Abstract
The four-chambered mammalian heart develops from two fields of cardiac progenitor cells (CPCs) distinguished by their spatiotemporal patterns of differentiation and contributions to the definitive heart [1–3]. The first heart field differentiates earlier in lateral plate mesoderm, generates the linear heart tube and ultimately gives rise to the left ventricle. The second heart field (SHF) differentiates later in pharyngeal mesoderm, elongates the heart tube, and gives rise to the outflow tract (OFT) and much of the right ventricle. Because hearts in lower vertebrates contain a rudimentary OFT but not a right ventricle [4], the existence and function of SHF-like cells in these species has remained a topic of speculation [4–10]. Here we provide direct evidence from Cre/Lox-mediated lineage tracing and loss of function studies in zebrafish, a lower vertebrate with a single ventricle, that latent-TGFβ binding protein 3 (ltbp3) transcripts mark a field of CPCs with defining characteristics of the anterior SHF in mammals. Specifically, ltbp3+ cells differentiate in pharyngeal mesoderm after formation of the heart tube, elongate the heart tube at the outflow pole, and give rise to three cardiovascular lineages in the OFT and myocardium in the distal ventricle. In addition to expressing Ltbp3, a protein that regulates the bioavailability of TGFβ ligands [11], zebrafish SHF cells co-express nkx2.5, an evolutionarily conserved marker of CPCs in both fields [4]. Embryos devoid of ltbp3 lack the same cardiac structures derived from ltbp3+ cells due to compromised progenitor proliferation. Additionally, small-molecule inhibition of TGFβ signaling phenocopies the ltbp3-morphant phenotype whereas expression of a constitutively active TGFβ type I receptor rescues it. Taken together, our findings uncover a requirement for ltbp3-TGFβ signaling during zebrafish SHF development, a process that serves to enlarge the single ventricular chamber in this species.
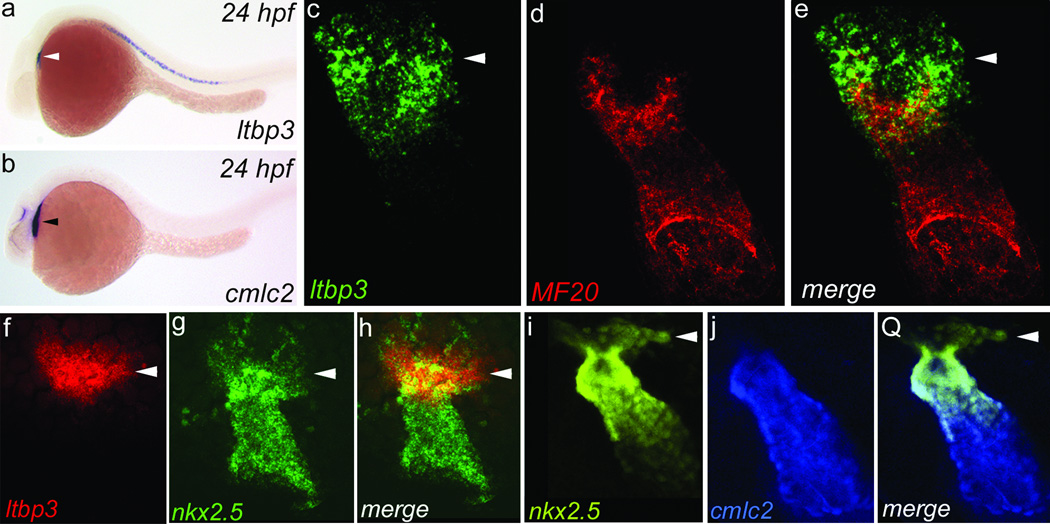
Using in situ hybridization, we discovered that zebrafish ltbp3 transcripts are expressed in cells at the outflow pole of the linear heart tube (Fig. 1a, b and Supplementary Fig. 1 and 2). Double marker analyses confirmed that a majority of ltbp3+ cells are non-overlapping with differentiated myocardium (Fig. 1c–e and Supplementary Fig. 2). Additionally, ltbp3+ cells are neither endothelial nor derived from the neural crest because ltbp3 expression remains robust in embryos lacking both cell types (Supplementary Fig. 3). Because the anatomical relationship of ltbp3+ cells to the heart tube is reminiscent of the anterior segment of the SHF in mice [3, 12], ltbp3+ cells were evaluated for co-expression of nkx2.5, an evolutionarily conserved marker of CPCs in both heart fields [4]. As reported previously, cells in the linear heart tube were positive for nkx2.5 [13]. Surprisingly, nkx2.5 expression also overlapped with ltbp3 transcripts demonstrating that an extra-cardiac population of ltbp3+, nkx2.5+ cells resides at the outflow pole of the zebrafish heart tube (Fig. 1f–h). This population was also readily identified in double transgenic Tg(nkx2.5BAC::ZsYellow); Tg(cmlc2::CSY) embryos expressing ZsYellow and AmCyan proteins from nkx2.5 and myocardial (cmlc2) promoters respectively (Fig. 1o–q). Lastly, these cells co-express zebrafish tgfβ3 transcripts (Supplementary Fig. 4) consistent with the demonstrated function of LTBP proteins as regulators of TGFβ signaling [11].
