Abstract
Eukaryotic genomes are packaged into chromatin, where diverse histone modifications can demarcate chromatin domains that are amenable or block gene expression. While silent chromatin has been associated with long noncoding RNAs (lncRNAs) for some time, new studies suggest that noncoding RNAs also modulate the active chromatin state. Divergent, antisense, and enhancer-like intergenic noncoding RNAs can either activate or repress gene expression by altering histone H3 lysine 4 methylation. An emerging class of enhancer-like lncRNAs may link chromosome structure to chromatin state and establish active chromatin domains. The confluence of several new technologies promises to rapidly expand this fascinating topic of investigation.
Introduction
Diverse sets of cellular mechanisms are employed to properly control gene expression under normal and stressed states. Organization of eukaryotic DNA into chromatin represents a significant layer of gene regulation, and active chromatin—representing sites that are available for transcription and other DNA-templated processes—are marked by specific histone variants and histone modifications [1–3]. For instance, histone H3 has many characterized sites for covalent modification; in particular, lysine 4 on its N-terminal tail (H3K4) can be mono-, di-, or trimethylated, which generally correlate with enhancers, active genes, and promoters, respectively[4–7]. Indeed, H3K4 methylation is required for the cellular memory of active gene state [8] and are mediated by a conserved family of histone methylases named SET1, Trithorax, and MLL in yeast, flies, and mammals, respectively [4]. Enhancer elements and promoters are dispersed throughout the genome, and yet histone methyltransferases (such as MLL and DOT family proteins [9–12]) and histone demethylases (LSD1, JARID1A, and UTX [13]) are able to localize to these specific regions and in a cell-type specific manner, affect their enzymatic function. Thus, the ubiquitous yet specific nature of these interactions creates an important biological paradox: how do these complexes know which histones to modify and which ones to leave alone?
Characterization of the chromatin landscape revealed that much of the genome is pervasively transcribed [14–16]. Initial efforts to explore the functional consequences of this transcription have revealed long noncoding RNAs (lncRNAs, defined as >200 nts in length) as mainly repressive players in gene regulation. Examples such as XIST, HOTAIR, and lincRNA-p21 are among the most well studied lncRNAs and have been shown to be involved with X-chromosome inactivation, breast cancer metastasis, and p53-depended gene repression, respectively [17–21]. These functions occur through interaction with chromatin complexes such Polycomb Repressive Complex 2 (PRC2) in the case of XIST and HOTAIR. These observations suggest that RNA can provide a gene-specific targeting mechanism to non-specific enzymatic activity, but until recently the ability for RNA to coordinate activation of gene expression has not been well explored. A notable exception is the roX RNAs in Drosophila, which mark the male X chromosome together with the Male Specific Lethal (MSL) complex to enhance transcriptional by two fold [22].
In this review, we focus on the identification and characterization of novel ncRNAs and how they act to affect active chromatin and gene expression. First, we discuss new methods that have uncovered novel ncRNAs “markers” of active gene states and regulatory elements. Second, we highlight recent mechanistic studies that elucidate the connection between ncRNAs and H3K4 methylation. Collectively, these studies suggest a general role in gene regulation where ncRNAs can mark and often modulate the active chromatin state in both positive and negative manners.
