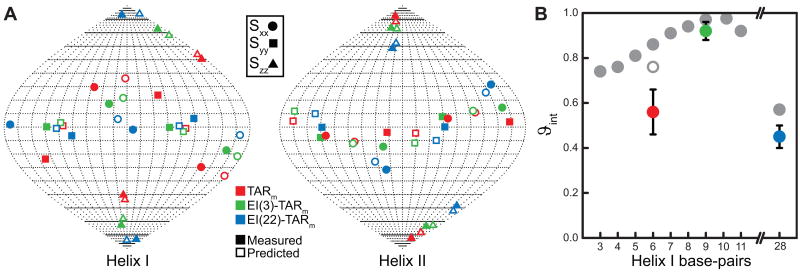
In Figure 3a, we use a Sauson-Flamsteed map to depict the best-fit principal orientational solutions (Sxx, Syy, and Szz) obtained for each helix in EI(3)-TARm. The orientational solutions are shown relative to a molecular frame in which the helix axis is oriented along the molecular z direction. For comparison, the corresponding order tensors reported previously for TARm [36] and EI(22)-TARm [20] are also shown. The estimated uncertainty in the Szz orientations determined for helix I/II using the program Aform-RDC [39] is 7°/6°, 2°/2°, and 2°/4° for TARm, EI(3)-TARm, and EI(22)-TARm, respectively. In TARm, Szz deviates by only ∼7° from the helix II axis but deviates by ∼40° from the helix I axis. As expected, elongation of helix I by three base-pairs tips Szz towards its helical axis such that Szz deviates by only ∼17° and is nearly fully aligned with the axis following a twenty-two base-pair elongation (∼7°). The three Szz directions in TARm, EI(3)-TARm, and EI(22)-TARm do not fall along a common plane, most likely because the A-form helices deviate from perfect cylindrical axial symmetry (Figure 3a). The three base-pair elongation has an opposite, albeit smaller effect on helix II. The angle between Szz and the helix II axis increases from ∼7° to ∼16° to ∼32° upon elongation by three and twenty-two base-pairs, respectively (Figure 3a). Changes are also observed in the Sxx and Syy orientations (Figure 3a) and the asymmetry parameter (η) (Figure S1a, S1b), indicating that elongation modulates all five elements of the order tensor.
Figure 3.
Measured and predicted alignment of variable elongated TAR. (a) Sauson-Flamsteed maps showing the best-fit measured (filled symbols) and PALES-predicted (open symbols) order tensor frames (Sxx, Syy, and Szz) for helices I and II. Solutions are depicted relative to a molecular frame in which the helix axis is oriented along the z direction. (b) Measured (in color) and PALES-predicted (in gray) ϑint values (ϑint = ϑII/ϑI) as a function of helix I length. Note that the value for EI(22)-TARm corresponds to ϑint = ϑI/ϑII. The open circle corresponds to the predicted ϑint value of TARm when the two terminal base-pairs are excluded.
Elongation of helix I leads to changes in relative degrees of helix order that mirror those observed for the principal Szz direction. In TARm, helix II dominates overall alignment, yielding an internal generalized degree of order ϑint = ϑI/ϑII = 0.56±0.1 that is consistent with large amplitude, inter-helical motions [36]. Elongating helix I by three base-pairs increases its level of order compared to helix II such that both helices now have comparable degrees of order (ϑint = ϑI/ϑII = 0.92±0.04). This, together with the similar angles between the Szz direction and the two helical axes (Figure 3a), strongly suggests that the three base-pair elongation equalizes contributions of the two helices to overall alignment and pushes TAR into the motional coupling limit. Extending the elongation of helix I to twenty-two base-pairs drives the system towards the decoupling limit in which helix I dominates alignment (ϑint = ϑII/ϑI = 0.45±0.05). Because RNA helices are often not coaxially stacked, moderate degrees of elongation are expected to modulate alignment. This was verified using PALES simulations on a variety of RNAs following elongation of the terminal helix by three base-pairs (data not shown). The combination of helix-elongation with measurements of magnetic field-induced RDCs [26,28] may make it possible to access the maximum of five linearly independent sets of RDCs in nucleic acids. Access to such measurements will make it possible to more fully characterize the structural dynamics of RNA.