Figure 1. ltbp3 and nkx2.5 transcripts mark extra-cardiac cells contiguous to the outflow pole of the zebrafish heart tube.
a,b, ltbp3+ cells (white arrowhead) visualized by in situ hybridization at 24 hours post-fertilization (hpf) reside dorsal to the cmlc2+ heart tube (black arrowhead; n=15+ embryos/group). c–e, Heart tube region in 24 hpf embryo co-stained with ltbp3+ riboprobe (green; arrowhead) and a muscle-specific antibody (MF20; red; n=3/3) that recognizes cardiomyocytes. f–h, Heart tube region in 24 hpf embryo co-stained with ltbp3 and nkx2.5 riboprobes highlighting ltbp3+, nkx2.5+ cells (arrowheads) at the outflow pole of the heart tube (n=9/9). i–k, Heart tube region in 26 hpf Tg(nkx2.5::ZsYellow); Tg(cmlc2::CSY) embryo highlighting non-myocardial nkx2.5+ cells (arrowheads).
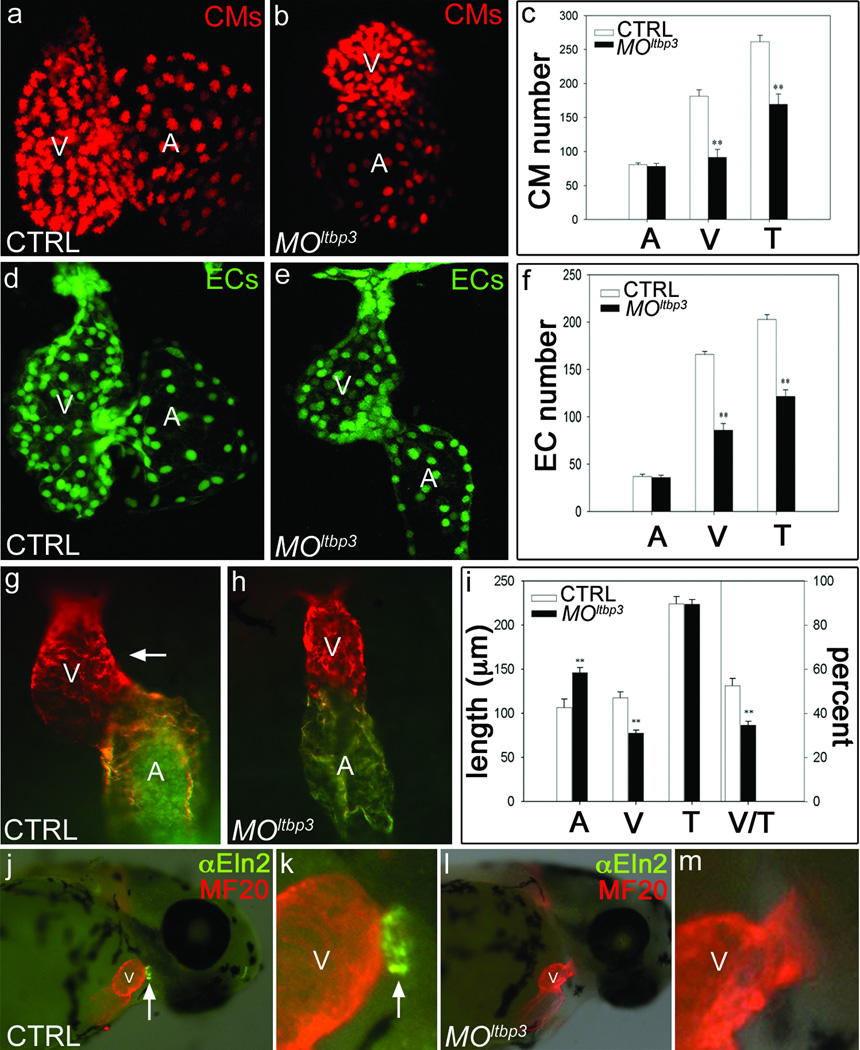
To determine if Ltbp3 is required for zebrafish cardiogenesis, we evaluated embryos injected with anti-sense ltbp3 morpholinos (MOltbp3; Supplementary Fig. 5) for chamber-specific defects in cardiomyocyte and endocardial cell number. Knocking down ltbp3 halved the number of ventricular cardiomyocytes and endocardial cells, while atrial cell numbers were unaffected (Fig. 2a–f). The ventricular deficit was evident earlier in development, soon after formation of the heart tube, as shortening of the ventricular segment accompanied by a defect in cardiac looping (Fig. 2g–i). MOltbp3 animals also lacked Eln2+ OFT smooth muscle precursor cells (Fig. 2j–m) [7, 14] homologous to SHF-derived smooth muscle surrounding the base of the aorticopulmonary trunk in higher vertebrates [10]. Lastly, ltbp3 morphants failed to form the ventral aorta (data not shown) and a majority of pharyngeal arch arteries (Supplementary Fig. 6). Taken together, these data demonstrate that knocking down ltbp3 causes multi-lineage cardiovascular defects in the pharyngeal arches, OFT, and ventricle.
Figure 2. Knocking down ltbp3 causes multi-lineage cardiovascular defects in the ventricle and OFT.
