Abstract
Centromeres are epigenetically defined chromatin domains marked by the presence of the histone H3 variant CENP-A. Here we review recent structural and biochemical work on CENP-A, and advances in understanding the mechanisms that propagate and read centromeric chromatin domains.
In the majority of eukaryotes, centromeres are not defined by a specific DNA sequence but instead by the presence of the centromeric histone CENP-A/CenH3 (CENP-A in humans, Cse4 in budding yeast, CID in Drosophila, HCP-3/CeCENP-A in C. elegans; referred to generally as CENP-A hereafter except where the specific species identity is important). CENP-A chromatin functions as both part of an epigenetic mark for centromere identity and a structural foundation for assembly of the kinetochore, the multi-protein machine that forms the primary attachment site on chromosomes for spindle microtubules during cell division.
The lack of a specific conserved centromeric DNA sequence had been long-suggested by the diversity of DNA sequences found at centromeres throughout evolution [1], as well as by the presence of monocentric and holocentric chromosome architecture in extant eukaryotes. The latter, documented to date in nematodes and several lower plant as well as insect species, is characterized by the presence of centromere activity along the length of the chromosome during mitosis [2]. The strongest functional evidence for sequence independence of centromere activity has been derived from studies of neocentromeres, which form rarely at non-centromeric sites on chromosomal loci that bear no sequence identity to the endogenous centromeric DNA in the same species [3]. A well-studied exception to the sequence-independence of centromeres is the budding yeast S. cerevisiae, where a specific 125 bp DNA sequence is sufficient for centromere activity—however, this centromere also assembles a single CENP-A nucleosome that is well-positioned on the CEN DNA sequence [4]. Interestingly, in the sleeping sickness pathogen Trypanosoma brucei and during meiosis in C. elegans, chromosome segregation may occur without CENP-A [5,6] but the mechanisms directing kinetochore assembly in these cases are not known.
Here, we will focus on the predominant CENP-A dependent centromere definition mechanism, reviewing recent work that has advanced our understanding of the structure and assembly of this specialized chromatin domain. In particular, we review recent structural efforts that have provided an atomic view of CENP-A-containing nucleosomes and pre-nucleosomal complexes, and biochemical/cell biological studies that have revealed how this specialized chromatin builds a kinetochore.
(Resolving) The Debate on DNA-Bound Structures Built from CENP-A
While the importance of chromatin containing CENP-A at centromeres is widely accepted, the same is not true for the architecture of the DNA-CENP-A assembly that is functionally relevant at centromeres. The initial discovery that CENP-A is a histone H3 variant and the mapping of its centromeric identity to its histone fold [7,8], led to the parsimonious hypothesis that CENP-A forms a structure similar to H3-containing nucleosomes and that differences in the CENP-A histone-fold underlie recognition by specialized trans-acting machinery. Initial in vitro reconstitution experiments in the presence of a DNA substrate were consistent with this view [9]. In subsequent years, reconstitution efforts in multiple species have consistently revealed that CENP-A forms robust octameric nucleosomes in vitro [10]. A recent experiment employing mixtures of histone H3 and the budding yeast CENP-A (Cse4) in the same assembly reactions showed that assembly of homotypic octamers containing only CENP-A or H3 is strongly favored [11] this result is consistent with relative paucity of H3 in CENP-A chromatin immunoisolations following extended micrococcal nuclease digestion [12]. The major (and consistent) difference observed between H3 nucleosomes and CENP-A nucleosomes in vitro, starting from the first study of CENP-A nucleosomes, has been the extent of DNA protection. CENP-A nucleosomes protect less DNA than H3 nucleosomes (~120 bp vs 145 bp) in nuclease digestion experiments [9].
The view that CENP-A forms octameric nucleosomes in vitro has been challenged in recent years by two alternative proposals: i) CENP-A assembles into half-nucleosomes (hemisomes) with right-handed DNA winding topology [13]; this proposal was based on reconstitutions with Drosophila CENP-A (CID), atomic force microscopy of Drosophila interphase CENP-A chromatin [14] and topological analysis of minichromosomes isolated from yeast; ii) Budding yeast CENP-A forms a distinct non-nucleosomal structure that lacks histones H2A and H2B; this proposal was based on formation of a hexameric complex of CENP-A/H4 and the CENP-A binding protein Scm3 in high salt, and the inability to detect H2A/H2B using chromatin immunoprecipitation in vivo at budding yeast centromeres [15]; for an in depth discussion of alternative proposals, see [16]. Subsequent in vitro efforts from multiple groups have argued against these views and provided additional evidence for an octameric CENP-A nucleosome with left-handed DNA winding topology regardless of whether the assembly is performed using dialysis from high salt or using a chaperone at physiological salt [11,17–22]. Thus, at least in vitro, it appears that CENP-A can form octameric nucleosomes closely resembling H3 nucleosomes. Whether an octameric nucleosome is assembled on budding yeast centromeric DNA in vitro is less clear; the highly AT-rich CEN DNA sequence disfavors octameric nucleosome assembly [22,23], but Cse4 is nonetheless able to form robust octamers on other DNA sequences [22]. A definitive answer in this system will likely require better understanding of the architectural contributions of other budding yeast-specific CEN DNA-binding proteins such as the CBF3 complex [24].
