SUMMARY
Abnormal inflammatory signaling activation occurs commonly in cancer cells. However, how it is initiated and maintained, and its roles in early stages of tumorigensis are largely unknown. Here we report that the monocyte-derived MCP-1-induced transformation of immortal breast epithelial cells is triggered by transient activation of MEK/ERK and IKK/NF-κB pathways, and maintained by constitutive activation of a feed-forward inflammatory signaling circuit composed of miR-200c, p65, JNK2, HSF1 and IL6. Suppression of miR-200c by IL6 constitutively activates p65/RelA and JNK2, and the latter phosphorylates and activates HSF1. In turn, HSF1 triggers demethylation of the IL6 promoter that facilitates the binding of p65 and c-Jun that together drives constitutive IL6 transcription. Importantly, this signaling circuit is manifest in human cancer cells and in a mouse model of ErbB2-driven breast cancer, where IL6 loss significantly impairs tumorigenesis. Therefore, targeting this signaling circuit represents an effective therapeutic avenue for breast cancer prevention and treatment.
INTRODUCTION
Tumorigenesis is a multi-step process whereby cells acquire a series of genetic and epigenetic alterations in key growth-regulatory genes such as oncogenes and tumor suppressors that provide them with proliferative and survival advantages over their normal neighbors (Fearon and Vogelstein, 1990; Hanahan and Weinberg, 2000). These alterations can occur in a cancer cell intrinsic fashion, or can be provoked by extrinsic signals from the tumor microenvironment. Indeed, both clinical studies and animal models have suggested that the host inflammatory response drives tumorigenesis and metastasis, and that high levels of inflammation connote poor prognosis (Ammirante et al., 2010; Balkwill and Mantovani, 2001; Coussens and Werb, 2002; Grivennikov et al., 2010; Luo et al., 2004).
About 25% of cancers appear due to chronic infection or other types of chronic inflammation, and almost all cancers trigger abnormal inflammatory signaling (Hussain and Harris, 2007). The cues that normally activate inflammatory signaling are widespread and range from microbial and viral toxins or antigens, allergens or toxic chemicals, and second messengers released in pathological states, such as autoimmune diseases and obesity (Schetter et al., 2010). Alterations in oncogenes or tumor suppressors in cancer cells also activate immune cells and inflammatory signaling through various means, including the release of reactive oxygen and nitrogen species, the aberrant production of inflammatory cytokines and chemokines, activation of cyclooxygenase-2 and nuclear factor kappa B (NF-κB) signaling effectors (Grivennikov et al., 2010; Schetter et al., 2010). Much progress has been made in elucidating the mechanisms by which inflammatory signaling drives tumor progression and metastasis. However, the mechanism by which inflammatory signaling contributes to the early stages of tumorigenesis, such as cell transformation, are largely unknown.
Here, we report that a constitutively activated, feed-forward inflammatory signaling circuit normally harnessed by miR-200c is established during epithelial cell transformation, and that this circuit plays crucial roles in epithelial cell transformation and mammary cell tumorigenesis.
RESULTS
Monocyte-derived MCP-1 triggers transformation of immortal breast epithelial cells
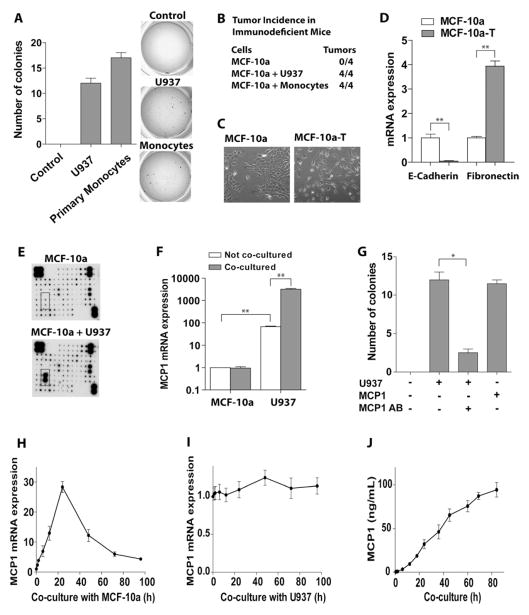
Monocytes and macrophages play important roles in promoting tumor progression and metastasis (Grivennikov et al., 2010; Qian and Pollard, 2010). However, their roles in initiating tumorigenesis are unclear. To test the potential roles of monocytes in the transformation of mammary epithelial cells, we employed a co-culture system. Human nontransformed MCF-10a cells were cultured in six-well dishes and treated with the carcinogen 7,12-dimethylbenz[α]anthracene (DMBA) for 24 hr. They were then co-cultured with human U937 monocytes grown in inserts having a 0.4 μM porous membrane. Thus, the two cell types shared the same culture medium but did not physically contact each other. After 72 hr of co-culture the monocytes were removed and the transformation status of MCF-10a cells was assessed by evaluation of anchorage independent growth in soft agar. Notably, co-culture with monocytes tripled the number of DMBA-treated MCF-10A colonies (Figure S1A). Interestingly, untreated MCF-10a cells co-cultured with U937 cells also formed colonies at a frequency similar to that of epithelial cells treated with DMBA alone (Figure 1A, S1A), indicating that co-culture with U937 cells alone is enough to transform MCF-10a cells. Notably, all mice injected with co-cultured MCF-10a cells developed tumors, whereas none were observed in mice injected with MCF-10a cells without co-culture (Figure 1B, S1B).
Figure 1. Monocyte co-culture induces oncogenic transformation of MCF-10a cells.
(A) MCF-10a cells were co-cultured with U937 monocytes or with normal primary monocytes for 72 hr. MCF-10a cells were then put into soft-agar and numbers of colonies were enumerated.
(B) Monocyte co-culture is sufficient to provoke tumorigenesis of MCF-10a cells. Xenograft tumor formation in Rag1-deficient mice. 5 × 106 MCF-10a cells that had been co-cultured with or without U937 cells or primary monocytes for 72 hr were injected into the right flanks of Rag1 mice. Number of animals injected with MCF-10a cells is shown.
(C) Monocyte co-culture induces morphology changes of MCF-10a cells. Phase contrast images of non-transformed MCF-10a cells and transformed (MCF-10a-T) cells are shown. MCF-10a-T cells were isolated from soft agar colonies that arose following co-culture with U937 cells. Representative images are shown.
(D) qRT-PCR of E-Cadherin and Fibronectin in MCF-10a and MCF-10a-T cells.
(E) Cytokines in medium from MCF-10a cells co-cultured with U937 for 24 hr were detected by Cytokine antibody arrays. (The expression of MCP-1 is marked in a square).
(F) MCP-1 mRNA expression in MCF-10a and U937 cells with or without 24 hr of co-culture. Note: the scale on y-axis is logarithmic.
(G) Soft agar colony assay of MCF-10a cells treated with recombinant MCP-1, or co-cultured with U937 cells, or treated with MCP-1 neutralizing antibody for 24 hr.
(H) Time course showing the induction of MCP-1 mRNA expression in U937 cells upon co-culture with MCF-10a cells.
(I) Time course showing MCP-1 mRNA levels in MCF-10a cells upon co-culture with U937 cells.
(J) Time course of MCP-1 protein levels (ELISA) in conditioned media from MCF-10a/U937 co-culture.