a–f, Cardiomyocyte (CM) and endocardial (EC) cell numbers in control and MOltbp3 atria (A) and ventricles (V) at 72 hpf in cmlc2::dsRed2-nuc and fli1::nEGFP embryos (n=6/group, T=total) g–i, Atrial (A) and ventricular (V) segment lengths in control and MOltbp3 embryos at 36hpf (n=9/group; white arrow highlights rightward looping of the ventricle in control embryos). Error bars in all graphs represent 1 s.d., ** p<0.01. j–m, 60 hpf control and MOltbp3 embryos stained with α-Eln2 antibodies (n=24/24 for WT; 28/31 for MOltbp3) that recognize OFT smooth muscle cell precursor cells (white arrow).
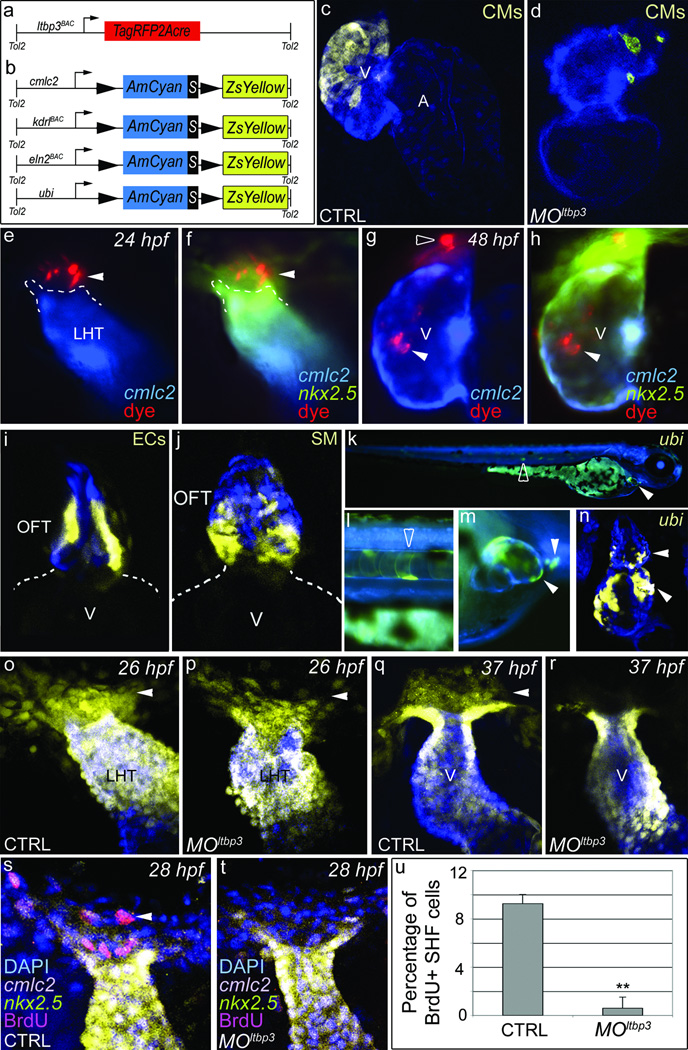
In mammals, the anterior segment of the SHF gives rise to the embryonic OFT and the right ventricular half of the common embryonic ventricle prior to septation [1–3]. Despite the fact that zebrafish embryos never septate their single ventricles, the MOltbp3 phenotype is remarkably similar to mouse anterior SHF mutants that die prior to septation with severe reductions in the primitive right ventricle and OFT [15–17]. Therefore, we tested the hypothesis that ltbp3+ cells represent a SHF-like population that gives rise to some or all of the structures missing in MOltbp3 embryos. To that end, we used Cre/Lox-mediate lineage tracing to irreversibly mark ltbp3+ cells and their descendents that assume myocardial, endocardial/endothelial, and smooth muscle cell fates. First, we derived a transgenic driver strain, Tg(ltbp3::TagRFP2Acre), that co-expresses a red fluorescent protein (TagRFP) and Cre recombinase in ltbp3+ cells (Fig. 3a and Supplementary Fig. 7). Secondly, we generated three lineage-restricted reporter strains that carry a unique Cre-responsive “color switching” cassette (AmCyan-Switch-ZsYellow; CSY) under transcriptional control of myocardial (cmlc2), endothelial (kdrl), and smooth muscle precursor cell (eln2) [7, 14] promoters (Fig. 3b and Supplementary Fig. 8). We also generated a ubiquitous reporter strain using the zebrafish ubiquitin promoter [18] (Fig. 3b and Supplementary Fig.8). For each reporter, we confirmed Cre-dependent AmCyan to ZsYellow “color switching” caused by excision of the floxed AmCyan-STOP sequence from the reporter cassette (Supplementary Fig. 8).