RNA Marks Enhancer Elements
Discovering novel ncRNAs is critical to expanding the catalog of known transcripts. The advent of GRO-Seq has recently enabled the systematic identification of nascent RNA transcripts across the genome [23]. Initial application of this method revealed many short RNA transcripts (less than 250nts) around gene promoters in both sense and antisense orientations, termed divergent transcripts, and characterized genome-wide polymerase pausing just downstream of the transcription start site [23]. More recently, GRO-Seq was used to characterize androgen-responsive transcription in prostate cancer cells. Interestingly enhancer-templated RNAs (eRNAs) were responsive to androgen treatment [24]. Active enhancer elements are marked with H3K4me1, histone H3 lysine 27 acetylation (H3K27ac) [25,26], depleted of H3K27me3, and have characteristic binding of p300 (histone acetyltransferase [27]) and Med12 (subunit of the mediator complex [28]). Analysis of chromatin modifications and transcription factor binding has recently allowed for identification of novel enhancer elements. Intriguingly, based on extensive correlation of active enhancers with local eRNAs versus histone modifications and regulatory protein (p300 or Med12) binding, several groups observed that a defining characteristic for functionally active enhancers is the production of divergent eRNAs [24,25,29]. The marking of enhancer elements by eRNAs as well as H3K4me1 hints at a scenario where eRNAs could interact with the chromatin to establish or maintain H3K4me1 or where the H3K4me1 promotes eRNA transcription. Looping of enhancer and promoter regions together offers a potential mechanism for eRNAs to affect coding-gene expression (Figure 1A). Further work will be needed to assign a specific function to eRNAs and define what role they play in establishing the chromatin signature at enhancers and promoters.
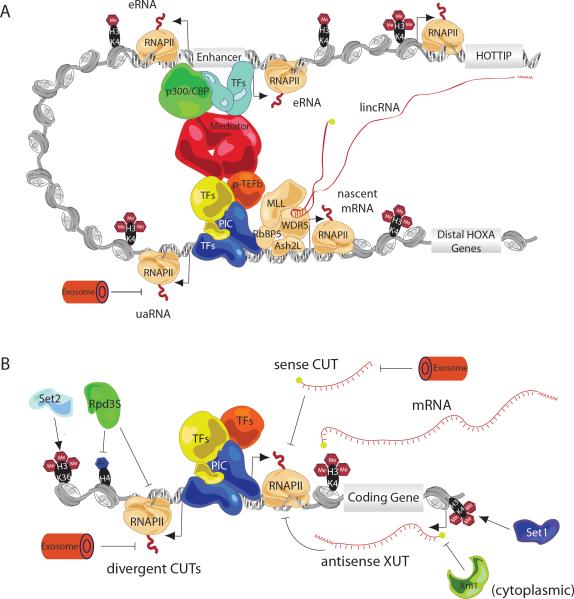
Figure 1. Schematic Diagram of Mammalian and Yeast lncRNA-chromatin Interactions.
A. Organization of a prototypical mammalian protein coding gene promoter, enhancer and lncRNA locus. The promoter of a protein-coding gene (e.g. distal HOXA genes) generates short sense and uaRNAs (degraded by the exosome) from divergently oriented sense and antisense RNAPII complexes. An enhancer element that regulates activity of the protein-coding gene forms a chromosomal loop to the promoter region through transcription factor and Mediator bridges. This enhancer generates eRNAs in a divergent manner marking it as a functional enhancer element. A lncRNA (HOTTIP) locus is brought in close proximity to protein-coding genes and the lncRNA transcript directly participates in targeting a chromatin modifying complex (MLL) to the promoter region to establish an active chromatin domain.
B. Organization of a prototypical yeast protein coding gene promoter and associated lncRNAs. The promoter of a protein-coding gene will produce sense and antisense (divergent) CUTs, which are substrates for the nuclear exosome. Sense CUTs are anti-correlated with mRNA levels. The upstream region of the promoter is often near the 3' end of another protein-coding gene and thus experiences Set2-mediated trimethylation of H3K36 and H4 deacetylaton by Rpd3S resulting in repression of the antisense CUT transcripts. Downstream of the promoter, antisense XUTs are activated by Set1 trimethylation of H3K4 and mediate mRNA repression. The XUT transcripts are degraded in the cytoplasm by the 5' to 3' exonuclease Xrn1.
LncRNAs at Cell Cycle Promoters
If enhancers can be transcribed, it may come as no surprise that promoters can also produce noncoding transcripts. A recent study systematically examined the transcriptional landscape of promoters encoding cell cycle genes over a large number of conditions, including phases of the cell cycle, oncogenic pathway activation, and stem cell differentiation [30]. This study revealed that cell cycle promoters, marked by domains of H3K4me3, indeed produce long noncoding transcripts (>200 nt) in a periodic fashion over the cell cycle, but in a manner that is out of phase with the expression of the neighboring coding genes. Thus, two parallel synchronous waves of gene expression—one coding and one noncoding—underlies human cell cycle progression. Some of these promoter lncRNAs are likely functional; one lncRNA, termed PANDA, is specifically induced by p53 during DNA damage and manages the balance between cell cycle arrest versus cell death. Interestingly, PANDA modulates cell death by binding to the transcription factor NF-YA and titrates it away from chromatin, indicating a trans rather than cis mode of action.