The results in A, D, F–J are shown as mean ± SD of one representative experiment (from three independent experiments) performed in triplicates. Statistically significant differences are indicated. (*) P < 0.05; (**) P < 0.01. See also Figure S1.
Importantly, co-culture with human primary monocytes, isolated from healthy donors, also induced MCF-10a cell transformation to a degree similar to that observed after co-culture with U937 cells (Figure 1A). These co-cultured cells also developed tumors when injected into Rag1-deficient mice (Figure 1B). Thus, transformation of MCF-10a breast epithelial cells is a shared property of human monocytes. Similarly, MCF-12a cells, another immortal human breast epithelial cell line, can be transformed upon co-culture with U937 or primary monocytes (Figure S1C).
To determine whether the duration of co-culture affected the transformation of MCF-10a cells, different periods of co-culture were tested. Co-culture for 24 hr was sufficient to trigger maximal transformation of MCF-10a cells (Figure S1D). Next, we purified transformed cells from isolated colonies in soft agar and cultured them in monolayer in basal medium without any transforming stimulus. These cells (coined MCF-10a-T cells) were permanently transformed even after several passages as shown by colony formation assays (Figure S1E). In contrast to parental MCF-10a cells, which had a tightly packed epithelial-like morphology, the transformed cells had a fibroblastic spindle-shaped morphology (Figure 1C) reminiscent of the epithelial mesenchymal transition (EMT). Indeed, MCF-10a-T cells expressed low level of the epithelial marker E-cadherin and high level of the mesenchymal marker fibronectin relative to parental MCF-10a cells (Figure 1D).
As described above, monocytes and MCF-10a cells were not in physical contact in the co-culture system. Thus, the culture medium that connected the two cells should be the mediator of transformation. To define the soluble factors that are present following co-culture with monocytes and that provoke the transformation of MCF-10a cells, Human Cytokine Antibody Arrays were used to detect the differentially expressed cytokines in the conditioned co-culture medium. These assays demonstrated that monocyte chemotactic protein-1 (MCP-1) was significantly increased in MCF-10a/U937 co-cultured medium compared with non-cocultured controls (Figure 1E). The basal levels of MCP-1 mRNA in U937 cells were much higher than those in MCF-10a cells, and MCF-10a/U937 co-culture promoted MCP-1 expression in U937 but not in MCF-10a cells. Thus, the marked increase of MCP-1 in co-culture medium is derived from monocytes (Figure 1F). Notably, anti-MCP-1 neutralizing antibody blocked U937 cells or monocytes-induced transformation of co-cultured MCF-10a cells (Figure 1G), while recombinant MCP-1 alone was sufficient to transform MCF-10a epithelial cells (Figure 1G).
Others have reported that tumor cells are the major source for MCP-1 in tumor progression and metastasis (Nam et al., 2006). To investigate whether transforming MCF-10a cells contribute to the accumulation of MCP-1 in co-culture medium, the levels of MCP-1 mRNA expressed in U937 versus MCF-10a cells were monitored during co-culture. MCP-1 was significantly induced in U937 cells and peaked at 24 hr of co-culture (Figure 1H), whereas levels in MCF-10a cells did not change (Figure 1I). MCP-1 protein levels gradually and continuously increased in co-cultured medium over time (Figure 1J). Further, the levels of the macrophage markers in U937 cells gradually increased during co-culture, indicating that monocytes undergo macrophage differentiation during co-culture (Figure S1F).
MCP-1 induced transformation is triggered by transient activation of MEK/ERK, IKK/NF-κB, and IL6 signaling in breast epithelial cells
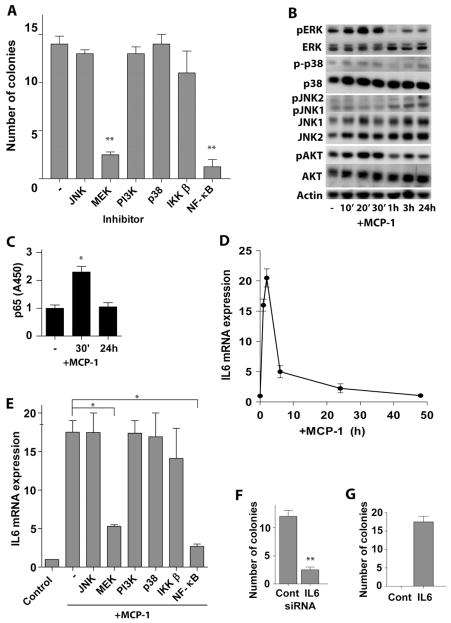
To define the mechanism by which MCP-1 triggers transformation, we initially assessed the effects of selective inhibitors that disable different signal pathways. MCF-10a cells were treated with these inhibitors for 2 hr and then co-cultured with U937 monocytes in medium containing the same inhibitor for 24 hr. MCF-10a were then assessed for anchorage independent growth in soft agar. Both MEK/ERK and NF-κB inhibitors significantly suppressed MCF-10a cell growth in soft agar, whereas an IKKβ inhibitor had moderate inhibitory effects. In contrast, JNK, PI3K, and p38 inhibitors had no such effect (Figure 2A). Similarly, MEK/ERK and NF-κB inhibitors significantly and the IKKβ inhibitor partially suppressed MCP-1-induced MCF-10a cell transformation (Figure S2A). The transient activation of MEK/ERK and p65/RelA in MCF-10a cells by MCP-1 was confirmed by western blot and ELISA analyses (Figure 2B, 2C). Finally, the effects of MCP-1 treatment on p65 activation were selective and there were no effects on other NF-κB family members (Figure S2B). Although p-AKT was transiently increased after MCP-1 treatment, PI3K inhibitor or silencing AKT in MCF-10a cells did not affect monocytes or MCP-1 –induced transformation (Figure S2C, S3C).
Figure 2. MCP-1-induced transformation of MCF-10a cells is initiated by transient activation of MEK/ERK, IKK/NF-κB, and IL6 signaling pathways.
(A) Soft agar colony assays of MCF-10a cells treated with indicated inhibitors (10μM) and co-cultured with U937 cells. Inhibitors were added 2 hr before and throughout co-culture. After 24 hr of co-culture, cells were placed into soft agar (without inhibitors).
(B) Western blot analysis of MCF-10a cells incubated with 50 ng/ml MCP-1 for the indicated intervals.
(C) NF-κB (p65) activity assay of MCF-10a cells treated with 50 ng/ml MCP-1 for the indicated intervals.
(D) Time course of IL6 mRNA expression in MCF-10a cells treated with 50 ng/ml MCP-1 for the indicated intervals.
(E) IL6 mRNA levels in MCF-10a cells treated with MCP-1 and the indicated inhibitors. Inhibitors were added in medium 2 hr before MCP-1 (50 ng/ml) treatment.
(F) Soft agar colony assay of MCF-10a cells transfected with IL6 siRNA or scrambled siRNA, followed by co-culture with U937 cells for 24 hr.
(G) Soft agar colony assay of MCF-10a cells treated with or without IL6 for 24 hr.
The results in A and C – G are shown as mean ± SD of one representative experiment (from three independent experiments) performed in triplicates. Statistically significant differences are indicated. (*) P < 0.05; (**) P < 0.01. See also Figure S2.