Atomic Structures of the CENP-A Nucleosome and CENP-A:H4 Pre-Nucleosomal Complex
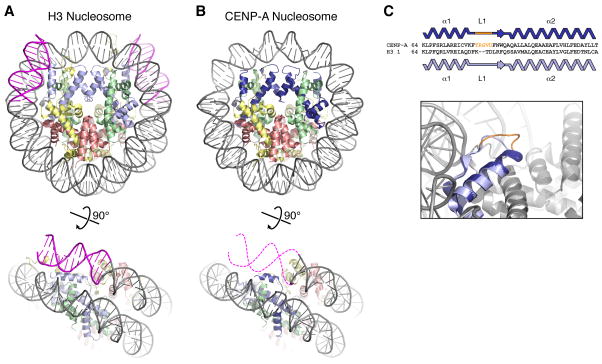
In the past two years, structural studies have shed much-needed light on CENP-A and CENP-A-DNA complexes, with the most significant advance being the crystal structure of a full human CENP-A nucleosome [20]. Comparison of CENP-A and H3 nucleosome structures that were determined with the same DNA sequence reveals almost perfect overlap, with a root mean square deviation of 0.6 Å comparing 663 of 706 carbon-α atoms in the octamer (Figure 1A, B). The primary difference between the two structures is that while the entire 147 bp of DNA is well-ordered in H3 nucleosome, only 121 bp of the same DNA fragment are ordered in the CENP-A nucleosome, with 13 bp on either end disordered (Figure 1B). This finding is consistent with biochemical experiments and may explain early observations that centromeric chromatin exhibits unusual nuclease sensitivity [25]. Another structural difference was that the Loop 1 region of CENP-A adopts a more extended and flexible conformation than in the canonical H3 nucleosome (Figure 1C) [20,21]. Overall, the CENP-A nucleosome structure has revealed remarkable similarity to H3 nucleosomes, with the major structural difference being less strong binding of DNA at the entry and exit sites.
Figure 1.
Canonical and centromeric nucleosomes. (A) Canonical H3-containing nucleosome (PBD ID 1KX5) [74], with histone H3 light blue, H4 green, H2A yellow, H2B red, and DNA gray (13 bp on each end colored magenta). (B) CENP-A-containing nucleosome (PDB ID 3AN2) [20], with CENP-A colored dark blue. Bottom: view of one DNA end, with the 13 bp of disordered DNA indicated by dashed magenta lines (figure adapted from [20]). (C) Loop 1 differences in CENP-A and H3 (adapted from [20]). Top: Sequence alignment showing the two-residue insertion in CENP-A loop 1. Bottom: closeup view of the loop 1 region, with CENP-A from [20] in dark blue and H3 from [74] in light blue. Shown in orange are CENP-A residues 79–83, which are ordered in the CENP-A monomer shown (potentially due to crystal packing interactions) and disordered in the second CENP-A monomer. An earlier CENP-A2:H42 tetramer structure also showed a distinct conformation of CENP-A loop 1 with high crystallographic B-factors indicating flexibility [21]. While mutation or truncation of loop 1 mildly affects centromere targeting of CENP-A in vivo [20], the mechanism of this defect remains unknown.
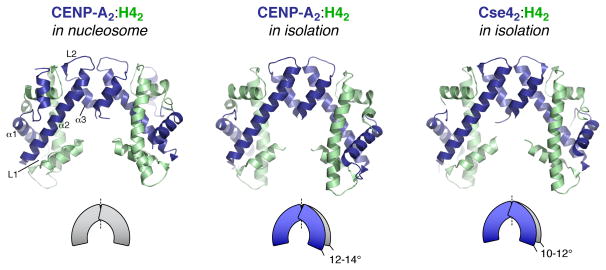
The nucleosome structure of CENP-A was preceded by structures of the human and budding yeast (Kluyveromyces lactis) CENP-A:H4 tetramers without bound DNA [21,26]. Both structures showed significant compaction (by 10–14°) of the tetramer relative to H32:H42 in the canonical H3 nucleosome structure (Figure 2; an atomic structure of the isolated H3:H4 tetramer has not been determined). This compaction was proposed to be a distinguishing property of CENP-A [21]. However, within the structure of the full centromeric nucleosome no difference is observed from H3, suggesting that the observed compaction may be due to the lack of bound DNA and/or histones H2A/H2B. The human CENP-A:H4 tetramer structure revealed an extensive network of hydrophobic residues anchoring the ends of the CENP-A:H4 interaction more tightly than the H3:H4 interaction [21]. This “hydrophobic stitching” likely explains the lower exchange in hydrogen-deuterium exchange assays for CENP-A nucleosomes compared to H3 nucleosomes [10]. As the rigid region includes the CENP-A α2 helix, which comprises part of the centromere targeting domain, this stitching may confer physical properties that are important for CENP-A function at centromeres in vivo.