Figure 3. ltbp3+ cells give rise to three cardiovascular lineages in the zebrafish ventricle and OFT.
a,b, Schematic of driver (A) and reporter (B) transgenes. c,d, Tg(ltbp3::TagRFP2Acre); Tg(cmlc2::CSY) control and MOltbp3 hearts at 72 hpf (n=>20/group; >90% exhibited ZsYellow distribution shown). e–h, 24 hpf Tg(nkx2.5BAC::ZsYellow); Tg(cmlc2::CSY) embryo injected with tracking dye (arrowheads) in non-myocardial nkx2.5+ region imaged again at 48 hpf. The dye tracked to the distal ventricle (7/16 embryos accurately injected), the myocardial segment of the OFT (5/16; not shown), and both structures (6/16) respectively. Black arrowhead highlights dye that failed to migrate. i, OFT region in Tg(ltbp3::TagRFP2Acre); Tg(kdrl::CSY) embryo at 6 days post fertilization (dpf) [n=16, >50% exhibited ZsYellow fluorescence in OFT endothelial cells (ECs)]. j, OFT region in Tg(ltbp3::TagRFP2Acre); Tg(eln2::CSY) embryo at 7 dpf [n=12, >80% of exhibited ZsYellow fluorescence in OFT smooth muscle (SM) precursors]. k–n, Tg(ltbp3::TagRFP2Acre); Tg(ubi::CSY) embryo at 4 dfp (k–m) and confocal section of the heart on 6 dpf (n) [n=12, >90% of embryos exhibited ZsYellow protein in the distal ventricle (white arrowhead) and OFT (white arrowhead)]. Open arrowheads highlight color switching in the ltbp3+ notochord. o–r, Heart tube regions in control and MOltbp3 Tg(nkx2.5::ZsYellow, cmlc2::CSY) embryos at 26 and 37 hpf. Arrows indicate non-myocardial nkx2.5+ cells present in all experimental groups except MOltbp3 animals at 37hpf (n=18/18 and 12/12 for control and morphants respectively). s-u, 28 hpf control and MOltbp3 Tg(nkx2.5::ZsYellow); Tg(cmlc2::CSY) embryos, processed for DAPI and BrDU staining. The percentages of BrdU+ cells in the ZsYellow+, AmCyan− region are shown in u (n=6 embryos/group, one s.d. is shown, ** p<0.01). V=ventricle, A=atrium, LHT=linear heart tube.
