Abstract
Diamond-Blackfan Anemia(DBA) is a congenital hypoproliferative macrocytic anemia; 5q-syndrome myelodysplastic syndrome(MDS) is an acquired hypoproliferative macrocytic anemia. Their common erythroid phenotype reflects a shared pathophysiology -- haploinsufficiency of one of many ribosomal proteins and somatic deletion of one allele of the ribosomal protein S14 gene, respectively. Although these abnormalities lead to defective ribosome biogenesis, why ribosomal protein hemizygosity results in anemia is not certain. Here, we characterize the hematopoietic phenotype of mice lacking one allele of the ribosomal protein S6 gene. The mice have an erythroid phenotype similar to both DBA and the 5q-syndrome and lenalidomide therapy improves their anemia.
Keywords: ribosomes, erythropoiesis
DBA is a macrocytic anemia with low CFU-E and variable deficiency of BFU-E[1, 2]. An elevated erythrocyte adenosine deaminase activity (eADA, elevated in >80% of patients) is an important supporting feature[3]. Granulocytopenia, thrombocytopenia, or thrombocytosis are occasionally seen at diagnosis or during follow-up[4]. 5q-syndrome is a subtype of myelodysplastic syndrome defined by the isolated interstitial deletion of chromosome 5q, which results in macrocytic anemia, variable neutropenia, and a normal or high platelet count associated with hypolobated megakaryocytes.
The erythroid phenotype of both diseases is linked to ribosomal protein haploinsufficiency and defective pre-ribosomal RNA processing or ribosome biogenesis[5]. Mutations in at least nine ribosomal protein genes have been identified in over 50% of DBA patients; Rps14 is haploinsufficient in 5q-syndrome. Both abnormalities impair erythroid differentiation in vitro[5] and in vivo[6]. Accumulating evidence suggests that haploinsufficiency of certain ribosomal proteins and/or defective ribosome biogenesis triggers p53 activation and cell cycle arrest and/or apoptosis[6-9]. Whether p53 activation is solely responsible for the anemia is debated and alternative or contributing physiologies remain open.
Our understanding is hindered by inadequate murine models. The initial Rps19 null mouse is lethal and heterozygous mice lack a DBA phenotype[10]. A chemical mutagenesis screen in mice identified a missense mutation of Rps19 in a mouse with a dominantly inherited dark skin phenotype[7]. While the mouse, like DBA patients, has a hypoproliferative, macrocytic anemia, the anemia is very mild, thus limiting this model’s utility. A mouse expressing a dominant negative (and not haploinsufficient) Rps19 allele exists[11]. Zebrafish models of Rps19 knockdown recapitulate the hematologic phenotype and result in malformations[8]. Mice engineered with hematopoietic-specific haploidy of a set of genes on 5q including Rps14 develop macrocytic anemia, prominent erythroid dysplasia and monolobated megakaryocytes consistent with the phenotype of 5q-syndrome, making this the most promising model for study[6], although the deletion of adjacent genes on 5q could impact hematopoiesis[12] and complicate studies.
We became aware of mice with postnatal deletion of Rps6, which encodes a 40S ribosomal subunit protein[13]. Embryos with haploinsufficiency of RPS6 are runted and die at gastrulation (≤ E8.5). Genetic inactivation of p53 bypasses this checkpoint, prolonging development until E12.5, at which point the embryos likely die from anemia[14]. Conditional deletion of the Rps6 gene in murine liver abrogates 40S ribosomal biogenesis and prevents hepatocytes from reentering cell cycle after partial hepatectomy[13]; conditional deletion of one Rps6 allele in murine T cells induces a p53-dependent checkpoint response that abolishes activated T cell proliferation[15]. The erythropoietic phenotype of mice lacking one Rps6 allele postnatally (Rps6flox/wild-type; Mx-cre) was very recently published. The animals recapitulate cardinal features of the 5q-syndrome, including macrocytic anemia, erythroid hypoplasia, and megakaryocyctic dysplasia with thrombocytosis[16]. Of note, RPS6 mutations have not been reported in DBA or MDS.