Figure 2.
CENP-A2/Cse42:H42 tetramer structures, in the context of the full nucleosome (left) (PDB ID 3AN2) [20], and in isolation (middle: human CENP-A2:H42 (PDB ID 3NQJ) [21], right: budding yeast (K. lactis) Cse42:H42 (PDB ID 2YFW) [26]), colored as in Figure 1. Secondary structure elements of one CENP-A monomer are labeled (left panel). Both tetramer structures determined without bound H2A:H2B and DNA show compaction of the tetramer by 10–14°, relative to the conformation of both the H32:H42 and CENP-A2:H42 tetramers in the context of the full nucleosome (illustration adapted from [21]).
Does the similarity of H3 and CENP-A nucleosome structures close the door on native CENP-A chromatin structures in cells adopting alternative states? Mutations in the budding yeast CENP-A (Cse4) based on interfaces in the canonical H3 nucleosome structure and suppression of the lethality of an Scm3 deletion by overexpression of Cse4 argue for an octamer in vivo in this organism [19]; however, the absence of H2A/H2b at CEN DNA by chromatin immunoprecipitation remains unexplained. In Drosophila and human cells, atomic force microscopy of digested CENP-A chromatin from asynchronous nuclei has revealed the presence of particles precisely half the height of H3 nucleosomes that were isolated in the same manner, and a single CENP-A molecule was detected by immunoEM in these particles [14,27]. One possible explanation for structural variation is assembly transition states representing pre and/or post DNA replication. Alternatively, in vivo factors, such as the CENP-A associated proteins known as the CCAN [28], may stabilize an intermediate state that is not favored in vitro. Additional means to directly assess the structure of CENP-A-DNA assemblies in cells, including imaging methods that do not require isolation and nuclease digestion, will hopefully resolve this debate.
Structural Analysis of the CENP-A Chaperone Scm3/HJURP
The question of how CENP-A is recognized and specifically deposited at centromeres has focused on the histone fold domain since early work [7]. Analysis of chimeric proteins revealed that Loop 1 and the α2 helix of CENP-A, termed the CENP-A targeting domain (CATD), are responsible for targeting CENP-A to centromeres [10,29]. While the importance of the CATD in centromere definition by CENP-A is well established, whether the H3-CATD chimera is sufficient for centromere function is less clear. In human cells, the H3-CATD chimera complements RNAi-based partial depletions of CENP-A [30], but genetic studies in yeast and plants suggest that additional parts of the CENP-A molecule are essential [30–32]. In particular, essential elements have been defined in the tail and the extreme C-terminus of budding yeast CENP-A; in Arabidopsis, introduction of a tailswap mutant (CENP-A histone-fold domain with the H3 tail) into a CENP-A null mutant does not inhibit mitotic divisions but leads to meiotic defects and the formation of haploid plants[32,33].
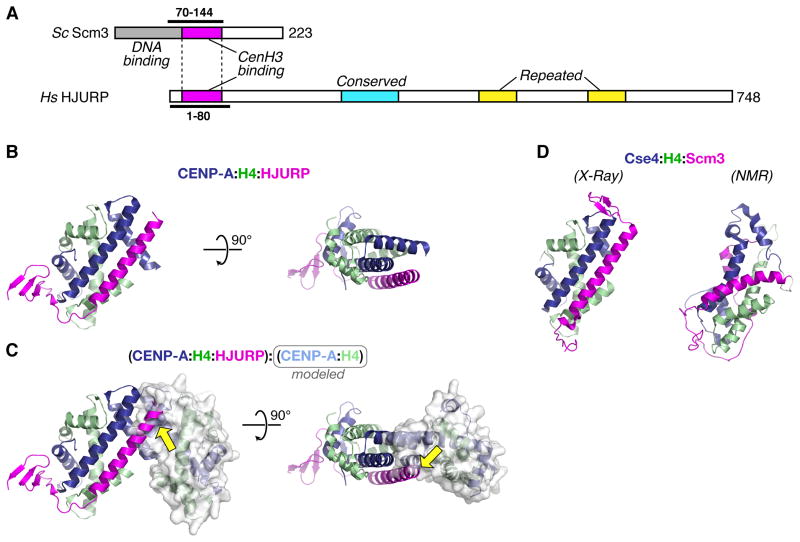
Understanding how the CATD is critical for centromeric targeting has been greatly advanced by the discovery of a conserved CENP-A specific chaperone family (HJURP in human cells and Scm3 in budding yeast) recognizing this region. HJURP/Scm3 chaperones are not ubiquitous – both Drosophila and C. elegans appear to lack them [34]. HJURP and Scm3 share a short region of homology that is sufficient for CENP-A recognition (Figure 3A) [15,17,35]. Three recent studies have now revealed the structural basis for CATD recognition by these chaperones: an x-ray crystal structure of HJURP bound to CENP-A:H4 (Figure 3B) [36], and both NMR and x-ray structures of budding yeast (NMR: S. cerevisiae, x-ray: K. lactis) Scm3 bound to Cse4:H4 (Figure 3C)[26,37]. The two x-ray structures, despite the extreme sequence divergence of Scm3 and HJURP, look similar (0.6 Å root mean square deviation between 148 common α-carbon atoms in the three chains) while the NMR structure is significantly different (Figure 3B, C)—the reasons for this are unclear but may reflect the different protein regions used in the NMR study or the fact that the three proteins were fused into a single chain (see [26,37,38] for debate on this issue). Here, for brevity, we will restrict our interpretations to the two structures that agree.