Double transgenic progeny from the driver and myocardial reporter strains expressed ZsYellow protein in approximately the distal half of the ventricle (Fig. 3c) demonstrating that myocardium in this segment of the ventricle descends from ltbp3+ progenitors. Myocardial cells in the proximal OFT also arise from ltbp3+ cells (Supplementary Fig. 9). Furthermore, MOltbp3 ventricles lacked ltbp3+ cell derived cardiomyocytes confirming that the distal ventricle is specifically affected in morphant embryos (Fig. 3d). In higher vertebrates, SHF cells labeled with a tracking dye after heart tube formation migrate into the OFT tract and ventricle [12, 19, 20]. Similarly, we injected CellTrackerRed into the non-myocardial (AmCyan−), nkx2.5+ (ZsYellow+) region of double transgenic Tg(nkx2.5BAC::ZsYellow); Tg(cmlc2::CSY) embryos at the heart tube stage and observed subsequent dye migration into the distal ventricle and OFT (Fig. 3 e–h). Taken together, these data demonstrate that ltbp3+ cells give rise to distal ventricular myocardium through late differentiation and accretion to the heart tube.
By crossing our driver line with the endothelial and smooth muscle reporter strains, we learned that ltbp3-expressing cells also give rise to endothelial and smooth muscle cells in the OFT (Fig. 3i, j). In smooth muscle reporter embryos, ZsYellow fluorescence was predominantly observed at the base of the OFT (Fig. 3j), suggesting that ltbp3+ cells make a regionalized contribution to OFT smooth muscle. We did not observe color switching in ventricular endocardium or pharyngeal arch artery endothelium suggesting that these cellular compartments do not derive from ltbp3+ cells (data not shown). Finally, lineage tracing ltbp3+ cells with the ubiquitous reporter corroborated the conclusions drawn from lineage-restricted reporters (Fig. 3k–n). Taken together, these data demonstrate that ltbp3+ cells, in addition to giving rise to ventricular myocardium, also give rise to three cardiovascular lineages in the OFT.
To elucidate the cellular mechanism(s) underlying the MOltbp3 phenotype, we evaluated MOltbp3 embryos, soon after formation of the heart tube, for cellular defects in the extra-cardiac population of ltbp3+, nkx2.5+ cells at its outflow pole. Although TagRFP-expressing ltbp3+ cells are too faint to visualize in Tg(ltbp3::TagRFP2Acre) embryos at this developmental stage, non-myocardial ZsYellow-expressing nkx2.5+ cells are readily detected in Tg(nkx2.5::ZsYellow); Tg(cmlc2::CSY) embryos (Fig. 1o–q). Shortly after heart tube formation, both control and MOltbp3 embryos harbored extra-cardiac nkx2.5+ cells demonstrating that progenitor specification is not compromised by loss of ltbp3 function (Fig. 3o, p). However, eleven hours later, this population was absent specifically in MOltbp3 animals (Fig. 3q, r). The absence of TUNEL staining suggested strongly that progenitor survival is not compromised (data not shown). By contrast, BrdU staining revealed that progenitor proliferation is significantly reduced in MOltbp3 embryos (Fig. 3s–u). Taken together, these data support a model in which ltbp3 functions in an autocrine fashion to stimulate proliferation or self-renewal of ltbp3+, nkx2.5+, progenitors to sustain adequate CPC numbers during their differentiation and migration from the SHF. Based on this model, we speculate that knocking down ltbp3 causes a premature depletion of CPCs from the SHF.