Here, we also characterize Rps6 heterozygously-deleted mice and confirm that the erythroid phenotype in these mice phenocopies 5q-syndrome MDS and DBA. In addition, we tested their erythroid response to DBA and 5q-syndrome MDS therapies.
Methods
Rps6flox/flox;Mx-cre and Rps6flox/flox [13] mice were a generous gift from George Thomas, University of Cincinnati. Animals were interbred to maintain a Rps6flox/flox;Mx-cre colony, which was bred to C57BL/6 mice for studies. Rps6flox/wildtype;Mx-cre were interbred to confirm that the hematopoietic parameters of Rps6flox/wildtype, Rps6wildtype/wildtype;Mx-cre and Rps6flox/wildtype were the same after poly(I)-poly(C) treatment. Subsequent studies used Rps6flox/wildtype as controls. To induce Mx-cre expression and deletion of the floxed-allele, 5-7 day-old pups were treated with 40μg of poly(I)-poly(C) (Amersham/GE Lifesciences, NJ) intraperitoneally every other day for three injections. Animals were sacrificed 5-6 weeks later. Single-cell suspensions of freshly prepared marrow or spleen were immunostained with anti-Ter119-APC(or-PE) and anti-CD-71-FITC (BD Pharmingen, CA) antibodies. Flow cytometry and GM-colony assays were performed as described[17]. To detect BFU-E and CFU-E colonies, 2×105 and 3×105 cells/plate, respectively were plated in duplicate in MethocultTM M3334 (StemCell Technologies, Canada) plus 100ng/ml mSCF according to manufacturer’s protocol. Additional cultures (± dexamethasone) were performed adapting the Narla methods[18]. Blood cell analyses used a Hemavet HV950FS analyzer (Drew Scientific, CT). Absolute reticulocyte counts were performed by Phoenix Central Laboratories, WA. Bert Glader’s laboratory performed eADA measurements (Stanford University, CA)[3]. Brian Kennedy, Buck Institute, kindly provided Rpl22 and Rpl29 haploinsufficient and null mice. Disruption of murine Rpl22 null was accomplished using an ES cell line (Bay Genomics) with a gene trap inserted between exons 3 and 4 to ablate Rpl22 expression (Stanfel et al., unpublished). Rpl29 null mice were previously generated [19].
Results and Discussion
Rps6 heterozygously-deleted mice develop a hypoproliferative, macrocytic anemia, granulocytopenia, thrombocytosis, and also lymphopenia (Table 1). Akin to DBA patients, eADA is elevated (2.30±0.11, n=6 vs. 1.13±0.02 in controls, n=6; mean±SEM, two-tailed Student’s t-test, p<8E−5).
Table 1. Hematopoietic parameters.
A. Rps6 heterozygously-deleted mice. CBC analyses of transgenic mice at 6-7 weeks of age, 5-6 weeks post deletion: mean±SEM, two-tailed Student’s t-test. The absolute reticulocyte count of heterozygously-deleted Rps6 vs. control mice is 32±2×104/μl, n=9 vs. 35±3×104/μl, n=9; mean±SEM, two-tailed Student’s t-test, p= 0.5. A normal absolute reticulocyte count in the deleted animals is inappropriately low given their anemia. Rps6flox/flox;Mxcre (n=3) died within 3 weeks of deletion with p(I)p(C) prior to analysis or had severe pancytopenia. B. Rpl29 and Rpl22 heterozygous mice. CBC analyses of adult mice, mean±SEM, two-tailed Student’s t-test (compared to controls). Studies are ongoing to determine if these Rpl mutants have tissue-specific ribosome biogenesis defects.