Divergent RNA at Promoters: Transcriptional start sites
Transcription around promoters has recently been reexamined studies in mammalian and yeast systems have described the process of divergent transcription where two distinct RNAPII complexes initiate in opposite directions to produce RNA transcripts [23,31–33]. Analysis of cryptic unstable transcripts (CUTs) and stable unannotated transcripts (SUTs) offered the first evidence of the widespread nature of divergent transcription in yeast. Divergent CUTs were found to be correlated with the expression of their sense protein-coding genes while sense CUTs were anti-correlated, suggesting that sense CUTs may interfere with transcription of the coding gene [32,33] (Figure 1B). A new method termed nascent transcript sequencing (NET-Seq), based on deep sequencing of nascent RNA fragments bound to RNA polymerase II, has revealed divergent transcripts in more detail [34]. This analysis revealed that, for most promoters, the ratio of antisense to sense transcription was less than 0.25, suggesting that while promoters have the capacity to generate divergent transcripts, at least in yeast, there is a strong directional preference towards the sense orientation. This directional bias was shown to be regulated by Rpd3S, an H4 deacetylase complex, which has activity at the 3' end of coding genes [34]. Interestingly, many of the divergent RNA transcripts in yeast overlap with the 3' ends of coding genes, suggesting that given the compact nature of the yeast genome, Rpd3S has evolved to control divergent RNA transcription to allow proper transcription of protein-coding genes (Figure 1B).
While divergent transcripts may be at the mercy of sense RNAs in yeast, the mammalian situation is more complicated. Divergent transcription was initially described in mouse embryonic stem cells (mESCs) and human lung fibroblasts, revealing that most gene exhibited divergent transcripts and that they were highly correlated with protein-coding gene expression [23,31]. Examination of the chromatin marks at these promoters revealed unexpected characteristics: the active H3K4me3 mark was colocalized to both the sense and antisense RNAPII regions whereas H3K79me2, a mark for productive transcriptional elongation, was only present downstream in the sense direction, suggesting a mechanism of divergent initiation but unidirectional elongation [35]. Divergent pausing of the RNAPII complexes was recently shown to occur at promoters with divergent RNAs suggesting the antisense RNAPII complex forms with many of the same factors as the sense complex [36]. Further work exploring the activity of the positive-transcriptional elongation factor b (pTEF-b), which is a master regulator of RNAPII pause release and productive elongation, on the antisense RNAPII complex revealed that the upstream-antisense RNAs (uaRNAs) are unexpectedly sensitive to its activity [37]. These studies have established divergent transcription as a pervasive and well-conserved process that yields divergent transcripts as well as divergent histone methylation marks (Figure 1A). In yeast, divergent transcripts appear to be involved in a repressive pathway while in mammalian cells no functional characteristics have yet to be assigned to the RNAs. The fact that uaRNAs in mammalian cells are regulated by the canonical transcription machinery but their expression tightly controlled could hint at some functional consequence of their transcription, perhaps as genomic anchors for recruitment of transcription regulatory factors.