To rationalize the observed trends in the alignment of TAR as a function of elongation, we used the three-state dynamical ensemble determined previously using RDCs [20] in conjunction with electrostatic-induced alignment simulations using the program PALES [23,41] to simulate TAR alignment in phage at varying lengths of helix I elongation. The three-state ensemble is a discrete approximation of what is likely a continuous distribution involving many conformations. While we do not expect to observe quantitative agreement between measured alignment parameters and values predicted using this three-state ensemble approximation, we would like to obtain insights into the general trends observed for orientation and alignment upon helix elongation. An elongated helix I was superimposed onto each of the three TAR conformers of the ensemble, and PALES was used to predict the RDCs for each conformer. Bulge residues were not included in the three-state dynamical ensemble, so they were not included in PALES calculations. The three sets of RDCs simulated for each conformer were then averaged and used to compute order tensors for each helix. As shown in Figure 3a, we observe very good agreement between measured and predicted Szz directions. The deviations range between 2-11° and 6-8°, for helices I and II, respectively. The simulations reproduce the observed non-planar approach of the Szz direction towards the helix I axis with increasing elongation. Interestingly, the Sxx and Syy directions (Figure 3a) and the asymmetry (Figure S1) are also reasonably well reproduced considering their much larger experimental uncertainty and given that exclusion of the bulge has a strong effect on these predicted values (data not shown). The simulations reproduce the increase in the helix I order compared to helix II upon helix I elongation (Figure 3b). Also, the simulated ϑint values reflect the observed trend. The larger discrepancy in TARm could arise from neglect of the bulge and fraying motions at terminal base-pairs which are likely to have a bigger effect on the small TARm compared to the other elongated constructs. Indeed, better agreement is observed for TARm when excluding the two terminal base-pairs in helix I (Figure 3b).
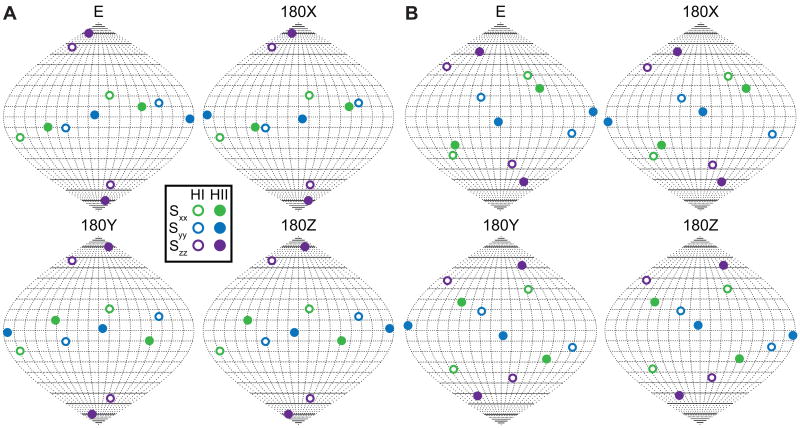
Independent RDC data sets are often used to overcome the 4n-1 fold orientational degeneracy arising from superposition of the order tensors of n domains [11,22]. One of the consequences of having differential motional averaging effects due to inter-domain motions is that two sets of RDCs may not be reconcilable with a single inter-domain orientation [42]. This is the case for the RDC datasets measured in TARm, EI(3)-TARm, and EI(22)-TARm. As shown in Figure 4a, none of the four inter-helix structures assembled using the RDCs measured in TARm satisfy the orientational solutions obtained using the EI(3)-TARm RDCs. Similarly, a common solution does not exist when using RDCs from EI(22)-TARm (Figure 4b). Thus, the inability to reconcile RDCs measured in differentially elongated RNAs in this manner may be an indication that inter-helix motions are present. However, other sources of experimental uncertainty should also be ruled out.
Figure 4.
Difficulty in resolving degeneracies in determining the average orientation of TAR helices using multiple RDC data sets. Shown are the experimental order tensor frames (Sxx, Syy, and Szz, in green, blue, and purple, respectively) determined for helix I (open circles) and II (filled circles) using (a) EI(3)-TARm and (b) EI(22)-TARm RDCs shown relative to the PAS of the order tensor determined using the TARm RDCs. The different orientational solutions were generated by rotating helix II in the TARm PAS 180° degrees about the x, y, and z axis.
3. Conclusion
In conclusion, we show that modest degrees of helix-elongation can be used to modulate both overall alignment of RNA and the degree of motional couplings in a semi-predictable manner. Our results underscore the importance of exercising caution in interpreting similar levels of order for two domains (ϑint ∼1) in terms of inter-domain rigidity. In general, motional couplings obscure inter-domain motion and will often result in underestimated dynamics. Our data suggest that even moderate degrees of helix elongation may be used to push an RNA system outside the motionally coupled regime, though the degree of elongation needed will obviously vary from RNA to RNA.