| A. | |||
|---|---|---|---|
| Rps6 hets (n = 18) | Controls (n= 24) | p | |
| WBC, k/μl | 3.7 ± 0.3 | 9.0 ± 0.4 | <4E-14 |
| ANC, k/μl | 0.9 ± 0.1 | 2.1 ± 0.1 | <4E-10 |
| ALC, k/μl | 2.4 ± 0.2 | 6.2 ± 0.3 | <1E-12 |
| RBC, m/μl | 5.6 ± 0.3 | 9.3 ± 0.1 | <3E-12 |
| HGB, g/dl | 9.2 ± 0.4 | 13.9 ± 0.2 | <2E-9 |
| HCT, % | 32.4 ± 1.4 | 45.2 ± 0.4 | <3E-8 |
| MCV, fL | 58.4 ± 0.8 | 48.7 ± 0.4 | <2E-11 |
| MCH, pg | 16.6 ± 0.3 | 15.0 ± 0.2 | <5E-5 |
| MCHC, % | 28.4 ± 0.3 | 30.4 ± 0.5 | 0.005 |
| RDW, % | 27.3 ± 1.1 | 19.1 ± 0.5 | <9E-7 |
| Platelets, k/μl | 1679.5 ± 81.9 | 900.5 ± 24.8 | <2E-8 |
| B. | ||||||
|---|---|---|---|---|---|---|
|
rpL29 hets (n = 4) |
Controls (n= 4) |
p |
rpL22 hets (n = 4) |
Controls (n= 4) |
p | |
| WBC, k/ul | 11.2 ± 1.0 | 12.1 ± 1.0 | 0.52 | 11.4 ± 0.8 | 9.6 ± 1.6 | 0.35 |
| ANC, k/ul | 2.2 ± 0.4 | 2.3 ± 0.3 | 0.86 | 2.1 ± 0.2 | 1.6 ± 0.2 | 0.13 |
| ALC, k/ul | 8.7 ± 0.7 | 9.4 ± 1.1 | 0.60 | 8.6 ± 0.7 | 7.5 ± 1.4 | 0.50 |
| RBC, m/ul | 10.7 ± 0.1 | 10.3 ± 0.3 | 0.26 | 10.1 ± 0.5 | 10.0 ± 0.3 | 0.87 |
| HGB, g/dl | 15.7 ± 0.2 | 14.9 ± 0.6 | 0.22 | 14.5 ± 0.6 | 14.9 ± 0.3 | 0.54 |
| HCT, % | 48.3 ± 0.8 | 45.9 ± 2.2 | 0.36 | 45.6 ± 1.7 | 46.1 ± 0.9 | 0.78 |
| MCV, fL | 45.1 ± 0.7 | 44.6 ± 0.9 | 0.66 | 45.2 ± 2.3 | 46.0 ± 1.4 | 0.76 |
| MCH, pg | 14.7 ± 0.1 | 14.5 ± 0.2 | 0.42 | 14.3 ± 0.1 | 14.9 ± 0.3 | 0.06 |
| MCHC, % | 32.6 ± 0.2 | 32.5 ± 0.4 | 0.87 | 31.8 ± 1.3 | 32.3 ± 0.5 | 0.72 |
| RDW, % | 17.8 ± 0.2 | 17.7 ± 0.4 | 0.87 | 18.7 ± 0.6 | 18.3 ± 0.4 | 0.60 |
| Platelets, k/ul | 846.5 ± 36.0 | 1129.5 ± 215.5 | 0.24 | 740.5 ± 57.3 | 855.8 ± 75.4 | 0.27 |
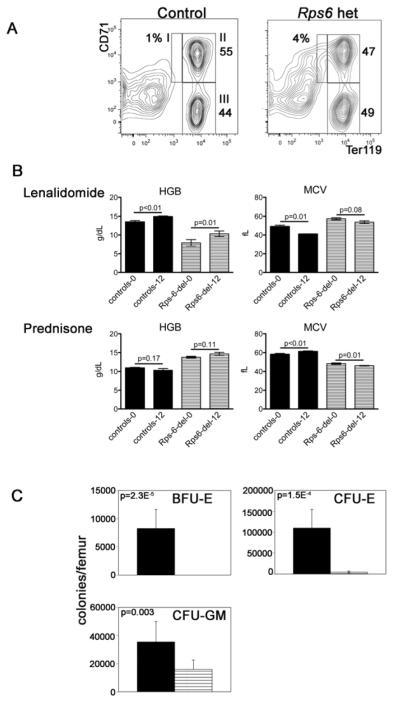
Flow cytometric analyses of marrow double-stained for Ter119 and CD71 demonstrate a relative expansion of proerythroblasts in mutant mice (population I, Figure 1A). In methylcellulose culture, CFU-E-derived colonies are markedly reduced in number and no BFU-E are detected. Thus, as in DBA, erythropoiesis is impaired both at[2] and before[1] the CFU-E stage. CFU-GM are also reduced (Figure 1C).