Interplay of ncRNAs and H3K4 methylation
Functional studies of ncRNAs in yeast have revealed genetic interactions between certain lncRNAs and the methylation status of H3K4. The PHO84 antisense transcript runs through the body and promoter of PHO84 gene, and can inhibit sense PHO84 transcription in a SET1-dependent manner in cis (the endogenous configuration) or in trans (when the antisense is transcribed from a plasmid) [38,39]. The Ty1 retrotransposon has an antisense CUT RNA named RTL whose expression is anti-correlated with the Ty1 transcript [40]. Interesting, silencing of the Ty1 transcript is accomplished in trans by the RTL RNA through the Set1 methyltransferase complex. More recent work has classified RTL RNA as a member of the Xrn1-sensitive unstable transcripts (XUTs) given their sensitivity to the cytoplasmic 5' to 3' exonuclease Xrn1 [41]. Many of these XUTs are antisense to coding genes are have properties similar to RTL. The widespread nature of these antisense XUTs in yeast and their connection to the Set1 complex suggest an important link between lncRNAs and H3K4 methylation in yeast, specifically that the antisense XUTs may be positively regulated by Set1 and in turn repress the sense transcript (Figure 1B). One aspect of the XUT mechanism that is not clear is how XUTs, which act at the chromatin level to affect gene expression, are signaled for exported from the nucleus for Xrn1 degradation. Focused studies exploring the specific mechanism for RNA-mediated gene control between cellular compartments will be of interest in the future.
While most of the mammalian lncRNAs characterized to date repress gene activity, recent studies revealed that perhaps many lncRNAs enhance gene expression. The GENCONE database identified numerous new lncRNAs, several of which resulted in examples of activating lncRNAs (ncRNA-a) [42]. Depletion of these lncRNAs resulted in decreased expression of nearby protein coding genes, and experiments fusing cDNAs encoding ncRNAs to reporter genes suggested several ncRNAs activate gene expression in cis in an enhancer-like manner, although the mechanism remains unclear. Separately, studies of a novel lncRNA termed HOTTIP has provided a paradigm for how enhancer-like ncRNAs may act. HOTTIP is located on the very 5' end of the HOXA homeotic gene cluster [43]. In cells where 5' HOXA genes are active, the locus adopts a highly compact form, looping to bring HOTTIP and the nascent HOTTIP RNA into proximity to multiple 5' HOXA genes. In turn, HOTTIP RNA directly binds to WDR5, a subunit of the WDR5-Ash2L-RbBP5 (WAR complex) associated with all MLL H3K4 methylases, and recruits it to 5' HOXA to enforce H3K4 methylation and gene expression (Figure 1A). Ectopic expression and RNA tethering experiments showed that HOTTIP can only act in cis, and thus is likely strictly dependent on the endogenous chromosome looping to dictate its target genes. Thus, lncRNAs such as HOTTIP mediate information transfer of spatial information in chromosomal looping into biochemical information in histone modifications. The recent identification of another lincRNA that activates HoxA6 and HoxA7 in cis via direct interaction with MLL1 suggests a common theme [44].
Broad domains of active chromatin are hallmarks of developmental genes under epigenetic maintenance [45]. Once a lncRNA targets the MLL complex, the relatively non-sequence specific DNA binding activity of Ash2L, newly recognized through structural studies, spreads the complex across chromatin. At the HOXA locus, point mutations that abolish Ash2L DNA-binding activity in vitro allows the complex to target to the HOTTIP element, but fails to spread properly across HOXA [46]. These finding support the notion that lncRNAs have the capacity to enhance gene expression by specifically nucleating H3K4 methylase complex at specific genes, which then spread or stabilize via a separate DNA binding activity in a manner dependent on chromosomal looping. We await future work that delineates in detail the RNA-protein interaction required to specify these events.
Establishing a more complete catalog of lncRNA transcripts will speed the discovery of novel ncRNAs with functional roles (such as HOTTIP), and recently work in mouse and human cells has revealed large numbers of previously unannotated lncRNAs [47,48]. Extensive deep sequencing of transcriptomes from diverse human tissues annotated more than 8000 lncRNAs, and importantly revealed that on average, lncRNAs are more tissues specific than protein-coding genes, suggesting that lncRNAs may actually play a significant role in determining cellular fate [48]. Guttman and colleagues characterized mESC-specific lncRNAs and through functional studies showed many of these lncRNA are regulated by pluripotency factors (Oct4, Sox2, and Nanog) and almost 30% of the ESC-specific lncRNAs appear to interact with at least one repressive chromatin protein complex [49]. Towards addressing the potential functions of lncRNAs at chromatin in an unbiased manner, a new method termed Chromatin Isolation by RNA Purification (ChIRP) was developed [50]. Analogous to ChIP-seq, ChIRP-seq provides genome-wide identification of the specific sites of RNA occupancy on chromatin, and has been applied to the telomerase RNA TERC, roX2, and HOTAIR. These lncRNAs bound hundreds to thousands of sites throughout the genome, had focal foot prints (similar to transcription factors) and sequence specificity in their localization. Together, shRNA screens and ChIRP should enable high throughput functional studies of the lncRNA landscape.