Figure 3.
Structural analysis of the CENP-A chaperone HJURP/Scm3. (A) Diagram of S. cerevisiae Scm3 and H. sapiens HJURP. Conserved domains identified in [34] are shown: CENP-A/Cse4 binding region (Scm3 residues 90–142, HJURP residues 16–68) magenta, HJURP conserved domain (228–304) cyan, HJURP repeated regions (411–462 and 556–608) yellow. The DNA-binding region of Scm3 (1–113) identified by Xiao et al [23] is shown in gray. Constructs used for x-ray crystallography are indicated by thick lines. (B) Side and top views of the CENP-A:H4:HJURP trimer structure (PDB ID 3R45) [36], with HJURP shown in magenta. (C) The CENP-A:H4:HJURP trimer aligned with a second CENP-A:H4 dimer (molecular surface displayed), showing how HJURP partially occludes the CENP-A:CENP-A interface in the tetramer (yellow arrows). (D) Two structures of the budding-yeast Cse4:H4:Scm3 trimer. Left: structure obtained by X-ray crystallography of the proteins from K. lactis (PDB ID 2YFV) [26]. Right: structure obtained by NMR of the S. cerevisiae proteins (PDB ID 2L5A) [37]. All structures are oriented the same relative to the left-hand copy of CENP-A/Cse4 in Figure 2.
The most striking feature revealed by the x-ray structures is that the region of homology between the two histone chaperones forms a long α-helix that binds antiparallel to the CENP-A/Cse4 α2 helix (Figure 3B, C)—this configuration of the complex is incompatible with both CENP-A:CENP-A tetramerization and with DNA binding [26,36]. Thus, both structures are of a heterotrimer containing one copy each of HJURP/Scm3, CENP-A/Cse4, and H4. The formation of a heterotrimer was confirmed by analytical ultracentrifugation using full-length proteins from both K. lactis and S. cerevisiae [26]. Scm3 is not observed to associate with the Cse4 loop 1 region, and while HJURP associates with this region of CENP-A, it does not contact residues that would provide significant specificity over H3 [36]. Thus, the critical recognition of CENP-A/Cse4 occurs in the α2 helix. Both structures show specific interactions between chaperone residues and CENP-A/Cse4-specific residues in this helix; mutating these residues disrupts in vitro binding [26,36].
Overall, the structures of Scm3/HJURP bound to CENP-A:H4 together with biochemical analysis have revealed a trimeric complex with the key recognition occurring via the α2 helix of the CATD. The CATD is also the recognition site for the E3 ubiquitin ligase Psh1 that functions to reduce CENP-A misincorporation at non-centromeric sites in budding yeast [39,40]. While there is no obvious sequence homology between Psh1 and Scm3, structural analysis of Psh1 may help reveal whether the recognition properties observed with Scm3 are also utilized in other contexts to recognize CENP-A.
Recent biochemical studies have shown that HJURP/Scm3 exhibits chaperone activity, promoting the formation of CENP-A:H4 nucleosomes in vitro [17,18,35]. Thus, an Scm3 molecule likely delivers a CENP-A:H4 dimer to the DNA for assembly. This result raises the question whether the CENP-A2:H42 tetramer purified following expression in bacteria is functionally relevant during assembly; the fact that the H3:H4 chaperone Asf1 also binds a dimer (albeit at different elements than α2; [41]), suggests that chaperone-bound physiologically relevant cargo during both CENP-A and H3 nucleosome assembly is a dimer. Once the CENP-A:H4 dimers are recruited to centromeric regions, they likely assemble first into CENP-A2:H42 tetramers bound to DNA. In the process of tetramerization and DNA binding, HJURP/Scm3 is ejected from the complex [22,23]. A higher-molecular weight complex of S. cerevisiae Scm3:Cse4:H4, potentially corresponding to a 2:2:2 heterohexamer, has been observed in vitro at high salt (2.0M NaCl) but not at lower salt (0.5M NaCl) [15,37]. This finding suggests the possibility of a transient hexameric complex existing as an assembly intermediate at centromeres.
