Abstract
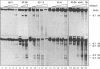
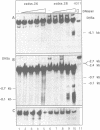
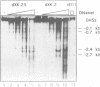
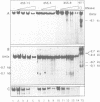
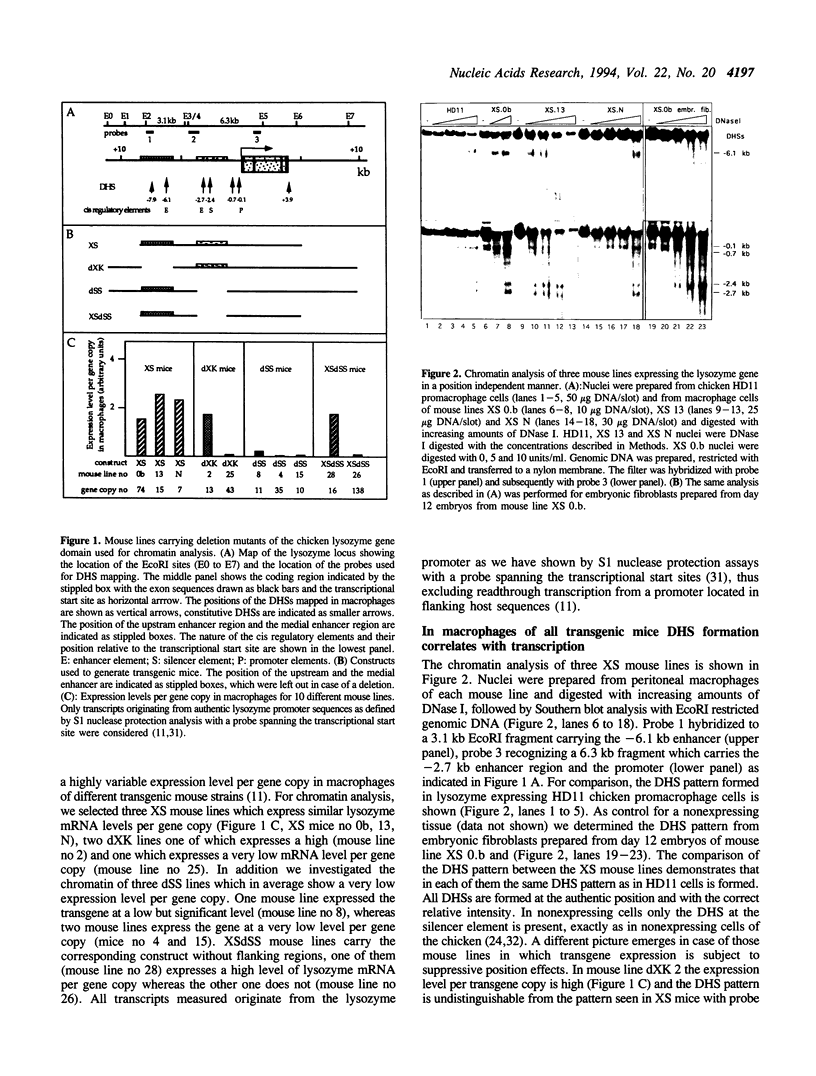
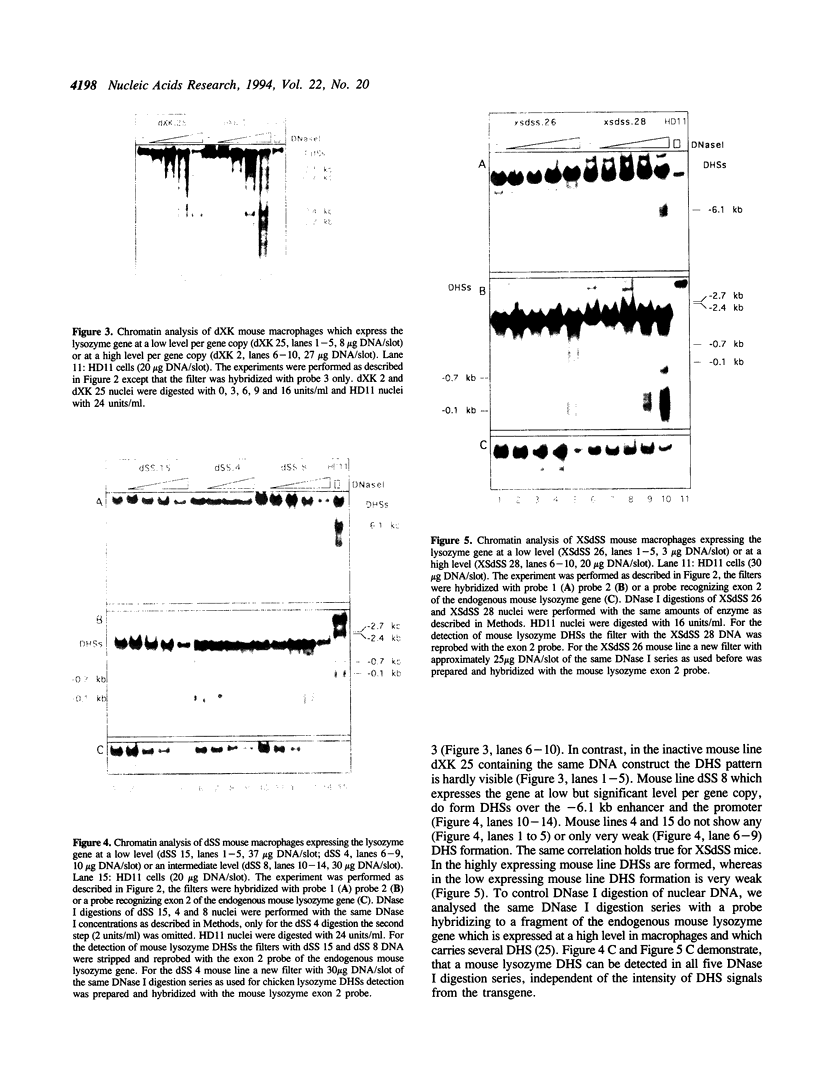
The complete chicken lysozyme gene locus is expressed copy number dependently and at a high level in macrophages of transgenic mice. Gene expression independent of genomic position can only be achieved by the concerted action of all cis regulatory elements located on the lysozyme gene domain. Position independency of expression is lost if one essential cis regulatory region is deleted. Here we compared the DNase I hypersensitive site (DHS) pattern formed on the chromatin of position independently and position dependently expressed transgenes in order to assess the influence of deletions within the gene domain on active chromatin formation. We demonstrate, that in position independently expressed transgene all DHSs are formed with the authentic relative frequency on all genes. This is not the case for position dependently expressed transgenes. Our results show that the formation of a DHS during cellular differentiation does not occur autonomously. In case essential regulatory elements of the chicken lysozyme gene domain are lacking, the efficiency of DHS formation on remaining cis regulatory elements during myeloid differentiation is reduced and influenced by the chromosomal position. Hence, no individual regulatory element on the lysozyme domain is capable of organizing the chromatin structure of the whole locus in a dominant fashion.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allshire R. C., Javerzat J. P., Redhead N. J., Cranston G. Position effect variegation at fission yeast centromeres. Cell. 1994 Jan 14;76(1):157–169. doi: 10.1016/0092-8674(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Bonifer C., Vidal M., Grosveld F., Sippel A. E. Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 1990 Sep;9(9):2843–2848. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch F. X., Udvarhelyi N., Venter E., Herold-Mende C., Schuhmann A., Maier H., Weidauer H., Born A. I. Expression of the histone H3 gene in benign, semi-malignant and malignant lesions of the head and neck: a reliable proliferation marker. Eur J Cancer. 1993;29A(10):1454–1461. doi: 10.1016/0959-8049(93)90020-g. [DOI] [PubMed] [Google Scholar]
- Cross M., Mangelsdorf I., Wedel A., Renkawitz R. Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6232–6236. doi: 10.1073/pnas.85.17.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester W. C., Takegawa S., Papayannopoulou T., Stamatoyannopoulos G., Groudine M. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987 Dec 23;15(24):10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P., Pruzina S., Antoniou M., Grosveld F. Each hypersensitive site of the human beta-globin locus control region confers a different developmental pattern of expression on the globin genes. Genes Dev. 1993 Jan;7(1):106–113. doi: 10.1101/gad.7.1.106. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Igo-Kemenes T., Nowock J., Strech-Jurk U., Theisen M., Sippel A. E. Alternative sets of DNase I-hypersensitive sites characterize the various functional states of the chicken lysozyme gene. Nature. 1984 Sep 13;311(5982):163–165. doi: 10.1038/311163a0. [DOI] [PubMed] [Google Scholar]