Figure 1. Hematologic characterization of Rps6 heterozygously-deleted mice.
(A) Representative flow cytometric analyses of whole bone marrow immunostained with antibodies to CD71 (transferrin receptor) and Ter119 (erythroid specific). The relative percentages of nucleated cells in each of the populations I to III are indicated. We found a statistically significant increase in the percentage of cells in population I in Rps6 heterozygously-deleted mice, 4.4%±1.0 vs. 1.2±0.1, p= 0.03, mean±SEM, two-tailed Student’s t-test, 4 mice in each group. Analyses of spleens showed similar findings. (B) Hemoglobin (HGB) and mean corpuscular values (MCV) of Rps6 heterozygously-deleted mice (striped, Rps-del-0 and Rps-del-12) and controls (solid, controls-0 and controls-12) treated with lenalidomide (top, rpS6 hets n= 7, controls n=5) or prednisone (bottom, rps6 hets n= 19, controls n= 8) for 12 weeks (baseline and 12-week values). The administered dose was based on pharmacokinetic studies in humans and rats[24] and communication with Celgene Corporation; it is estimated to achieve an ~2.2μM concentration of drug in the mice, equivalent to the therapeutic concentration achieved with a 25 mg/day dose of lenalidomide to patients with multiple myeloma[25]. Prednisone was dosed in their daily ad libitum food supply at approximately 2 mg prednisone/kg body weight per day (assuming a 20 gram mouse eating 4 grams diet/day). We excluded the possibility that the improvement in hemoglobin with lenalidomide reflects a preferential expansion of normal (undeleted) cells in the lenalidomide cohort by Southern blot (SK data not shown). A cohort of Rps6 heterozygously-deleted and control mice, age-matched to the animals treated with prednisone or lenalidomide were followed with monthly CBC analyses (n=3 in each group); in these animals the hemoglobin was not statistically different between the start and end of 12 weeks of monitoring (data not shown). We also analyzed the MCV and hemoglobin data in the prednisone-treated cohort as individual mice and observed no response to therapy. Our treatment studies are limited by not following drug levels in animals.
(C) Hematopoietic colony assays in Rps6 heterozygously-deleted mice (striped, n=6) and controls (solid, n=6). Mean±SEM, two-tailed Student’s t-test. That CFU-E were present while BFU-E were not detected suggests that our culture conditions did not optimally support BFU-E to CFU-E differentiation (since the detection of BFU-E in this assay requires that the plated BFU-E differentiate through the CFU-E-stage for enumeration), which has been reported in colony assays of DBA patients[26].
Mice with constitutive deletion and haploinsufficiency of Rpl29 or Rpl22 were also studied (Table 1). Murine embryonic fibroblasts derived from rpL29 null mice demonstrate a cell cycle delay and the animals exhibit a global skeletal growth defect[19] and αβ-T cell development is specifically impaired at a p53-dependent checkpoint in rpL22 null mice[20]. Hematologic parameters are normal in these animals. Interestingly, DBA patients have short stature. To date, the only reported genotype-phenotype associations in DBA are mutations in RPL5 with craniofacial clefting and RPL11 with thumb abnormalities[21]. These animal models (with nonerythroid phenotypes) demonstrate that ribosomal protein haploinsufficiency results in tissue-specific phenotypes. Animal models with haploinsufficiency of different ribosomal proteins are thus relevant platforms to study p53 dosage, tissue-specific ribosomal protein expression, and disease modifiers; these may also expand our clinical recognition of diseases due to ribosomal protein haploinsufficiency.