Steps in Propagation of CENP-A Domains in Vivo
Centromeric CENP-A is proposed to be partitioned to sister chromatids during DNA replication and replenished later in the cell cycle. Intervening “gaps” between CENP-A nucleosomes following replication are likely filled by nucleosomes containing histone H3.1 and H3.3, a variant normally associated with non-replicative chromatin assembly. New CENP-A loading occurs soon after anaphase in mammalian cells and Drosophila embryos ([42,43]; see [44] for evidence that loading occurs in metaphase in cultured Drosophila cells); in plants, loading occurs in G2, whereas in fission yeast it occurs during both S phase and G2 [45,46]. A reduction in H3.3 levels at centromeres in G1 of human cells suggests that H3.3 acts as a “placeholder” and is replaced by CENP-A nucleosomes during replenishment [47].
The loading of new CENP-A nucleosomes must require CENP-A:H4-HJURP complexes to target specifically to centromeres through protein-protein/DNA interactions. In the case of budding yeast, Scm3 is recognized by the CBF3 complex, which specifically binds to CEN DNA [48]. As CBF3 is not conserved outside of budding yeasts, targeting of CENP-A:H4-HJURP likely occurs by an interaction with other proteins that recognize existing centromeric chromatin domains. The Mis18 complex (comprised of Mis18 and Knl2/MI8BP1 [49,50]) is the primary candidate for this function, but the mechanism by which this complex recognizes centromeric chromatin and recruits pre-nucleosomal CENP-A complexes is currently unclear (only a weak Scm3-Mis18 interaction has been observed in fission yeast [51]. However, consistent with this view, ectopically targeting HJURP to non-centromeric DNA promotes de novo CENP-A incorporation and kinetochore assembly without a requirement for Mis18/Knl2 [18]. Work in Xenopus egg extracts has shown that the centromere protein CENP-C, which is closely associated with CENP-A nucleosomes, can bind to and recruit Knl2 (and thus Mis18 [52]); however, this does not appear to be the case in C. elegans embryos [49]. Knl2 itself contains a conserved Myb-type DNA binding domain, raising the possibility that its ability to directly bind DNA plays a critical role [49]. Interestingly, the Mis18 and Knl2 family proteins are not always present together—Mis18 is not found in C. elegans, while Knl2 appears to be absent from fission yeast, and both proteins are absent in Drosophila, which instead relies on a protein named Cal1 for CENP-A assembly [53,54].
Analysis of Knl2 in human cells has revealed an additional maintenance step during centromere propagation, where the small GTPase Cdc42 helps ensure that newly incorporated CENP-A is retained at centromeres [55]. A recent study of H3 nucleosome assembly in vitro has identified a non-nucleosomal histone-DNA intermediate with the dimensions of a nucleosome that is converted by an ATPase chromatin motor into nucleosomes [56]—whether such an intermediate exists for CENP-A nucleosomes and whether specific mechanisms, such as the Cdc42 GTPase cycle, are involved in stabilizing such an intermediate are avenues for future investigation.
The rules by which CENP-A incorporation are guided to a specific chromosomal site are likely to involve both recognition of pre-existing CENP-A nucleosomes and other cues that reflect specific chromatin states [57]. In support of this, de novo CENP-A incorporation is observed at non-centromeric sites in Drosophila cells following ectopic targeting of the heterochromatin protein HP1 [58]. De novo CENP-A incorporation and neocentromerization is also rapidly observed on naked DNA injected into the germline of the holocentric nematode C. elegans [59]; however, this reaction appears to be independent of HP1 family proteins. In mammals, the alpha-satellite sequence binding CENP-B protein controls de novo CENP-A chromatin formation depending on the chromatin context [60]. Mechanistic insights into de novo CENP-A chromatin domain formation are likely to reveal principles that are utilized in the context of pre-existing CENP-A nucleosomes to propagate centromere identity [61,62] and enable artificial centromere engineering.
Reading CENP-A Chromatin to Build a Kinetochore
Two major advances in understanding how CENP-A chromatin templates assembly of the kinetochore have emerged from biochemical work in Xenopus egg extracts and through ectopic targeting of proteins to non-centromeric chromosomal loci in mammalian cells [63,64]. In vitro analysis had revealed that CENP-C, a widely conserved centromeric protein, specifically recognizes a short C-terminal tail sequence that is specific to CENP-A [65]. CENP-A nucleosome arrays built on DNA attached to beads recruit kinetochore proteins in Xenopus egg extracts. Remarkably, a chimeric H3 with the C-terminal CENP-C binding motif is sufficient for this recrutiment activity. This result points to a critical role for the C-terminal CENP-A tail in kinetochore assembly, via its recognition by CENP-C [63]. Surprisingly, this tail sequence is not conserved outside of vertebrates, although the central importance of CENP-C in kinetochore assembly is widely conserved. Understanding how CENP-C recognizes CENP-A nucleosomes outside of vertebrates will be important to address in future work.