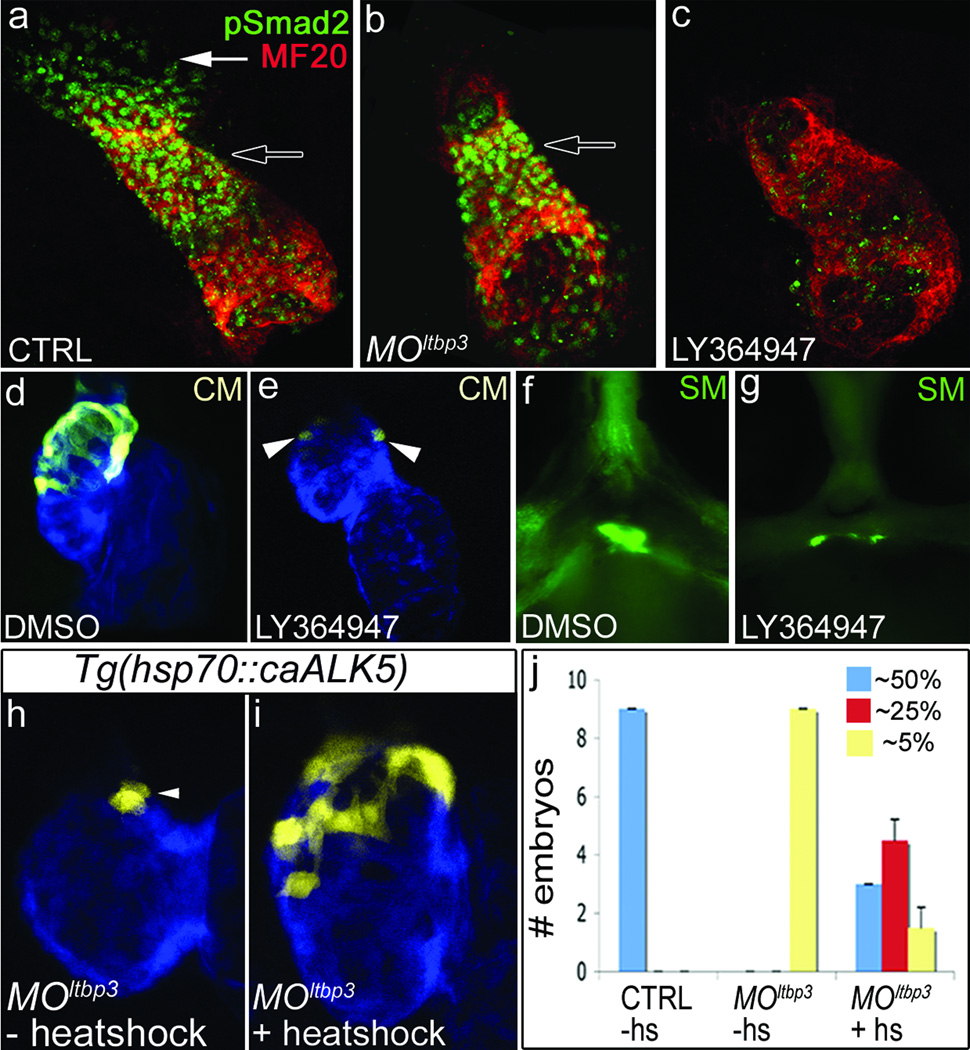
LTBP proteins are secreted with TGFβ ligands in a large latent complex (LLC) that becomes anchored to the extracellular matrix prior to ligand activation [11]. Because LTBP proteins are also required for secretion of TGFβ ligands, we used three approaches to test the hypothesis that defective TGFβ signaling underlies the cardiovascular phenotypes seen in MOltbp3 embryos. First, we evaluated the linear heart tube and surrounding regions for phosphorylated pSmad2 epitopes, a hallmark of active TGFβ signaling [21]. TGFβ signaling was observed in the heart tube proper and in extra-cardiac cells approximating the location of ltbp3+, nkx2.5+ progenitors (Fig. 4a). Loss of ltbp3 function eliminated pSmad2 epitopes specifically in the extra-cardiac population leaving signaling in the heart tube unaffected. These data demonstrate that morphant embryos exhibit a highly localized defect in TGFβ signaling specifically in the SHF (Fig. 4B). Next, we evaluated whether small molecule inhibition of TGFβ signaling would phenocopy MOltbp3 animals. To that end, we used a selective inhibitor (LY-364947; [22]) of the TGFβ type I receptor ALK5 that effectively abrogated TGFβ signaling in zebrafish embryos (Fig 4c). LY-364947-treated animals phenocopied ltbp3 morphants as evidenced by reductions in ltbp3+ cell derived myocardium and OFT smooth muscle (Fig. 4d–g). Using LY-364947, we also determined the developmental time window, 26–36 hours post-fertilization during which TGFβ signaling is required for the formation of the distal ventricle (Supplementary Fig. 10), the same time window during which nkx2.5+ progenitors disappear in MOltbp3 embryos (Fig. 3o–r). Lastly, we attempted to rescue MOltbp3 embryos by inducing ubiquitous expression of a constitutively active TGFβ-type I receptor (caALK5), the same receptor targeted by LY-364947, predicted to be downstream of ltbp3 and TGFβ ligands (Supplementary Fig. 9). In a majority of embryos, heat-shock inducible expression of caALK5 partially or fully rescued the loss of distal myocardium seen in MOltbp3 embryos (Fig. 4h–j). Taken together, these results suggest that attenuation of TGFβ signaling underlies the cardiovascular phenotypes present in MOltbp3 embryos.