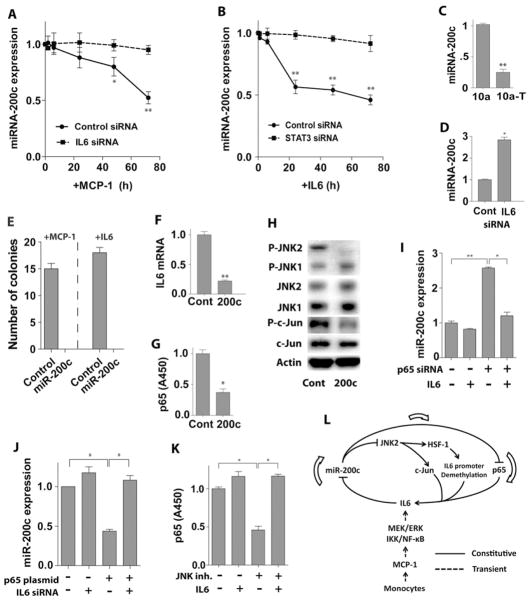
Given these findings we assessed the expression of MEK/ERK and p65 target genes that are important in tumor development in co-cultured or MCP-1 treated MCF-10a cells. IL6 expression was significantly transiently increased in MCF-10a cells in both scenarios (Figure 2D, S2D), while the expression of TNFα, I L8, and IL1β were, surprisingly, suppressed (Figure S2E – S2G). IL6 protein levels in culture medium also increased after MCP-1 treatment (Figure S2H). Further, MEK/ERK or NF-κB inhibitors, but not JNK, p38, PI3K inhibitors or AKT siRNA, blocked MCP-1-induced IL6 expression in MCF-10a cells (Figure 2E, S2I). Importantly, knockdown of IL6 in MCF-10a cells repressed U937 monocyte-induced transformation (Figure 2F, S3C), and recombinant IL6 alone was sufficient to trigger the transformation of MCF-10a breast epithelial cells (Figure 2G). Thus, monocyte-derived MCP-1-induced transformation of MCF-10a cells is initiated by MEK/ERK and IKK/NF-κB activation, and is driven by IL6.
Constitutive activation of JNK2, p65, and IL6 is essential for breast epithelial cell transformation and for the maintenance of the transformed state
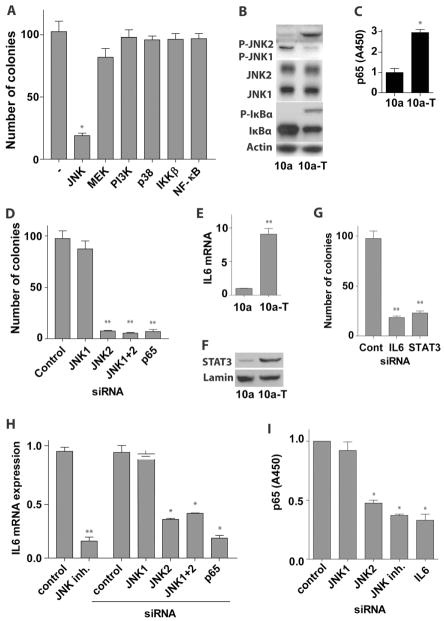
Transient activation of MEK/ERK and IKK/NF-κB signaling is initiated during MCP-1-induced MCF-10a cells transformation. To test whether these pathways were also required to sustain the malignant phenotype of transformed cells, MCF-10a-T cells were seeded into soft agar containing selective inhibitors that disable these pathway s. Incontrast to co-culture experiments, a JNK inhibitor blocked colony growth of MCF-10a-T cells whereas MEK/ERK inhibitors had modest effects and those targeting IKKβ and NF-κB inhibitors had none at all (Figure 3A). In addition, similar to PI3K inhibitor, knockdown of AKT1 and AKT2 in MCF-10a-T cells by siRNAs did not affect colony formation in soft agar (Figure S3A).
Figure 3. Constitutive activation of JNK2 and p65, and of IL6 expression in transformed MCF-10a cells.
(A) Soft agar colony assay of transformed MCF-10a-T cells. Different inhibitors as indicated (10μM) were added in the soft agar and medium.
(B) Western blot analysis of MCF-10a (10a) and MCF-10a-T (10a-T) cells with indicated antibodies.
(C) NF-κB (p65) activity analysis in MCF-10a (10a) or MCF-10a-T (10a-T) cells.
(D) Soft agar colony assay of MCF-10a-T cells transfected with indicated siRNAs. Cells were transfected in monolayer culture and placed in soft agar 48 hr after transfection.
(E) IL6 mRNA levels in MCF-10a (10a) versus MCF-10a (10a-T) cells.
(F) Western blot analysis of nuclear extract of MCF-10a (10a) and MCF-10a-T (10a-T) cells with indicated antibodies.
(G) Soft agar colony assay of MCF-10a-T cells transfected with IL6 or STAT3 siRNA. Cells were transfected in monolayer culture and placed into soft agar 48 hr after transfection.
(H) IL6 mRNA expression in MCF-10a-T cells treated with 10μM JNK inhibitor (SP600125) or transfected with indicated siRNAs for 72 hr.
(I) NF-κB (p65) activity analysis of MCF-10a-T cells transfected with indicated siRNAs or treated with 10μM JNK inhibitor (SP600125) for 72 hr.
The results in A, C – E, and G – I are shown as mean ± SD of one representative experiment (from three independent experiments) performed in triplicates. Statistically significant differences are indicated. (*) P < 0.05; (**) P < 0.01. See also Figure S3.
Western blot analyses demonstrated a switch in JNK isoform activation triggered by transformation, where JNK1 was active in non-transformed MCF-10a cells and JNK2 was constitutively active in transformed MCF-10a-T cells (Figure 3B), suggesting that JNK1 and JNK2 may play opposite roles in MCF-10a cell transformation. Supporting this, silencing JNK2 but not JNK1 suppressed colony formation of MCF-10a-T cells (Figure 3D, S3C). Interestingly, p65 but not p50, p52, c-Rel, and RelB was constitutively activated in MCF-10a-T cells (Figure 3C, S3B), and silencing p65 significantly reduced colony formation in soft agar (Figure 3D). Further, the NF-κB suppressor, IκBα, was phosphorylated and down-regulated in MCF-10a-T cells (Figure 3B). It should be noted that the constitutive activation of p65 in MCF-10a-T cells is not contradictory with our results showing that IKKβ and NF-κB inhibitors did not suppress colony formation of MCF-10a-T cells (Figure 3A) because these inhibitors do not repress constitutively activated NF-κB (Pierce et al., 1997). The expression of IL6 was also much higher in MCF-10a-T cells than MCF-10a cells (Figure 3E), whereas there were no changes in the expression of TNFα, IL1β, and IL8 (Figure S3E – S3G). Moreover, the IL6 target STAT3 was activated in MCF-10a-T cells (Figure 3F), and inhibiting IL6 or silencing STAT3 repressed colony formation of MCF-10a-T cells in soft agar (Figure 3G). In addition, knockdown of p65 or JNK2, but not JNK1 suppressed IL6 expression in MCF-10a-T cells (Figure 3H, S3D, S3C), while knockdown of IL6 or JNK2, but not JNK1 impaired p65 activity (Figure 3I, S3C). Thus, a feed-forward IL6-JNK2-p65 pathway is constitutively activated in MCF-10a-T cells and is essential for transformation and maintenance of the transformed state.