Efforts to understand the interplay between ncRNAs and chromatin have established that both the RNA and chromatin can affect one another. While features of the active chromatin state have been well studied, the interactions between RNA and active are only beginning to be explored. Given the pervasive nature of transcription and the near ubiquitous nature of histone methylation, future work is likely to uncover mechanisms that employ both ncRNAs and the state of chromatin methylation to regulate gene expression.
Table 1.
Selected characteristics of lncRNAs in Mammals and Yeast
| Capped? | Spliced? | Average Length | Poly-A Tail? | Degradation Pathway | Function | |
|---|---|---|---|---|---|---|
| Mammalian lncRNAs | ||||||
| lincRNA | Yes [51]* | Yes [48] | 1000nts [48] | Yes [48] | lincRNA-specific pathway unknown | Act as Signals, Decoys, Guides and Scaffolds to the chromatin [17] |
| eRNA [29] | Unknown | No | <2000nts | No | eRNA-specific pathway unknown | Unknown, however their transcription is highly correlated with active enhancers [24] |
| uaRNA [37] | Yes | No | 50–1000nts | No | Exosome, a 3' → 5' exonuclease | Unknown, however they are likely the precursors to TSSa-RNAs and may act as chromatin tethers |
| Yeast lncRNAs | ||||||
| CUTs | Yes [52] | No | 100–3000nts (11) | Yes [52]** | Nrd1-exosome-TRAMP complex (nuclear) [53,54] | Unknown however sense and antisense (divergent) CUTs are negatively and positively, respectively, correlated with sense mRNA transcription [32,33] |
| XUTs [41] | Unknown*** | No | Not Calculated | Yes | Xrnl, a 5' → 3' exonuclease (cytoplasmic) | Antisense XUTs (the majority of the XUTs identified) act in opposition of the transcription of the sense gene |
LincRNA = Long intergenic noncoding RNA, eRNA = enhancer RNA, uaRNA = upstream-antisense RNA, CUT = cryptic unstable transcript, XUT = Xrn1-sensitive unstable transcript
Indirectly determined through identification of lincRNAs through CAGE analysis
Short poly-A tail added through the TRAMP complex as part of CUT degradation
RNA degradation by Xrn1 is only possible post cap removal, thus if they are initially capped they must undergo a decapping step prior to Xrn1-mediated degradation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000;34:77–137. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T. Translating the Histone Code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nature Structural & Molecular Biology. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 6.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 8.Muramoto T, MUller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 Is Required for Inheritance of Active Transcriptional States. Current Biology. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Sims RJ, III, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends in Genetics. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nature Structural & Molecular Biology. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 11.Min J, Feng Q, Li Z, Zhang Y, Xu R-M. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 12.Park G, Gong Z, Chen J, Kim J-E. Characterization of the DOT1L Network: Implications of Diverse Roles for DOT1L. Protein J. 2010;29:213–223. doi: 10.1007/s10930-010-9242-8. [DOI] [PubMed] [Google Scholar]
- 13.Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 15.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 16.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Wang KC, Chang HY. Molecular Mechanisms of Long Noncoding RNAs. Molecular Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JT. The X as model for RNA's niche in epigenomic regulation. Cold Spring Harb Perspect Biol. 2010;2:a003749. doi: 10.1101/cshperspect.a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sass GL, Pannuti A, Lucchesi JC. Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc Natl Acad Sci USA. 2003;100:8287–8291. doi: 10.1073/pnas.1332749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011 doi: 10.1038/nature10006. doi:10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study confirms the observation that enhancer RNAs (eRNAs, transcribed from enhancer elements) are widely transcribed and provided further evidence that the production of eRNAs marks fully functional enhancers.