Figure 4. Diminished TGFβ signaling underlies the MOltbp3 cardiovascular phenotypes.
a–c, Heart tube regions in 24 hpf control, MOltbp3, and LY364947-treated embryos co-stained with a pSmad2 antibody and a muscle-specific antibody (MF20) that recognizes cardiomyocytes (n =20/20 for a, n=11/11 for b, n=15/15 for c). Black and white arrows highlight the heart tube and extra-cardiac pSmad2+ cells respectively. d, e, 72 hpf hearts in Tg(ltbp3::TagRFP2Acre); Tg(cmlc2::CSY) embryos treated with DMSO or LY364947 (n=8/8 per group); CM=cardiomyocytes. f, g, OFT region in 60 hpf embryos treated with DMSO or LY364947 and immunostained for Eln2 epitopes (n=20/20). SM=smooth muscle precursors. h–j, Control and MOltbp3 Tg(ltbp3::TagRFP2Acre); Tg(cmlc2::CSY); Tg(hsp70::caALK5) embryos were heat shocked (+hs) or not (−hs) and evaluated visually at 72hpf. Graph in (k) indicates the number of embryos in each experimental group that expressed ZsYellow protein in the distal ~50% (full rescue), ~25% (partial rescue), or ~5% (no rescue) of the ventricle. Shown in (j) is a heart that is partially rescued.
Because ltbp3+ cells differentiate after formation of the heart tube in pharyngeal mesoderm, elongate the tube at the outflow pole, and give rise to three cardiovascular lineages in the OFT and myocardium in the distal ventricle, they exhibit defining characteristics of the mouse anterior SHF. As such, ltbp3+ cells would also be predicted to encompass the zebrafish equivalents of the avian secondary and anterior heart fields [1, 19, 23].
Our findings are consistent with a recent study demonstrating that pre-existing ventricular cardiomyocytes do not give rise to new cardiomyocytes during elongation of the zebrafish heart tube [9]. The same authors report that the zebrafish homolog of Islet1, a well-known marker of mammalian SHF cells, is dispensable for formation of the distal ventricle, herein reported to derive from the zebrafish SHF. Thus, it appears that the critical role of Islet1, during anterior SHF development, has become minimized or replaced by another factor specifically in the fish lineage. Alternatively, Islet1 function during mammalian anterior SHF development might have co-evolved with the four-chambered heart [24].
Although zebrafish do not have a right ventricle, ltbp3+ cells accrete the distal ventricular segment to the zebrafish heart tube much like SHF cells accrete the primitive right ventricle to the mouse heart tube prior to septation. In mouse, the SHF-derived segment becomes a distinct chamber after septation. In zebrafish, the SHF-derived segment instead augments the singular ventricular chamber, a trait that may have conferred an evolutionary advantage to a common ancestor of both zebrafish and mammals.
Methods Summary
Single and double, fluorescent and non-fluorescent, in situ hybridization and immunohistochemical stainings were performed using standard protocols. To analyze the ltbp3 loss of function phenotype, we injected anti-sense ltbp3 morpholinos into one-cell stage WT and transgenic embryos. For genetic lineage tracing, a transgenic driver strain expressing Cre recombinase in ltbp3+ cells and four Cre-responsive “color switching” reporter strains were generated using standard methods. The driver strain was crossed individually to each of the reporter strains and their double transgenic progenies were analyzed for ZsYellow protein fluorescence using confocal microscopy. To follow the migration of zebrafish SHF cells, a tracking dye was injected into the ZsYellow+, AmCyan− region of Tg(nkx2.5::ZsYello); Tg(cmlc2::CSY) embryos at 24hpf, the embryos were imaged immediately, and then again at 48 and/or 72hpf. A transgenic strain carrying a cDNA encoding a constitutively active human TGFβ type I receptor (caALK5) under control of the zebrafish heat shock promoter was generated and used to rescue the myocardial defect in ltbp3 morphant embryos.