Proteomic analysis of CENP-A nucleosomes identified an extensive set of proteins (collectively referred to as the CCAN) specifically associated with centromeres throughout the cell cycle [12,28,66]. Of these components, CENP-N specifically recognizes the CATD of CENP-A; however, the contribution of this recognition to kinetochore assembly is currently unclear [63,67]. The CENP-T/W complex has been proposed to bind H3 nucleosomes adjacent to CENP-A nucleosomes to direct kinetochore assembly [68] and CENP-T changes its conformation when kinetochores are attached and under tension [69]. Ectopic targeting experiments in human cells suggest that kinetochore assembly is directed by a cooperative mechanism, involving CENP-C recognition of the C-terminal tail of CENP-A, and CENP-T recognition of H3 nucleosomes adjacent to CENP-A [64]. Some species, such as D. melanogaster and C. elegans, appear to lack the CCAN and rely exclusively on CENP-C for kinetochore assembly [70–72]. How CENP-T/W recognize H3 nucleosomes in proximity to CENP-A nucleosomes but not elsewhere in the genome is an important question to address in future work.
In addition to the molecular mechanisms underlying recognition of CENP-A chromatin, the organization of this chromatin to form a surface for kinetochore assembly is critical for chromosome segregation. Superresolution imaging has suggested a complex layered organization of CENP-A and H3 nucleosomes, with binding of CENP-C forming a platform for kinetochore assembly [73].
Conclusion
Here, we have summarized recent advances in centromere biology. This is an exciting time when the convergence of structural/biochemical studies and in vivo work is providing a glimpse of how centromeric chromatin is defined, propagated and read to direct chromosome segregation. Many questions remain in each area and the future holds great promise for converting initial insights into deeper mechanistic understanding.
Acknowledgments
Work in the Desai lab is supported a grant from the NIH (GM074215). K.C. and A.D. receive salary support from the Ludwig Institute for Cancer Research. P.S.M. is the Canada Research Chair in Cell Division and Chromosomal Organization and supported by research grants from the CIHR (MOP-106548) and CCSRI (700824).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choo KH. Centromerization. Trends Cell Biol. 2000;10:182–188. doi: 10.1016/s0962-8924(00)01739-6. [DOI] [PubMed] [Google Scholar]
- 2.Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. doi: 10.1023/B:CHRO.0000036588.42225.2f. [DOI] [PubMed] [Google Scholar]
- 3.Voullaire LE, Slater HR, Petrovic V, Choo KH. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? Am J Hum Genet. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 4.Cole HA, Howard BH, Clark DJ. The centromeric nucleosome of budding yeast is perfectly positioned and covers the entire centromere. Proc Natl Acad Sci U S A. 2011;108:12687–12692. doi: 10.1073/pnas.1104978108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowell JE, Cross GA. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J Cell Sci. 2004;117:5937–5947. doi: 10.1242/jcs.01515. [DOI] [PubMed] [Google Scholar]
- 6.Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer DK, O’Day K, Wener MH, Andrews BS, Margolis RL. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol. 1987;104:805–815. doi: 10.1083/jcb.104.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci U S A. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black BE, Foltz DR, Chakravarthy S, Luger K, Woods VL, Jr, Cleveland DW. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. doi: 10.1038/nature02766. [DOI] [PubMed] [Google Scholar]
- 11.Kingston IJ, Yung JS, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2010;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- *13.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. This paper proposed an alternative right-handed topology for DNA bound to CENP-A nucleosomes based on in vitro analysis of Drosophila CENP-A (CID) and supercoiling of yeast minichromosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 16.Black BE, Cleveland DW. Epigenetic Centromere Propagation and the Nature of CENP-A Nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *17.Shuaib M, Ouararhni K, Dimitrov S, Hamiche A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc Natl Acad Sci U S A. 2010;107:1349–1354. doi: 10.1073/pnas.0913709107. This paper was the first to demonstrate that HJURP functions as a CENP-A chaperone. See also refs 18 & 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *18.Barnhart MC, Kuich PH, Stellfox ME, Ward JA, Bassett EA, Black BE, Foltz DR. HJURP is a CENP-A chromatin assembly factor sufficient to form a functional de novo kinetochore. J Cell Biol. 2011;194:229–243. doi: 10.1083/jcb.201012017. This paper demonstrates that HJURP targeting is sufficient to assemble CENP-A chromatin in vivo. Additionally, the authors show that CENP-A nucleosomes assembled in vitro with HJURP have similar properties as H3 nucleosomes. Finally, this paper shows that HJURP requires the Mis18-Knl2 complex for centromere localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *19.Camahort R, Shivaraju M, Mattingly M, Li B, Nakanishi S, Zhu D, Shilatifard A, Workman JL, Gerton JL. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. This paper tests whether budding yeast CENP-A (Cse4) forms octameric nucleosomes. In addition to in vitro biochemistry and in vivo mutagenesis of Cse4, the authors report that overexpression of Cse4 suppresses the lethality of an Scm3 deletion, thereby arguing against Scm3 being an integral component of a centromeric non-nucleosomal structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **20.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. This paper reports the structure of the full CENP-A nucleosome particle. The key finding, besides a shorter wrapped DNA length, is the striking similarity to canonical H3 nucleosomes. [DOI] [PubMed] [Google Scholar]