JNK2-dependent activation of HSF1 promotes IL6 expression via suppressing methylation of the IL6 promoter
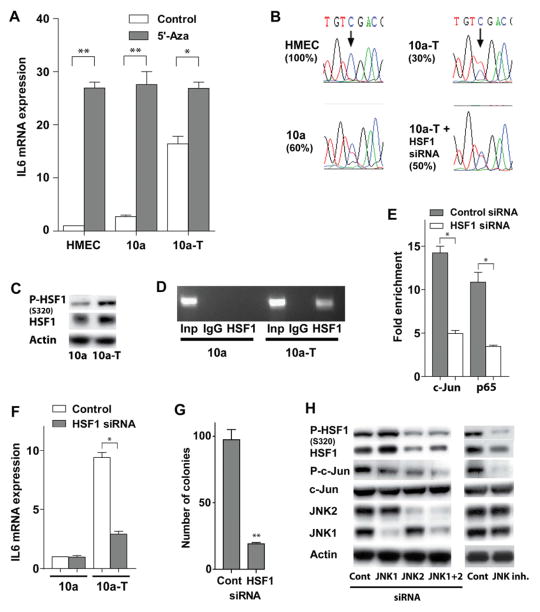
The IL6 promoter was hypomethylated in MCF-10a-T cells and hypermethylated in primary human mammary epithelial cells (HMEC) and MCF-10a cells (Figure 4A, 4B). The difference in methylation was at the cytosine -1099, whereas other CpG islands were completely mehtylated in all tested cells. The -1099C is the only CpG island in IL6 promoter related to IL6 expression (Nile et al., 2008). It was previously reported that the binding of HSF1 to IL6 promoter opens its chromatin structure and facilitate the binding of other transcription factors, which enhances lipopolysaccharide (LPS)-induced IL6 expression (Inouye et al., 2007). Activated HSF1 is phosphorylated on serine-320 (Murshid et al., 2010). We found that the levels of phospho-S320 HSF1 were much higher in MCF-10a-T versus MCF-10a cells, as were the levels of total HSF1 (Figure 4C). Further, chromatin immunoprecipitation (ChIP) analyses demonstrated that HSF1 was bound to the endogenous IL6 promoter in MCF-10a-T cells, but not in MCF-10a cells (Figure 4D). Notably, HSF1 knockdown increased the methylation of IL6 promoter in MCF-10a-T cells (Figure 4B), suggesting that the binding of HSF1 to the IL6 promoter prevents its methylation. Furthermore, HSF1 facilitated the binding of c-Jun and p65 to the IL6 promoter in MCF-10a-T cells (Figure 4E). Finally, HSF1 knockdown suppressed IL6 expression in MCF-10a-T but not MCF-10a cells (Figure 4F) and, accordingly, inhibited MCF-10a-T cell colony formation in soft agar (Figure 4G, S4C).
Figure 4. JNK2-dependent activation of HSF1 promotes IL6 expression via suppressing methylation of the IL6 promoter.
(A) IL6 mRNA levels in primary human mammary epithelial cells (HMEC), MCF-10a, and MCF-10a-T (10a-T) cells treated with 10μM 5-aza-2′deoxycytidine DNA methytransferase inhibitor for 4 days.
(B) Sequence of bisulfite converted IL6 promoter genomic DNA in HMEC, MCF-10a, and MCF-10a-T, and MCF-10a-T cells transfected with HSF1 siRNA. The arrow indicates the Cytosine at position -1099 that is differentially methylated.
(C) Western blot analysis of total and pHSF (S320) levels in MCF-10a (10a) and MCF-10a-T (10a-T) cells.
(D) ChIP assay of HSF1 bound to the IL6 promoter in MCF-10a versus MCF-10a-T cells.
(E) ChIP assay of c-Jun or p65 bound to the IL6 promoter in MCF-10a-T cells transfected with HSF1 siRNA or scrambled siRNA. The data are presented as fold enrichment of precipitated DNA from chromatin incubated with c-Jun or p65 antibody versus IgG.
(F) IL6 mRNA expression in MCF-10a (10a) or MCF-10a-T (10a-T) cells transfected with HSF1 siRNA or scrambled siRNA for 72 hr.
(G) Soft agar colony assay of MCF-10a-T cells transfected with HSF1 siRNA or scrambled siRNA. Cells were transfected in monolayer culture and placed in soft agar 48 hr after transfection.
(H) Western blot analysis of MCF-10a-T cells transfected with indicated siRNAs or treated with 10 μM JNK inhibitor (SP600125) for 72 hr.
The histograms in A and E – G show the mean ± SD of one representative experiment (from three independent experiments) performed in triplicates. Statistically significant differences are indicated. (*) P < 0.05; (**) P < 0.01. See also Figure S4.
HSF1 can be phosphorylated by JNK (Park and Liu, 2001). We found that treatment of MCF-10a-T cells with the JNK inhibitor SP600125, or silencing JNK2 but not JNK1 blocked S320 phosphorylation of HSF1, and also reduced total HSF1 protein levels (Figure 4H), but did not affect levels of HSF1 transcripts (Figure S4A, S4B). Finally, as expected, treatment with JNK inhibitor or silencing JNK2 impaired c-Jun phosphorylation in MCF-10a-T cells. Together, these findings demonstrate that constitutively active JNK2 in MCF-10a-T cells induces phosphorylation and activation of HSF1, which then binds to and facilitates demethylation of the IL6 promoter to facilitate the binding of p65 and c-Jun and constitutive transcription of IL6.
IL6-directed suppression of miR-200c activates JNK2 and p65
To test if miRNAs contribute to MCP-1-induced transformation of MCF-10a cells, we performed miRNA array analysis in MCF-10a cells co-cultured with U937 cells. The expression of 4 miRNAs, miR-200c, Let-7b, miR-34b, and miR-146b, were changed >2-fold in co-cultured MCF-10a cells. Real time PCR analyses confirmed a gradual suppression of miR-200c during co-culture or following MCP-1 treatment (Figure 5A, S5A), whereas the expression levels of Let-7b, miR-34b and miR-146b remained unchanged (Figure S5B – S5D). Silencing IL6 blocked the suppression of miR-200c expression by MCP-1 treatment (Figure 5A), and treatment of MCF-10a cells with IL6 suppressed miR-200c levels, which was blocked by silencing STAT3 (Figure 5B). The expression of miR-200c was much lower in transformed MCF-10a-T cells versus MCF-10a cells (Figure 5C) and again these effects were selective as there were no differences in Let-7b, miR-34b, and miR-146b levels (Figure S5B – S5D). Moreover, silencing IL6 induced miR-200c expression in MCF-10a-T cells (Figure 5D). Importantly, stable overexpression of miR-200c in MCF-10a cells inhibited colony formation of MCF-10a cells treated with MCP-1 or IL6 or co-cultured with monocytes (Figure 5E and S5E), while a miR-200c inhibitor completely blocked colony formation of MCF-10a-T cells (Figure S5F). Thus, miR-200c plays a critical role in the suppression of breast epithelial cell transformation. Notably, overexpression of miR-200c in MCF-10a-T cells inhibited IL6 expression, p65 activation, and JNK2 phosphorylation (Figure 5F-5H). Thus, suppression of miR-200c by IL6 signaling drives constitutive activation of JNK2 and p65 in transformed breast epithelial cells.