Full Methods and any associated references are available in the online version of the paper at www.natre.com/nature
Supplementary Material
Acknowledgements
We thank M. Whitman for advice on pSmad2 staining, D. Hami and M. Kirby for providing Eln2 anti-sera and their immunohistochemistry protocol, R. Cornell for providing tfAP2a and tfAP2c morpholinos, M. Whitman, A. Srinivasan, D. Langenau, E. Provost, S. Leach, R. Anderson, D. Stainier, I.Woods, and A. Schier for providing plasmids. J.W. Xiong for providing clom39 fish, B. Barut and L. Zon for providing BACs, and the MGH Nephrology Division for access to their confocal microscopy facilities. S.C., R.E.P., and W.H. were supported by NIH grant R01 ES012716 from the National Institute of Environmental Health Sciences. C.M. received support through an EMBO long-term fellowship and a HFSP long-term fellowship. This work was funded by the Cardiovascular Research Center at Massachusetts General Hospital, a Claflin Distinguished Scholar Award and Harvard Stem Cell Institute Seed Grant to C.E. Burns, and by awards from the National Heart Lung and Blood Institute (5R01HL096816), American Heart Association (Grant in Aid # 10GRNT4270021), and Harvard Stem Cell Institute (Seed Grant) to C.G. Burns.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Y.Z. performed the majority of the experiments and analyzed data; T.C., K.N., P.O, Y.L., A.G, and S.S. performed experiments and analyzed data; C.M. provided the ubiquitin promoter prior to publication; S.C., R.P., W.H. and C.G.B. discovered that ltbp3 transcripts are upregulated in a cardiac fraction on day 3 post fertilization; C.G.B performed experiments including BAC recombineering; C.E.B. and C.G.B. co-directed the study, analyzed data, and wrote the paper with input from all authors.
Author Information The zebrafish ltbp3 cDNA sequence was deposited into GenBank under accession number JF731042.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol. 2009;336(2):137–144. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rochais F, Mesbah K, Kelly RG. Signaling pathways controlling second heart field development. Circ Res. 2009;104(8):933–942. doi: 10.1161/CIRCRESAHA.109.194464. [DOI] [PubMed] [Google Scholar]
- 3.Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- 4.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313(5795):1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meilhac SM, et al. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6(5):685–698. doi: 10.1016/s1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 7.Grimes AC, et al. Solving an enigma: arterial pole development in the zebrafish heart. Dev Biol. 2006;290(2):265–276. doi: 10.1016/j.ydbio.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 8.Brade T, et al. The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev Biol. 2007;311(2):297–310. doi: 10.1016/j.ydbio.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 9.de Pater E, et al. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136(10):1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimes AC, et al. Phylogeny informs ontogeny: a proposed common theme in the arterial pole of the vertebrate heart. Evol Dev. 2010;12(6):552–567. doi: 10.1111/j.1525-142X.2010.00441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280(9):7409–7412. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- 12.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 2001;1(3):435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 13.Chen JN, et al. Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development. 1997;124(21):4373–4382. doi: 10.1242/dev.124.21.4373. [DOI] [PubMed] [Google Scholar]
- 14.Miao M, et al. Differential expression of two tropoelastin genes in zebrafish. Matrix Biol. 2007;26(2):115–124. doi: 10.1016/j.matbio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Ilagan R, et al. Fgf8 is required for anterior heart field development. Development. 2006;133(12):2435–2445. doi: 10.1242/dev.02408. [DOI] [PubMed] [Google Scholar]
- 16.Prall OW, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Both I, et al. Foxh1 is essential for development of the anterior heart field. Dev Cell. 2004;7(3):331–345. doi: 10.1016/j.devcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Mosimann C, et al. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2010;138(1):169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldo KL, et al. Conotruncal myocardium arises from a secondary heart field. Development. 2001;128(16):3179–3188. doi: 10.1242/dev.128.16.3179. [DOI] [PubMed] [Google Scholar]
- 20.Zaffran S, et al. Right ventricular myocardium derives from the anterior heart field. Circ Res. 2004;95(3):261–268. doi: 10.1161/01.RES.0000136815.73623.BE. [DOI] [PubMed] [Google Scholar]
- 21.Todorovic V, et al. Long form of latent TGF-beta binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development. 2007;134(20):3723–3732. doi: 10.1242/dev.008599. [DOI] [PubMed] [Google Scholar]
- 22.Li HY, et al. Dihydropyrrolopyrazole transforming growth factor-beta type I receptor kinase domain inhibitors: a novel benzimidazole series with selectivity versus transforming growth factor-beta type II receptor kinase and mixed lineage kinase-7. J Med Chem. 2006;49(6):2138–2142. doi: 10.1021/jm058209g. [DOI] [PubMed] [Google Scholar]
- 23.Mjaatvedt CH, et al. The outflow tract of the heart is recruited from a novel heart-forming field. Dev Biol. 2001;238(1):97–109. doi: 10.1006/dbio.2001.0409. [DOI] [PubMed] [Google Scholar]
- 24.Kang J, et al. Isl1 is a direct transcriptional target of Forkhead transcription factors in second-heart-field-derived mesoderm. Dev Biol. 2009;334(2):513–522. doi: 10.1016/j.ydbio.2009.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.