- **21.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. This paper reports a three-dimensional structure of the CENP-A:H4 tetramer, which closely resembles the H3:H4 tetramer in the nucleosome, with the exception of a slight compaction and extended hydrophobic interactions. See Ref 20 that suggests the compaction likely reflects absence of H2A/H2B and DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *22.Dechassa ML, Wyns K, Li M, Hall MA, Wang MD, Luger K. Structure and Scm3-mediated assembly of budding yeast centromeric nucleosomes. Nat Commun. 2011;2:313. doi: 10.1038/ncomms1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *23.Xiao H, Mizuguchi G, Wisniewski J, Huang Y, Wei D, Wu C. Nonhistone Scm3 binds to AT-rich DNA to organize atypical centromeric nucleosome of budding yeast. Mol Cell. 2011;43:369–380. doi: 10.1016/j.molcel.2011.07.009. These two papers and characterize the effects of the budding yeast Scm3 chaperone on Cse4 nucleosome assembly and DNA binding in vitro, and both show that the AT-rich CEN DNA sequence disfavors octameric nucleosome assembly. The two papers come to differing conclusions regarding the composition of the budding yeast centromeric nucleosome: Dechassa et al argue for an octameric nucleosome, while Xiao et al draw on chromatin immunoprecipitation data to argue for a nucleosome missing H2A/H2B, and potentially containing stably associated Scm3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho US, Corbett KD, Al-Bassam J, Bellizzi JJ, 3rd, De Wulf P, Espelin CW, Miranda JJ, Simons K, Wei RR, Sorger PK, et al. Molecular structures and interactions in the yeast kinetochore. Cold Spring Harb Symp Quant Biol. 2010;75:395–401. doi: 10.1101/sqb.2010.75.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- **26.Cho US, Harrison SC. Recognition of the centromere-specific histone Cse4 by the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:9367–9371. doi: 10.1073/pnas.1106389108. Three papers together (Refs 26, 36, and 37) illustrate the structural basis for how the HJURP/Scm3 chaperone specifically recognizes CENP-A/Cse4. The papers also illustrate that CENP-A recognition is conserved despite the sequence divergence between budding yeast Scm3 and metazoan HJURP. Note that the structure in Ref 37, which employs NMR, differs significantly from Ref 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *27.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci U S A. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. This paper employs atomic force microscopy to show that CENP-A chromatin isolated following nuclease digestion from human cultured cells has particles with half the height of particles present in H3 chromatin. Immuoelectron microscopy revealed that these particles contain a single CENP-A molecule. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 29.Vermaak D, Hayden HS, Henikoff S. Centromere targeting element within the histone fold domain of Cid. Mol Cell Biol. 2002;22:7553–7561. doi: 10.1128/MCB.22.21.7553-7561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *32.Ravi M, Kwong PN, Menorca RM, Valencia JT, Ramahi JS, Stewart JL, Tran RK, Sundaresan V, Comai L, Chan SW. The rapidly evolving centromere-specific histone has stringent functional requirements in Arabidopsis thaliana. Genetics. 2010;186:461–471. doi: 10.1534/genetics.110.120337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Ravi M, Chan SW. Haploid plants produced by centromere-mediated genome elimination. Nature. 2010;464:615–618. doi: 10.1038/nature08842. This pair of papers utilizes a null mutant of CENP-A in Arabidopsis to show that the tail domain is important for meiotic segregation and that the H3-CATD chimera is not sufficient for centromere activity. A tailswap mutant (CENP-A with the H3 tail) generates haploid plants, which has potential practical implications in crop engineering. [DOI] [PubMed] [Google Scholar]
- *34.Sanchez-Pulido L, Pidoux AL, Ponting CP, Allshire RC. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. This paper presents elegant bioinformatic analysis that revealed a common ancestry of Scm3 and HJURP despite their significant primary sequence divergence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shivaraju M, Camahort R, Mattingly M, Gerton JL. Scm3 is a centromeric nucleosome assembly factor. J Biol Chem. 2011;286:12016–12023. doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Hu H, Liu Y, Wang M, Fang J, Huang H, Yang N, Li Y, Wang J, Yao X, Shi Y, et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 2011;25:901–906. doi: 10.1101/gad.2045111. See ref 26 for description. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **37.Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011;472:234–237. doi: 10.1038/nature09854. See ref 26 for description. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng H, Zhou Z, Zhou BR, Bai Y. Structure of the budding yeast Saccharomyces cerevisiae centromeric histones Cse4-H4 complexed with the chaperone Scm3. Proc Natl Acad Sci U S A. 2011;108:E596. doi: 10.1073/pnas.1109548108. author reply E597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *39.Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *40.Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. These 2 papers report the discovery of an E3 ubiquitin ligase in budding yeast that specifically recognizes the CATD and through ubiquitin-mediated proteolysis restricts Cse4 incorporation at non-centromeric sites in the genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17:237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 43.Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takayama Y, Sato H, Saitoh S, Ogiyama Y, Masuda F, Takahashi K. Biphasic incorporation of centromeric histone CENP-A in fission yeast. Mol Biol Cell. 2008;19:682–690. doi: 10.1091/mbc.E07-05-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lermontova I, Schubert V, Fuchs J, Klatte S, Macas J, Schubert I. Loading of Arabidopsis centromeric histone CENH3 occurs mainly during G2 and requires the presence of the histone fold domain. Plant Cell. 2006;18:2443–2451. doi: 10.1105/tpc.106.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G phase. Nucleus. 2011;2:146–157. doi: 10.4161/nucl.2.2.15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 49.Maddox PS, Hyndman F, Monen J, Oegema K, Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 51.Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52.Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol. 2011;194:855–871. doi: 10.1083/jcb.201106079. This paper utilizes frog egg extracts to show that CENP-C binds to Knl2 and suggests that this binding may be important for centromere propagation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schittenhelm RB, Althoff F, Heidmann S, Lehner CF. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J Cell Sci. 2010;123:3768–3779. doi: 10.1242/jcs.067934. [DOI] [PubMed] [Google Scholar]
- 54.Erhardt S, Mellone BG, Betts CM, Zhang W, Karpen GH, Straight AF. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183:805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **55.Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS. A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP-A. Nat Cell Biol. 2010;12:1186–1193. doi: 10.1038/ncb2129. This paper demonstrates a CENP-A maintenance step in human cells and details the dynamics of CENP-A loading. The authors show that CENP-A loading occurs over the course of G1, after which a small GTPase pathway is required to ensure newly loaded CENP-A is retained. [DOI] [PubMed] [Google Scholar]
- *56.Torigoe SE, Urwin DL, Ishii H, Smith DE, Kadonaga JT. Identification of a rapidly formed nonnucleosomal histone-DNA intermediate that is converted into chromatin by ACF. Mol Cell. 2011;43:638–648. doi: 10.1016/j.molcel.2011.07.017. This paper reports the formation of a non-nucleosomal intermediate in chromatin assembly that is converted into a nucleosome by the action of an ATPase motor. This finding has potential implications for understanding nucleosome assembly in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *58.Olszak AM, van Essen D, Pereira AJ, Diehl S, Manke T, Maiato H, Saccani S, Heun P. Heterochromatin boundaries are hotspots for de novo kinetochore formation. Nat Cell Biol. 2011;13:799–808. doi: 10.1038/ncb2272. [DOI] [PubMed] [Google Scholar]
- *59.Yuen KW, Nabeshima K, Oegema K, Desai A. Rapid De Novo Centromere Formation Occurs Independently of Heterochromatin Protein 1 in C. elegans Embryos. Curr Biol. 2011 doi: 10.1016/j.cub.2011.09.016. These papers address formation of de novo centromeric chromatin in Drosophila & C. elegans. Olszak et al. show that CENP-A can be targeted to ectopic loci in Drosophila cells by local concentration of heterochromatin protein 1 (HP1). Yuen et al. show formation of CENP-A chromatin and kinetochore assembly on DNA injected into the C. elegansgermline; this reaction does not require HP1 family proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- *61.Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. Embo J. 2010;30:328–340. doi: 10.1038/emboj.2010.329. This paper reports the use of an artificial chromosome system to tether and manipulate the chromatin state at centromeres. The authors demonstrate that removal of a specific modification (H3K4me2) by tethering of a demethylase reduces HJURP targeting and leads to eventual centromere inactivation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **63.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **64.Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. These two papers elucidate how CENP-A chromatin directs kinetochore assembly. Gascoigne et al. demonstrate that ectopic targeting of CENP-T and CENP-C to non-centromeric loci in human cells recruits kinetochore proteins to those loci in the absence of CENP-A nucleosomes. Guse et al. use CENP-A nucleosome arrays in frog egg extracts to show that CENP-C recognition of the CENP-A C-terminal tail is the key determinant directing kinetochore assembly. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *65.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. This paper defines the recognition site on CENP-A nucleosomes for CENP-C. Using an in vitro approach, the authors find that CENP-C recognizes the extreme C-terminal tail of CENP-A. This work is developed further in Ref. 63, which shows sufficiency of this recognition mechanism for kinetochore assembly in frog egg extracts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 67.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- *69.Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. This paper utilizes immunoelectron microscopy to visualize architectural changes in the inner kinetochore following microtubule attachment in vivo. The authors suggest that CENP-T is a flexible tether that changes conformation when placed under tension by microtubule attachments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 72.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A. 2010;107:10484–10489. doi: 10.1073/pnas.1002325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1. 9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]