Figure 5. IL6-directed suppression of miR-200c leads to activation of p65 and JNK2.
(A) Time course of miR-200c suppression in MCF-10a cells transfected with IL6 siRNA or scrambled siRNA for 24 hr, followed by treatment with 50 ng/ml MCP-1.
(B) Time course of miR-200c suppression in MCF-10a cells transfected with STAT3 siRNA or scrambled siRNA for 24 hr, followed by treatment with 5 ng/ml IL6.
(C) MiR-200c levels in MCF-10a (10a) versus MCF-10a-T (10a-T) cells.
(D) MiR-200c levels in MCF-10a-T cells transfected with IL6 siRNA or scrambled siRNA for 48 hr.
(E) Soft agar colony assay of MCF-10a (Control) or MCF-10a cells stably transfected with miR-200c (200c). Cells were treated with 50 ng/ml MCP-1 or IL6 for 24 hr in monolayer culture and then placed in soft agar.
(F) IL6 mRNA levels in MCF-10a-T cells transfected with a control (Cont) or a miR-200c expression plasmid (200c).
(G) NF-κB (p65) activity analysis in MCF-10a-T cells transfected with a control (Cont) or a miR-200c expression plasmid (200c).
(H) Western blot analysis of JNK activation in MCF-10a-T cells transfected with a control (Cont) or a miR-200c expression plasmid (200c).
(I) MiR-200c levels in MCF-10a-T cells transfected with p65 siRNA or scrambled siRNA for 24 hr, followed by treatment with IL6 or vehicle for 24 hr.
(J) MiR-200c levels in MCF-10a cells transfected with control or p65 expression plasmid for 24 hr, followed by transfection with IL6 siRNA or scrambled siRNA for 48 hr.
(K) NF-κB (p65) activity analysis in MCF-10a-T cells treated with 10μM JNK inhibitor (SP600125) or vehicle for 24 hr, followed by treatment with IL6 or vehicle for 24h (JNK inhibitor was included during IL6 treatment).
(L) Schematic overview of monocyte-induced cell transformation, demonstrating that monocyte-derived MCP-1 induced transformation of MCF-10a cells is triggered by transient activation of MEK/ERK and IKK/NF-κB, and maintained by constitutive activation of an inflammatory signaling circuit consisting of miR-200c, p65, JNK2, HSF1 and IL6.
The results in A – G and I – K are shown as mean ± SD of one representative experiment (from three independent experiments) performed in triplicates. Statistically significant differences are indicated. (*) P < 0.05; (**) P < 0.01. See also Figure S5.
Given these findings, the interplay between IL6, miR-200c, JNK2, HSF1, and p65 in breast cell transformation was investigated. IL6 treatment of MCF-10a cells suppressed miR-200c levels (Figure 5B), while IL6 knockdown in transformed MCF-10a-T cells increased miR-200c levels (Figure 5D). Similarly, p65 knockdown in MCF-10a-T cells induced miR-200c expression, yet this response was suppressed by IL6 (Figure 5I). Further, the induction of miR-200c following knockdown of IL6 could not be overcome by overexpression of p65 in MCF-10a-T cells (Figure S5G), and the down-regulation of miR-200c by p65 overexpression could be blocked by IL6 siRNA in MCF-10a cells (Figure 5J). Thus, IL6-induced suppression of miR-200c is not directly mediated by p65. IL6 activates JNK2 and p65 in MCF-10a cells and silencing JNK2 reduces the activity of p65 (Figure 3H). Though JNK inhibition or HSF1 knockdown impairs p65 activity in MCF-10a-T cells this was overcome by IL6 treatment (Figure 5K, S5H). Thus, IL6 directed down-regulation of miR-200c leads to activation of p65, JNK2, and HSF1, which constitute the inflammatory autoregulatory loop that drives cell transformation (Figure 5L).
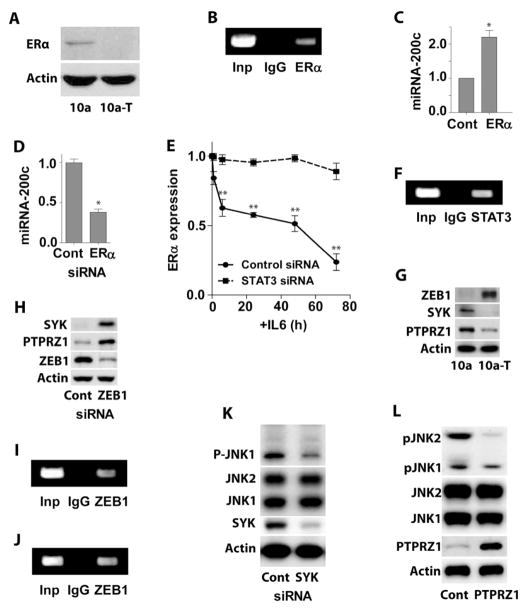
IL6 inhibits miR-200c expression via STAT3-induced down-regulation of estrogen receptor alpha (ERα)
The expression of miR-200c was notably higher, while the expression of IL6 was significantly lower in two estrogen receptor alpha (ERα) positive human breast cancer cell lines as compared with three ERα negative cell lines (Figure S7A), suggesting that ERα might be involved in the IL-6-induced down-regulation of miR-200c. MCF-10a cells are generally considered ERα negative. However, it has been reported that they express low levels of ERα (Yusuf and Frenkel, 2010). Using western blot we confirmed the presence of low but detectable expression level of ERα in MCF-10a, but not in MCF-10a-T cells (Figure 6A). We also found that ERα bound to the miR-200c promoter in MCF-10a cells (Figure 6B). Over-expression of ERα in MCF-10a-T cells resulted in higher levels of miR-200c (Figure 6C), while silencing ERα in MCF10a cells led to down-regulation of miR-200c (Figure 6D). Importantly, treatment of MCF-10a cells with IL6 led to down-regulation of ERα (Figure 6E), however, this down-regulation was blocked by silencing STAT3. ChIP assays demonstrated that STAT3 bound to the ERα promoter in MCF-10a-T cells (Figure 6F). Altogether, our results suggest that IL6 inhibits miR-200c expression via STAT3-induced down-regulation of ERα.
Figure 6. IL6-induced miR-200c suppression is mediated by STAT3-induced down-regulation of ERα, while miR-200c-induced suppression of JNK2 and p65 is via PTPRZ1.
(A) Western blot analysis of estrogen receptor α (ERα) expression in MCF-10a (10a) and MCF-10a-T (10a-T) cells.
(B) ChIP assay of ERα bound to the miR-200c promoter in MCF-10a cells.
(C) MiR-200c levels in MCF-10a-T cells transfected with ERα expression plasmid or scrambled plasmid for 48 hr.
(D) MiR-200c levels in MCF-10a cells transfected with ERα siRNA or scrambled siRNA for 48 hr.
(E) Time course of ERα expression in MCF-10a cells transfected with STAT3 siRNA or scrambled siRNA for 24 hr, followed by treatment with 5 ng/ml IL6.
(F) ChIP assay of STAT3 bound to the ERα promoter in MCF-10a-T cells.