Supplementary Material
Acknowledgments
We thank members of the Al-Hashimi lab for insightful comments and help, and Dr. Alex Kurochkin for maintenance of the NMR instruments. This work was supported by funding from the NIH (RO1 AI066975-01) to H.M.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Nuclear magnetic dipole interactions in field-oriented proteins - information for structure determination in solution. Proc Natl Acad Sci USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 3.Tolman JR, Ruan K. NMR residual dipolar couplings as probes of biomolecular dynamics. Chem Rev. 2006;106:1720–36. doi: 10.1021/cr040429z. [DOI] [PubMed] [Google Scholar]
- 4.Bouvignies G, Markwick PR, Blackledge M. Simultaneous definition of high resolution protein structure and backbone conformational dynamics using NMR residual dipolar couplings. Chemphyschem. 2007;8:1901–9. doi: 10.1002/cphc.200700353. [DOI] [PubMed] [Google Scholar]
- 5.Getz M, Sun X, Casiano-Negroni A, Zhang Q, Al-Hashimi HM. NMR studies of RNA dynamics and structural plasticity using NMR residual dipolar couplings. Biopolymers. 2007;86:384–402. doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- 6.Bothner-By AA. Magnetic field induced alignment of molecules. In: Grant DM, Harris RK, editors. Encyclopedia of Nuclear Magnetic Resonance. Wiley; Chichester: 1995. pp. 2932–2938. [Google Scholar]
- 7.Bax A, Grishaev A. Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol. 2005;15:563–70. doi: 10.1016/j.sbi.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Clore GM, Starich MR, Gronenborn AM. Measurement of residual dipolar couplings of macromolecules aligned in the nematic phase of a colloidal suspension of rod-shaped viruses. J Am Chem Soc. 1998;120:10571–10572. [Google Scholar]
- 9.Hansen MR, Hanson P, Pardi A. Filamentous bacteriophage for aligning RNA, DNA, and proteins for measurement of nuclear magnetic resonance dipolar coupling interactions. Meth Enzymol. 2000;317:220–240. doi: 10.1016/s0076-6879(00)17017-x. [DOI] [PubMed] [Google Scholar]
- 10.Ruckert M, Otting G. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J Am Chem Soc. 2000;122:7793–7797. [Google Scholar]
- 11.Ramirez BE, Bax A. Modulation of the alignment tensor of macromolecules dissolved in a dilute liquid crystalline medium. J Am Chem Soc. 1998;120:9106–9107. [Google Scholar]
- 12.Ruan K, Briggman KB, Tolman JR. De novo determination of internuclear vector orientations from residual dipolar couplings measured in three independent alignment media. J Biomol NMR. 2008;41:61–76. doi: 10.1007/s10858-008-9240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peti W, Meiler J, Bruschweiler R, Griesinger C. Model-free analysis of protein backbone motion from residual dipolar couplings. J Am Chem Soc. 2002;124:5822–33. doi: 10.1021/ja011883c. [DOI] [PubMed] [Google Scholar]
- 14.Bouvignies G, Markwick P, Bruschweiler R, Blackledge M. Simultaneous determination of protein backbone structure and dynamics from residual dipolar couplings. J Am Chem Soc. 2006;128:15100–1. doi: 10.1021/ja066704b. [DOI] [PubMed] [Google Scholar]
- 15.Clore GM, Schwieters CD. Amplitudes of protein backbone dynamics and correlated motions in a small alpha/beta protein: correspondence of dipolar coupling and heteronuclear relaxation measurements. Biochemistry. 2004;43:10678–91. doi: 10.1021/bi049357w. [DOI] [PubMed] [Google Scholar]
- 16.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–5. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 17.Tolman JR. A novel approach to the retrieval of structural and dynamic information from residual dipolar couplings using several oriented media in biomolecular NMR spectroscopy. J Am Chem Soc. 2002;124:12020–30. doi: 10.1021/ja0261123. [DOI] [PubMed] [Google Scholar]
- 18.Meiler J, Prompers JJ, Peti W, Griesinger C, Bruschweiler R. Model-free approach to the dynamic interpretation of residual dipolar couplings in globular proteins. J Am Chem Soc. 2001;123:6098–6107. doi: 10.1021/ja010002z. [DOI] [PubMed] [Google Scholar]
- 19.Briggman KB, Tolman JR. De novo determination of bond orientations and order parameters from residual dipolar couplings with high accuracy. J Am Chem Soc. 2003;125:10164–5. doi: 10.1021/ja035904+. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–7. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Throolin R, Pitt SW, Serganov A, Al-Hashimi HM. Probing motions between equivalent RNA domains using magnetic field induced residual dipolar couplings: accounting for correlations between motions and alignment. J Am Chem Soc. 2003;125:10530–1. doi: 10.1021/ja0363056. [DOI] [PubMed] [Google Scholar]
- 22.Al-Hashimi HM, Valafar H, Terrell M, Zartler ER, Eidsness MK, Prestegard JH. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J Magn Reson. 2000;143:402–406. doi: 10.1006/jmre.2000.2049. [DOI] [PubMed] [Google Scholar]