- 25.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogryzko V, Schiltz R, Russanova V, Howard B. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996 doi: 10.1016/s0092-8674(00)82001-2. no volume. [DOI] [PubMed] [Google Scholar]
- 28.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011 doi: 10.1038/ng.848. doi:10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 33.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper describes a new method, NET-Seq, which can be used to profile nascent RNAs from RNA polymerase II. Here, NET-Seq is initially used in yeast to characterize divergent transcription and reveal other principal characteristics of RNA polymerase II transcription.
- 35.Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 36.Rahl PB, Lin CY, Seila AC, Flynn RA, Mccuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci USA. 2011;108:10460–10465. doi: 10.1073/pnas.1106630108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes & Development. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes & Development. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011 doi: 10.1038/nature10118. doi:10.1038/nature10118. [DOI] [PubMed] [Google Scholar]; ** This study expands on previous work to characterizing antisense transcripts in yeast. Here, the authors detail the degradation pathway for a novel class of antisense RNAs, XUTs, in yeast. Many XUTs are shown to anticorrelate with their cognate sense transcript.
- 42.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper identifies a novel lincRNA, HOTTIP, that acts to coordinate positional identity through direct interaction with MLL, a histone methyltransferase, by activating distal HOXA genes.
- 44.Bertani S, Sauer S, Bolotin E, Sauer F. The Noncoding RNA Mistral Activates Hoxa6 and Hoxa7 Expression and Stem Cell Differentiation by Recruiting MLL1 to Chromatin. Molecular Cell. 2011;43:1040–1046. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Aiden AP, Rivera MN, Rheinbay E, Ku M, Coffman EJ, Truong TT, Vargas SO, Lander ES, Haber DA, Bernstein BE. Wilms Tumor Chromatin Profiles Highlight Stem Cell Properties and a Renal Developmental Network. Stem Cell. 2010;6:591–602. doi: 10.1016/j.stem.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Wan B, Wang KC, Cao F, Yang Y, Protacio A, Dou Y, Chang HY, Lei M. Crystal structure of the N-terminal region of human Ash2L shows a winged-helix motif involved in DNA binding. EMBO Rep. 2011;12:797–803. doi: 10.1038/embor.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & Development. 2011 doi: 10.1101/gad.17446611. doi:10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper catalogs over 8000 lincRNAs from human cell lines and tissues, which allows for future functional studies of the novel lincRNAs. Analysis in the paper revealed an unexpectedly high tissue-specificity for lincRNAs suggesting a role for the lincRNAs in cell-type specificity.
- 49.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011 doi: 10.1038/nature10398. doi:10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This study defines mouse ES cell specific lincRNAs and explores their functional characteristics. Many of the identified lincRNAs are regulated by ES cell transcription factors and some of the lincRNAs interact with chromatin modifying proteins.
- 50.Chu C, Qu K, Zhong F, Artandi SE, Chang HY. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Molecular Cell. 2011 doi: 10.1016/j.molcel.2011.08.027. doi:10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study develops a method for genome-wide identification of RNA-chromatin interaction sites and reveals that long noncoding RNAs have qualities more akin to transcription factors than histone modification marks.
- 51.The FANTOM Consortium The Transcriptional Landscape of the Mammalian Genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 52.Wyers F, Rougemaille M, Badis G, Rousselle J-C, Dufour M-E, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B. Cryptic Pol II Transcripts Are Degraded by a Nuclear Quality Control Pathway Involving a New Poly(A) Polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 53.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription Termination and Nuclear Degradation of Cryptic Unstable Transcripts: A Role for the Nrd1-Nab3 Pathway in Genome Surveillance. Molecular Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 54.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of Cryptic Unstable Transcripts Is Directed by Yeast RNA-Binding Proteins Nrd1 and Nab3. Molecular Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]