- Fritton H. P., Sippel A. E., Igo-Kemenes T. Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res. 1983 Jun 11;11(11):3467–3485. doi: 10.1093/nar/11.11.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler A., Joyner A. L., Rossant J., Skarnes W. C. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989 Apr 28;244(4903):463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- Gottschling D. E., Aparicio O. M., Billington B. L., Zakian V. A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990 Nov 16;63(4):751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Greaves D. R., Wilson F. D., Lang G., Kioussis D. Human CD2 3'-flanking sequences confer high-level, T cell-specific, position-independent gene expression in transgenic mice. Cell. 1989 Mar 24;56(6):979–986. doi: 10.1016/0092-8674(89)90631-4. [DOI] [PubMed] [Google Scholar]
- Grez M., Land H., Giesecke K., Schütz G., Jung A., Sippel A. E. Multiple mRNAs are generated from the chicken lysozyme gene. Cell. 1981 Sep;25(3):743–752. doi: 10.1016/0092-8674(81)90182-3. [DOI] [PubMed] [Google Scholar]
- Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987 Dec 24;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988 Dec 23;55(6):1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Hauser H., Graf T., Beug H., Greiser-Wilke I., Lindenmaier W., Grez M., Land H., Giesecke K., Schütz G. Structure of the lysozyme gene and expression in the oviduct and macrophages. Haematol Blood Transfus. 1981;26:175–178. doi: 10.1007/978-3-642-67984-1_29. [DOI] [PubMed] [Google Scholar]
- Hebbes T. R., Clayton A. L., Thorne A. W., Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994 Apr 15;13(8):1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A., Berkenstam A., Strömstedt P. E., Gustafsson J. A., Sippel A. E. A progesterone responsive element maps to the far upstream steroid dependent DNase hypersensitive site of chicken lysozyme chromatin. EMBO J. 1988 Jul;7(7):2063–2073. doi: 10.1002/j.1460-2075.1988.tb03046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs D. R., Wood W. G., Jarman A. P., Sharpe J., Lida J., Pretorius I. M., Ayyub H. A major positive regulatory region located far upstream of the human alpha-globin gene locus. Genes Dev. 1990 Sep;4(9):1588–1601. doi: 10.1101/gad.4.9.1588. [DOI] [PubMed] [Google Scholar]
- Jantzen K., Fritton H. P., Igo-Kemenes T. The DNase I sensitive domain of the chicken lysozyme gene spans 24 kb. Nucleic Acids Res. 1986 Aug 11;14(15):6085–6099. doi: 10.1093/nar/14.15.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Forrester W. C., Qiu R. G., Grosschedl R. The immunoglobulin mu enhancer core establishes local factor access in nuclear chromatin independent of transcriptional stimulation. Genes Dev. 1993 Oct;7(10):2016–2032. doi: 10.1101/gad.7.10.2016. [DOI] [PubMed] [Google Scholar]