- 23.Zweckstetter M, Bax A. Predicition of sterically induced alignment in a dilute liquid crystalline phase; aid to protein structure determination by NMR. J Am Chem Soc. 2000;122:3791–3792. [Google Scholar]
- 24.Zweckstetter M, Hummer G, Bax A. Prediction of charge-induced molecular alignment of biomolecules dissolved in dilute liquid-crystalline phases. Biophys J. 2004;86:3444–60. doi: 10.1529/biophysj.103.035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu B, Petersen M, Girard F, Tessari M, Wijmenga SS. Prediction of molecular alignment of nucleic acids in aligned media. J Biomol NMR. 2006;35:103–115. doi: 10.1007/s10858-006-9004-2. [DOI] [PubMed] [Google Scholar]
- 26.Al-Hashimi HM, Majumdar A, Gorin A, Kettani A, Skripkin E, Patel DJ. Field- and phage-induced dipolar couplings in a homodimeric DNA quadruplex, relative orientation of G·(C-A) triad and G-tetrad motifs and direct determination of C2 symmetry axis orientation. J Am Chem Soc. 2001;123:633–640. doi: 10.1021/ja003379y. [DOI] [PubMed] [Google Scholar]
- 27.Vermeulen A, McCallum SA, Pardi A. Comparison of the global structure and dynamics of native and unmodified tRNAval. Biochemistry. 2005;44:6024–33. doi: 10.1021/bi0473399. [DOI] [PubMed] [Google Scholar]
- 28.Latham MP, Hanson P, Brown DJ, Pardi A. Comparison of alignment tensors generated for native tRNA(Val) using magnetic fields and liquid crystalline media. J Biomol NMR. 2008;40:83–94. doi: 10.1007/s10858-007-9212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–6. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 30.Yao L, Bax A. Modulating protein alignment in a liquid-crystalline medium through conservative mutagenesis. J Am Chem Soc. 2007;129:11326–7. doi: 10.1021/ja073937+. [DOI] [PubMed] [Google Scholar]
- 31.Pitt SW, Zhang Q, Patel DJ, Al-Hashimi HM. Evidence that electrostatic interactions dictate the ligand-induced arrest of RNA global flexibility. Angew Chem Int Ed Engl. 2005;44:3412–5. doi: 10.1002/anie.200500075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casiano-Negroni A, Sun X, Al-Hashimi HM. Probing Na+-induced changes in the HIV-1 TAR conformational dynamics using NMR residual dipolar couplings: new insights into the role of counterions and electrostatic interactions in adaptive recognition. Biochemistry. 2007;46:6525–35. doi: 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Al-Hashimi HM. Extending the NMR spatial resolution limit for RNA by motional couplings. Nat Methods. 2008;5:243–5. doi: 10.1038/nmeth.1180. [DOI] [PubMed] [Google Scholar]
- 34.Dethoff EA, Hansen AL, Musselman C, Watt ED, Andricioaei I, Al-Hashimi HM. Characterizing complex dynamics in the transactivation response element apical loop and motional correlations with the bulge by NMR, molecular dynamics, and mutagenesis. Biophys J. 2008;95:3906–15. doi: 10.1529/biophysj.108.140285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struc Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 36.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 37.Ennifar E, Nikulin A, Tishchenko S, Serganov A, Nevskaya N, Garber M, Ehresmann B, Ehresmann C, Nikonov S, Dumas P. The crystal structure of UUCG tetraloop. J Mol Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- 38.Al-Hashimi HM, Gorin A, Majumdar A, Gosser Y, Patel DJ. Towards structural Genomics of RNA: Rapid NMR resonance assignment and simultaneous RNA tertiary structure determination using residual dipolar couplings. J Mol Biol. 2002;318:637–649. doi: 10.1016/S0022-2836(02)00160-2. [DOI] [PubMed] [Google Scholar]
- 39.Musselman C, Pitt SW, Gulati K, Foster LL, Andricioaei I, Al-Hashimi HM. Impact of static and dynamic A-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J Biomol NMR. 2006;36:235–49. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 40.Bailor MH, Musselman C, Hansen AL, Gulati K, Patel DJ, Al-Hashimi HM. Characterizing the relative orientation and dynamics of RNA A-form helices using NMR residual dipolar couplings. Nat Protoc. 2007;2:1536–46. doi: 10.1038/nprot.2007.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zweckstetter M. NMR: prediction of molecular alignment from structure using the PALES software. Nat Protoc. 2008;3:679–90. doi: 10.1038/nprot.2008.36. [DOI] [PubMed] [Google Scholar]
- 42.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and dynamic analysis of residual dipolar coupling data for proteins. J Am Chem Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 43.Sun X, Zhang Q, Al-Hashimi HM. Resolving fast and slow motions in the internal loop containing stem-loop 1 of HIV-1 that are modulated by Mg2+ binding: role in the kissing-duplex structural transition. Nucleic Acids Res. 2007;35:1698–713. doi: 10.1093/nar/gkm020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.