(G) Western blot analysis of MCF-10a (10a) and MCF-10a-T (10a-T) cells with indicated antibodies.
(H) Western blot analysis of SYK and PTPRZ1 expression in MCF-10a-T cells transfected with control or ZEB1 siRNA.
(I) ChIP assay of ZEB1 bound to the SYK promoter in MCF-10a-T cells.
(J) ChIP assay of ZEB1 bound to the PTPRZ1 promoter in MCF-10a-T cells.
(K) Western blot analysis of pJNK in MCF-10a cells transfected with control or SYK siRNA.
(L) Western blot analysis of pJNK in MCF-10a-T cells transfected with control or PTPRZ1 expression plasmid.
The histograms in C – E show the mean ± SD of one representative experiment (from three independent experiments) performed in triplicates. Statistically significant differences are indicated. (*) P < 0.05; (**) P < 0.01. See also Figure S6.
Suppression of JNK2 and p65 activation by miR-200c is mediated by protein tyrosine phosphatase Z1 (PTPRZ1)
To investigate how miR-200c differentially regulates the phosphorylation of JNK1 and JNK2, we screened a group of kinases and phosphatases, and found that the expression of spleen tyrosine kinase (SYK) and PTPRZ1 was significantly decreased in MCF-10a-T cells versus MCF-10a cells (Figure 6G). Over-expression of miR-200c in MCF-10a-T cells increased (Figure S6A), while inhibition of miR-200c in MCF-10a cells decreased SYK and PTPRZ1 expression (Figure S6B). MiR-200c represses the expression of ZEB1 (Gregory et al., 2008). We found that ZEB1 expression was significantly higher in MCF-10a-T cells than in MCF-10a cells (Figure 6G). Importantly, silencing ZEB1 increased SYK and PTRPZ1 expression in MCF-10a-T cells (Figure 6H), suggesting that ZEB1 represses SYK and PTPRZ1. Both SYK and PTPRZ1 promoters contain conserved putative ZEB1 binding sites (Figure S6C, S6D). ChIP assay showed ZEB1 bound to SYK and PTPRZ1 promoters in MCF-10a-T cells (Figure 6I, 6J). It has been reported that the SYK activates JNK1 (Cambien et al., 1999). Consistently, silencing SYK in MCF-10a cells decreased p-JNK1 (Figure 6K). Importantly, over-expression of PTPRZ1 in MCF-10a-T cells significantly reduced p-JNK2 (Figure 6L).
There was no difference in total p65 protein expression between MCF-10a and MCF-10a-T cells (Figure S6E). However, the expression of nuclear p65 protein was significantly higher in MCF-10a-T cells (Figure S6F). In addition, IκBα was constitutively phosphorylated and down-regulated in MCF-10a-T cells (Figure 3B). Interestingly, overexpression of PTPRZ1 in MCF-10a-T cells decreased phosphorylation and increased expression of IκBα (Figure S6G). These results suggest that PTPRZ1 is also involved in the de-phosphorylation of IκBα, resulting in accumulation of p65 in the nucleus of MCF-10a-T cells.
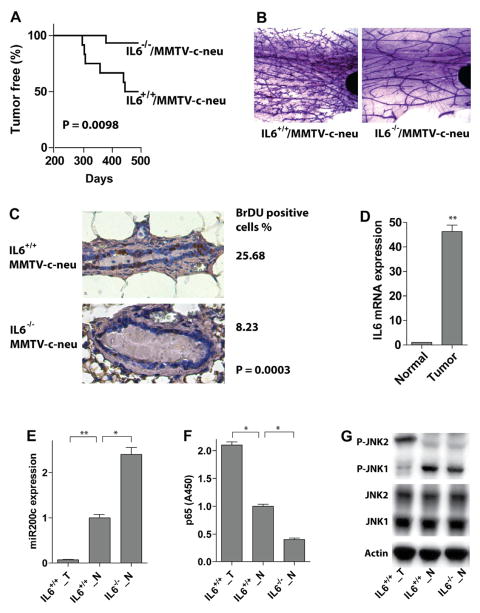
Loss of IL6 disables the inflammatory signaling circuit and impairs Neu-driven mouse mammary tumorigenesis
To test the contribution of IL6 to breast tumorigenesis in vivo, we crossed IL6−/− mice with MMTV-Neu (ErbB2) transgenics to generate IL6−/−;MMTV-Neu and IL6+/+;MMTV-Neu mice. Mammary tumor development was followed in these two cohorts. 50% of the IL6+/+;MMTV-Neu mice developed tumors while only 6.7% of IL6−/−;MMTV-Neu mice had tumors (these have been monitored for >one year, Figure 7A). A comparison of pre-neoplastic alterations such as hyperplastic alveolar nodules by mammary gland whole mounts demonstrated that IL6+/+;MMTV-Neu mammary glands exhibited pronounced lobular hyperplasia and marked increases in numbers of terminal branches and alveoli, whereas the mammary glands of IL6−/−;MMTV-Neu had markedly reduced hyperplastic and dysplastic lesions (Figure 7B). To compare the proliferation rate of mammary epithelial cells in the two cohorts, mammary glands were collected 8 hr after BrdU injection. 26% of mammary epithelial cells from IL6+/+;MMTV-Neu mice were BrdU-positive, while only about 8% cells were positive in IL6−/−;MMTV-Neu mice (Figure 7C).
Figure 7. IL6 loss turns off the inflammatory signaling circuit and impairs mammary tumor development in MMTV-c-Neu mouse model.
(A) Tumor incidence in IL6+/+/MMTV-c-Neu (n = 12) and IL6−/−/MMTV-c-Neu (n = 15) mice. The P-value was calculated using the log-rank test.
(B) The staining of the whole mount mammary gland from 6-month old mice with indicated genotypes is shown.
(C) Proliferation rate of mammary epithelial cells analyzed by BrdU staining of paraffin-embedded sections of mammary glands from 6-month old mice (shown is a representative image of one mouse from three analyzed mice for each group). The percentage of BrdU positive cells is displayed at right and the significance was calculated using the Fisher’s exact test.
(D) IL6 mRNA levels in normal mammary epithelial (Normal) cells or tumor cells (Tumor) isolated from mammary glands or tumors of one-year old IL6+/+/MMTV-c-Neu mice.
(E) MiR-200c levels in normal mammary epithelial cells (IL6+/+_N) or tumor cells (IL6+/+_T) isolated from mammary glands or tumors of one-year old IL6+/+/MMTV-c-Neu mice or of one-year old IL6−/−/MMTV-c-Neu mice (IL6−/−_N).
(F) NF-κB (p65) activity analysis in normal mammary epithelial cells (IL6+/+_N) or tumor cells (IL6+/+_T) isolated from mammary glands or tumors of one-year old IL6+/+/MMTV-c-Neu mice or of one-year old IL6−/−/MMTV-c-Neu mice (IL6−/−_N).
(G) Western blot analysis of normal mammary epithelial cells (IL6+/+_N) or tumor cells (IL6+/+_T) isolated from mammary glands or tumors of one-year old IL6+/+/MMTV-c-Neu mice or of one-year old IL6−/−/MMTV-c-Neu mice (IL6−/−_N).
The histograms in D – F show the mean ± SD of the measurements of expression/activity in one representative mouse (from three analyzed mice) performed in triplicates. Statistically significant differences are indicated. (*) P < 0.05; (**) P < 0.01.