The erythroid phenotype of Rps6 heterozygously-deleted mice provided the rationale to treat the animals with corticosteroids, the mainstay of therapy for DBA, and lenalidomide, which results in a red cell and cytogenetic response in 5q-syndrome MDS and has not been clinically tested in DBA.
Corticosteroids improve the hemoglobin within 2-4 weeks in ~70-80% of DBA patients; we are unaware of well-designed clinical studies using corticosteroids in 5q-syndrome MDS. There was no improvement in the hemoglobin and minimal alteration of red cell size in Rps6 heterozygously-deleted mice during 12 weeks of prednisone therapy (Figure 1). Our finding of no increase in the numbers of BFU-E, CFU-E, or Ter119+ cells when Rps6 heterozygously-deleted (n=5) or control (n=5) murine marrow cells were cultured in the presence of 100nM dexamethasone (JA and KS, data not shown) is consistent with the in vivo observations, yet contrast studies of normal human progenitor cells and cells expressing RPS14 or RPS19 shRNA[18].
We next tested whether the macrocytic anemia in Rps6 heterozygously-deleted mice responds to lenalidomide (Revlimid®). Patients with the 5q-syndrome have a striking response to lenalidomide with 70% of patients achieving transfusion independence[22]. To date, no clinical trials or animal models of DBA have tested this application. The hemoglobin increased in control mice and markedly increased in Rps6 heterozygously-deleted mice after 12 weeks of 3mg/kg/day of lenalidomide by oral gavage (Figure 1B, 13.5±0.4 to 14.9±0.2, p<0.01 and 7.9g/dL±0.9 to 10.3±0.8, p=0.01, respectively; mean±SEM, Student’s t-test, paired). Additionally, the MCV decreased with therapy in both groups (49.1fL±1.4 to 41.1±0.2, p=0.01 and 57.4±1.1 to 53.77±1.4, p=0.08, respectively). These data coupled with data in an erythroid culture of human CD34+ progenitor cells expressing shRNA against RPS19 or RPS24 treated with lenalidomide[18] suggest that lenalidomide improves hemoglobinization. Lenalidomide may have additive effects to steroids in improving erythropoiesis in DBA and 5q-syndrome patients[18]. As erythropoiesis improved in control mice, it is unclear if the improvement in Rps6 heterozygously-deleted mice is specific. The lack of understanding of the mode of action of lenalidomide in 5q-syndrome MDS, the suggestion that the erythroid response may be mediated in part by haploinsufficiency of two phosphatase genes located on 5q[23] independent of a ribosome biogenesis defect, and the drug’s clinical risk of causing neutropenia and thrombocytopenia underscore the importance of animal models for testing its therapeutic efficacy in DBA. Our studies establish Rps6 heterozygously-deleted mice as one available model.
Acknowledgements
Special thanks to Bert Glader for measuring eADA levels in the mice and Marilyn Sanchez-Bonilla for technical assistance.
Support and financial disclosure declaration
This work was supported by grants from the National Institutes of Health (K08 DK075422-05, P30 CA147883-01, R01 31823, and R01 AG033373-01A1, P20 RR016458) and the Ellison Medical Foundation (AG-IA-0052-05). Lenalidomide was a gift from Celgene Corporation, San Diego, CA, and the authors have no competing interests to declare.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: Siobán Keel, Susan Phelps, Kathleen Sabo, Monique O’Leary, Catherine B. Kirn-Safran, and Janis L. Abkowitz-all none.