- Jung A., Sippel A. E., Grez M., Schütz G. Exons encode functional and structural units of chicken lysozyme. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5759–5763. doi: 10.1073/pnas.77.10.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. Y., Hayes J. J., Pruss D., Wolffe A. P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993 Jan 15;72(1):73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- Lowrey C. H., Bodine D. M., Nienhuis A. W. Mechanism of DNase I hypersensitive site formation within the human globin locus control region. Proc Natl Acad Sci U S A. 1992 Feb 1;89(3):1143–1147. doi: 10.1073/pnas.89.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möllers B., Klages S., Wedel A., Cross M., Spooncer E., Dexter T. M., Renkawitz R. The mouse M-lysozyme gene domain: identification of myeloid and differentiation specific DNasel hypersensitive sites and of a 3'-cis acting regulatory element. Nucleic Acids Res. 1992 Apr 25;20(8):1917–1924. doi: 10.1093/nar/20.8.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., Grussenmeyer T., Strech-Jurk U., Theisen M., Stief A., Sippel A. E. Macrophage differentiation at the genome level: studies of lysozyme gene activation. Bone Marrow Transplant. 1990 Jan;5 (Suppl 1):13–14. [PubMed] [Google Scholar]
- O'Kane C. J., Gehring W. J. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987 Dec;84(24):9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phi-Van L., von Kries J. P., Ostertag W., Strätling W. H. The chicken lysozyme 5' matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990 May;10(5):2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M., Lee E., Westphal H., Felsenfeld G. An enhancer/locus control region is not sufficient to open chromatin. Mol Cell Biol. 1993 Jul;13(7):3990–3998. doi: 10.1128/mcb.13.7.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M., Lee E., Westphal H., Felsenfeld G. Site-independent expression of the chicken beta A-globin gene in transgenic mice. Nature. 1990 Dec 20;348(6303):749–752. doi: 10.1038/348749a0. [DOI] [PubMed] [Google Scholar]
- Renauld H., Aparicio O. M., Zierath P. D., Billington B. L., Chhablani S. K., Gottschling D. E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993 Jul;7(7A):1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Shaffer C. D., Wallrath L. L., Elgin S. C. Regulating genes by packaging domains: bits of heterochromatin in euchromatin? Trends Genet. 1993 Feb;9(2):35–37. doi: 10.1016/0168-9525(93)90171-D. [DOI] [PubMed] [Google Scholar]
- Stief A., Winter D. M., Strätling W. H., Sippel A. E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989 Sep 28;341(6240):343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- Taylor I. C., Workman J. L., Schuetz T. J., Kingston R. E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991 Jul;5(7):1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- Theisen M., Stief A., Sippel A. E. The lysozyme enhancer: cell-specific activation of the chicken lysozyme gene by a far-upstream DNA element. EMBO J. 1986 Apr;5(4):719–724. doi: 10.1002/j.1460-2075.1986.tb04273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C., Bellen H. J., Gehring W. J. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- al-Shawi R., Kinnaird J., Burke J., Bishop J. O. Expression of a foreign gene in a line of transgenic mice is modulated by a chromosomal position effect. Mol Cell Biol. 1990 Mar;10(3):1192–1198. doi: 10.1128/mcb.10.3.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]