Given the circuit established in human breast cancer we assessed IL6 and miR-200c levels, and p65 and JNK activity in mammary epithelial cells purified from mammary glands or mammary tumors of both cohorts. The expression of IL6, activity of p65, and phosphorylation of JNK2 was increased, while miR-200c levels were reduced in mammary tumor cells versus normal mammary epithelial cells from IL6+/+;MMTV-Neu mice. Furthermore, p65 activity was decreased and miR-200c levels increased in normal mammary epithelial cells from IL6−/−;MMTV-Neu versus IL6+/+;MMTV-Neu mice (Figure 7D – 7G). Collectively, these findings suggest that the IL6-miR-200c-JNK2-p65 circuit is also manifest in Neu-driven mammary tumorigenesis.
The constitutively active inflammatory signaling circuit is manifest in human cancer cells
The expression of miR-200c in both human breast and prostate cancer cells was inversely correlated with IL6 expression level, NF-κB activity, and JNK2 phosphorylation (Figure S7A, S7B). Knockout of IL6, HSF1, JNK2, or p65 in MDA-MB-231 breast cancer cells by siRNA repressed colony formation in soft agar (Figure S7C). Silencing IL6 induced miR-200c expression, while overexpression of miR-200c repressed IL6 expression in MDA-MB-231 cells (Figure S7D, S7E). These results suggest the IL6-miR-200c-JNK2-p65 circuit is manifest in human cancer cells.
DISCUSSION
The mechanisms by which inflammation contributes to tumor progression and metastasis are rapidly coming into focus (Grivennikov et al., 2010; Luo et al., 2007; Qian and Pollard, 2010). However, how the abnormal and constitutive inflammatory signaling is initiated and maintained in transforming cells and its role in the early stages of tumorigenesis, such as cell transformation, are largely unknown. Recently, Iliopoulos et al reported that transient activation of oncogene Src is sufficient to induce neoplastic transformation, which is maintained through a positive feedback loop involving NF-κB, Lin28, Let-7, and IL6 (Iliopoulos et al., 2009; Iliopoulos et al., 2010). Here, using a co-culture model system we have shown that monocyte-derived MCP-1 directly acts as a tumor-initiating cytokine that installs a feed-forward IL6-dependent circuit in immortal breast epithelial cells that is necessary for the initiation and maintenance of the transformed state. Our findings also underscore the interplay of signals between MCF-10a cells and monocytes in this response, where co-culture triggers massive increases in MCP-1 production by monocytes that then binds to its receptors on breast epithelial cells to initiate MEK/ERK, IKK/NF-κB, and IL6 signaling, and where the latter is sustained through the establishment of a feed-forward loop that drives IL6 transcription and that is normally harnessed by miR-200c. We also demonstrated that this autoregulatory loop is established and maintained by IL6-directed suppression of miR-200c expression and that signals downstream of this regulatory RNA bifurcate to regulate p65 activity versus that of a JNK2-to-HSF1 pathway, which are required to demethylate the IL6 promoter to facilitate p65/c-Jun-directed transcription of IL6 (Figure 5L). Therefore, our findings provide an important mechanism linking inflammation to cell transformation.
Although a prolonged inflammatory response can contribute to increased DNA mutation rates and overall genetic instability (Hussain and Harris, 2007), in our co-culture transformation model, the phenotypic switch from non-transformed to transformed MCF-10a cells occurs very quickly. Thus, it is unlikely that the transiently activated inflammatory signaling induced by MCP-1 causes gene mutations or changes in DNA sequence. Hence, the transformation of MCF-10a cells induced by monocytes is most likely driven by the accumulation of epigenetic alterations (switch). Our findings suggest that this epigenetic switch is a constitutively activated inflammatory signaling circuit that is triggered by a transient inflammatory signal. As a transient inflammatory signal is clearly insufficient to trigger such an epigenetic and phenotypic switch in normal cells, we believe that MCF-10a cells have already accumulated some genetic and epigenetic alterations that provide the basis for the oncogenic transformation. Therefore, our model on one hand demonstrates that inflammatory signaling triggers epigenetic alterations that lead to cancer, and on the other hand it does support the ‘multiple-hits’ hypothesis of tumorigenesis in which the accumulation of a sequential genetic alteration of key growth-regulatory genes is required for tumor initiation and genesis (Fearon and Vogelstein, 1990; Hanahan and Weinberg, 2000). However, instead of counting only genetic alterations as ‘hits’, our model suggests that both genetic and epigenetic alterations can be accumulated to form the ‘multiple-hits’ that collectively drive cell transformation and tumorigenesis. Therefore, it is speculated that like MCF-10a cells, “normal” cells in human body accumulated with pre-cancerous mutations will be at high-risk for inflammatory cytokine-driven oncogenic transformation. Importantly, our study showed that the constitutively activated inflammatory signaling circuit does not only exist in monocytes-induced cell transformation, but is also manifest in breast and prostate cancer cells and in oncogene Neu-induced mouse mammary tumorigenesis. In MMTV-Neu mammary tumor model, IL6 inflammatory signaling circuit is triggered by c-neu oncogene itself or/and by the other extrinsic signaling from microenvironment. In either case this is a demonstration of genetic change cooperating with epigenetic switch in tumor initiation.
NF-κB transcription factors play important roles in the regulation of innate and adaptive immune responses, inflammation, and cell survival (Ghosh and Karin, 2002). In normal cells, NF-κB binds to and is negatively regulated by inhibitors of kappa B (IκB) in the cytoplasm, and this interaction is disabled through IκB kinase (IKK)-dependent phosphorylation and subsequent degradation of IκB (Karin and Ben-Neriah, 2000). Liberated NF-κB dimers then enter the nucleus where they induce the transcription of many genes (Luo et al., 2005). In contrast, the constitutive activation of NF-κB, a phenomenon common in most cancers, is not necessarily through IKK (Karin et al., 2002). Our results demonstrate that the initial activation of p65 by MCP-1 is triggered by a transient activation of IKK. However, the maintenance of p65 activity is not dependent on IKK, but rather relies on IL6-directed repression of miR-200c. MiR-200c down-regulates PTPRZ1 that contributes to constitutive phosphorylation of IκBα, resulting in constitutive activation of p65.
The JNK family contains three members, JNK1, JNK2 and JNK3 (Gupta et al., 1996). A major JNK target is the AP1 transcription factor complex that is comprised of Fos and Jun family members. As JNKs have both oncogenic and tumor suppressor functions their roles in cancer are complex (Wagner and Nebreda, 2009). Our study demonstrates a clear switch from JNK1-to-JNK2 activation during mammary epithelial cell transformation and, accordingly, that silencing JNK2 but not JNK1 impairs tumorigenesis. These findings are in accord with those showing opposite roles for JNK1 and JNK2 in tumor development, where for example loss of JNK1 augments the development of skin cancer whilst deletion of JNK2 impairs tumorigenesis (Chen et al., 2001).