References
- [1].Nathan DG, Clarke BJ, Hillman DG, Alter BP, Housman DE. Erythroid precursors in congenital hypoplastic (Diamond-Blackfan) anemia. J Clin Invest. 1978;61:489–498. doi: 10.1172/JCI108960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ohene-Abuakwa Y, Orfali KA, Marius C, Ball SE. Two-phase culture in Diamond Blackfan anemia: localization of erythroid defect. Blood. 2005;105:838–846. doi: 10.1182/blood-2004-03-1016. [DOI] [PubMed] [Google Scholar]
- [3].Glader BE, Backer K, Diamond LK. Elevated erythrocyte adenosine deaminase activity in congenital hypoplastic anemia. N Engl J Med. 1983;309:1486–1490. doi: 10.1056/NEJM198312153092404. [DOI] [PubMed] [Google Scholar]
- [4].Willig TN, Niemeyer CM, Leblanc T, et al. DBA group of Societe d’Hematologie et d’Immunologie Pediatrique (SHIP), Gesellshaft fur Padiatrische Onkologie und Hamatologie (GPOH), and the European Society for Pediatric Hematology and Immunology (ESPHI) Identification of new prognosis factors from the clinical and epidemiologic analysis of a registry of 229 Diamond-Blackfan anemia patients. Pediatr Res. 1999;46:553–561. doi: 10.1203/00006450-199911000-00011. [DOI] [PubMed] [Google Scholar]
- [5].Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q-syndrome. Nat Med. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- [9].Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Matsson H, Davey EJ, Draptchinskaia N, et al. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24:4032–4037. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond-Blackfan anemia. Blood. 2010 doi: 10.1182/blood-2010-03-275776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Starczynowski DT, Kuchenbauer F, Argiropoulos B, et al. Identification of miR-145 and miR-146a as mediators of the 5q-syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- [13].Volarevic S, Stewart MJ, Ledermann B, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- [14].Panic L, Tamarut S, Sticker-Jantscheff M, et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26:8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19:3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McGowan KA, Pang WW, Bhardwaj R, et al. Reduced ribosomal protein gene dosage and p53 activation in low risk myelodysplastic syndrome. Blood. 2011 doi: 10.1182/blood-2010-11-318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Keel SB, Doty RT, Yang Z, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319:825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- [18].Narla A, DS McAuley, JR, Al-Shahrour F, Hurst S, McConkey M, Neuberg D, Ebert BL. Dexamethasone and lenalidomide have distinct funcitonal effects on erythropoiesis. Blood. 2011 doi: 10.1182/blood-2010-11-318543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kirn-Safran CB, Oristian DS, Focht RJ, Parker SG, Vivian JL, Carson DD. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn. 2007;236:447–460. doi: 10.1002/dvdy.21046. [DOI] [PubMed] [Google Scholar]
- [20].Anderson SJ, Lauritsen JP, Hartman MG, et al. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- [21].Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83:769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].List A, Dewald G, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355:1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- [23].Komrokji RS, Lancet JE, List AF. Lenalidomide in myelodysplastic syndromes: an erythropoiesis-stimulating agent or more? Curr Hematol Malig Rep. 2010;5:9–14. doi: 10.1007/s11899-009-0036-z. [DOI] [PubMed] [Google Scholar]
- [24].Dredge K, Horsfall R, Robinson SP, et al. Orally administered lenalidomide (CC-5013) is anti-angiogenic in vivo and inhibits endothelial cell migration and Akt phosphorylation in vitro. Microvascular research. 2005;69:56–63. doi: 10.1016/j.mvr.2005.01.002. [DOI] [PubMed] [Google Scholar]
- [25].Chen N, Lau H, Kong L, et al. Pharmacokinetics of lenalidomide in subjects with various degrees of renal impairment and in subjects on hemodialysis. Journal of clinical pharmacology. 2007;47:1466–1475. doi: 10.1177/0091270007309563. [DOI] [PubMed] [Google Scholar]
- [26].Abkowitz JL, Sabo KM, Nakamoto B, et al. Diamond-blackfan anemia: in vitro response of erythroid progenitors to the ligand for c-kit. Blood. 1991;78:2198–2202. [PubMed] [Google Scholar]