In the innate immune system select mi-RNAs are regulated by inflammatory stimuli and some have been shown to mediate inflammatory stimuli (Moschos et al., 2007). In breast epithelial cells IL6 suppresses miR-200c expression, and this repression is persistent and necessary to sustain the feed-forward loop that drives transformation and maintains the transformed state. In accord with its key roles in suppressing epithelial cell transformation, miR-200c expression is suppressed in cancer stem cells and enforced miR-200c expression impairs the tumorigenic potential of extant cancer cells and increases their sensitivity to anti-tumor drugs (Cochrane et al., 2009; Shimono et al., 2009). Thus, agents that disable miR-200c repression could represent broad-spectrum therapeutics.
Our studies suggest that ERα is involved in the IL6-induced down-regulation of miR-200c. Our findings showing ERα has negative role in cell transformation is not contradictory to, but rather consisting with the clinical characteristics of breast cancer. Although ER promotes tumor growth in ER positive breast cancers, ER negativity is one of the important molecular markers for aggressive breast cancer and for identification of breast cancer patients at high risk for recurrence (Sheikh et al., 1994). In addition, the inflammatory breast cancer, a particularly aggressive form of breast cancer is usually ER negative (Robertson et al., 2010).
EXPERIMENTAL PROCEDURES
RNA isolation and real time PCR
RNA was isolated using the miRVANA isolation kit (Amgen) according to manufacturer’s recommendations including DNAse treatment. For mRNA analyses 1μg of total RNA was transcribed to cDNA using the Sigma Enchanced Avian Reverse Transcriptase (Sigma) and quantitative real time PCR (qRT-PCR) was performed using SYBR Green (Quanta) in the IQ5 BioRad thermocycler. MicroRNA expression was determined by qRT-PCR using the mercury LNA Universal RT microRNA PCR kit and microRNA LNA™ PCR primer sets specific for analyzed microRNAs. The mRNA and miRNA data were normalized for the amount of GAPDH and 5S RNA, respectively using the ΔΔCt method. The data is presented as relative expression with the conrol set to 1. The experiments were performed in triplicates and repeated for three times. The data are presented as mean ± SD. Oligonucleotides used for qRT-PCR are shown in Table S1.
Mammary tumorigenesis transgenic mouse models
IL6−/− mice (Kopf et al., 1994) were crossed to N-Tg(MMTV-Neu) 202Mul/J mice (JAX Lab) to generate IL6+/−/MMTV-c-Neu mice that were then intercrossed for more than six generations. IL6+/−/MMTV-c-neu mice were then used for generating IL6+/+/MMTV-c-Neu and IL6−/−/MMTV-c-Neu mice. Animals were observed for tumor development daily. All mice were maintained under specific pathogen-free conditions and experimental protocols were approved by the Scripps Florida IACUC following guidelines from the National Institute of Health.
Statistical analysis
Differences between groups were examined for statistical significance using the Student’s t-test. All P-values are two-tailed. Statistical significance of BrdU staining and tumor incidence in mice was calculated using the Fisher’s exact test and the Log-rang Mantel-Cox test, respectively. Data were considered significant when P < 0.05.
Supplementary Material
HIGHLIGHTS.
Monocyte-derived MCP-1 triggers cell transformation via MEK/ERK and IKK/NF-κB
Transformation requires a constitutively active feed-forward IL6 signaling pathway
miR-200c, p65, JNK, HSF1, and c-Jun direct the IL6 inflammatory signaling circuit
Targeting this IL6 pathway disables development and maintenance of breast cancer
Acknowledgments
We thank Dr. John Cleveland for critical review of the manuscript. This work was supported by grants from the United States Department of Defense (W81XWH-09-1-0533), National Institute of Health (1R01CA140956-01) and The Florida Department of Health (09BB-13) to J.L.L., and by a postdoctoral fellowship from the Frenchman’s Creek Women For Cancer Research (to M.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ammirante M, Luo JL, Grivennikov S, Nedospasov S, Karin M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature. 2010;464:302–305. doi: 10.1038/nature08782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- Cambien B, Millet MA, Schmid-Antomarchi H, Brossette N, Rossi B, Schmid-Alliana A. Src-regulated extracellular signal-related kinase and Syk-regulated c-Jun N-terminal kinase pathways act in conjunction to induce IL-1 synthesis in response to microtubule disruption in HL60 cells. J Immunol. 1999;163:5079–5085. [PubMed] [Google Scholar]
- Chen N, Nomura M, She QB, Ma WY, Bode AM, Wang L, Flavell RA, Dong Z. Suppression of skin tumorigenesis in c-Jun NH(2)-terminal kinase-2-deficient mice. Cancer Res. 2001;61:3908–3912. [PubMed] [Google Scholar]
- Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther. 2009;8:1055–66. doi: 10.1158/1535-7163.MCT-08-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M. Missing pieces in the NF-kB puzzle. Cell. 2002;109(Suppl):S81–96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, Nakai A. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J Biol Chem. 2007;282:33210–33217. doi: 10.1074/jbc.M704471200. [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-kB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. NF-kB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Kopf M, Baumann H, Freer G, Freudenberg M, Lamers M, Kishimoto T, Zinkernagel R, Bluethmann H, Kohler G. Impaired immune and acute-phase responses in interleukin-6-deficient mice. Nature. 1994;368:339–342. doi: 10.1038/368339a0. [DOI] [PubMed] [Google Scholar]
- Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death--a new approach to cancer therapy. J Clin Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Inhibition of NF-kappaB in cancer cells converts inflammation- induced tumor growth mediated by TNFalpha to TRAIL-mediated tumor regression. Cancer Cell. 2004;6:297–305. doi: 10.1016/j.ccr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Luo JL, Tan W, Ricono JM, Korchynskyi O, Zhang M, Gonias SL, Cheresh DA, Karin M. Nuclear cytokine-activated IKKalpha controls prostate cancer metastasis by repressing Maspin. Nature. 2007;446:690–694. doi: 10.1038/nature05656. [DOI] [PubMed] [Google Scholar]
- Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshid A, Chou SD, Prince T, Zhang Y, Bharti A, Calderwood SK. Protein kinase A binds and activates heat shock factor 1. PLoS One. 2010;5:e13830. doi: 10.1371/journal.pone.0013830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam JS, Kang MJ, Suchar AM, Shimamura T, Kohn EA, Michalowska AM, Jordan VC, Hirohashi S, Wakefield LM. Chemokine (C-C motif) ligand 2 mediates the prometastatic effect of dysadherin in human breast cancer cells. Cancer Res. 2006;66:7176–7184. doi: 10.1158/0008-5472.CAN-06-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- Park J, Liu AY. JNK phosphorylates the HSF1 transcriptional activation domain: role of JNK in the regulation of the heat shock response. J Cell Biochem. 2001;82:326–338. doi: 10.1002/jcb.1163. [DOI] [PubMed] [Google Scholar]
- Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, Krishnamurthy S, Le-Petross H, Bidaut L, Player AN, et al. Inflammatory breast cancer: the disease, the biology, the treatment. CA Cancer J Clin. 2010;60:351–375. doi: 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MS, Garcia M, Pujol P, Fontana JA, Rochefort H. Why are estrogen-receptor-negative breast cancers more aggressive than the estrogen-receptor-positive breast cancers? Invasion Metastasis. 1994;14:329–336. [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Yusuf R, Frenkel K. Morphologic transformation of human breast epithelial cells MCF-10A: dependence on an oxidative microenvironment and estrogen/epidermal growth factor receptors. Cancer Cell Int. 2010;10:30. doi: 10.1186/1475-2867-10-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.