Abstract
Nucleic acid amplification for the enteropathogens Cryptosporidium and Giardia is complicated by low target template concentrations and PCR inhibitors. In this work we designed dual capture oligonucleotides for both Cryptosporidium and Giardia 18S rRNA targets which when utilized during DNA extraction from stool improved the limit of detection of our multiplex PCR assay by 1-2 logs, to as little as 10 cysts. When applied to clinical specimens, the method improved the real-time PCR CT by an average of 10.7 9.7 cycles. This work provides a highly sensitive protocol for Cryptosporidium and Giardia when limit of detection is of utmost importance.
Keywords: Cryptosporidium, Giardia, PCR, fecal
Giardia and Cryptosporidium are major diarrheal pathogens worldwide [1, 2]. Detection in clinical specimens usually entails microscopy or antigen detection using multiple stool examinations [3, 4]. Several PCR-based methods for detection of these protozoa have been reported and are increasingly seeing use [5, 6]. We and other groups have reported PCR assays that amplify the 18S rRNA gene of these parasites [7, 8], a high copy target that improves sensitivity. However PCR for these parasites remains challenged by low template concentrations, difficulty in extracting DNA from cysts, DNA stability in stool, and inhibitors present in the specimen. Indeed PCR could be an appealing alternative to existing immunomagnetic separation-microscopy methods used on water [9], however in water sediments the sensitivity of PCR can fall 1,000-fold [10].
For these reasons we embarked on this study to improve the sensitivity of detection of Cryptosporidium and Giardia in difficult specimens. We previously showed that capturing PCR target through an oligonucleotide probe could enhance detection of Giardia up to 16-fold in stool [7]. Such capture methodologies have been used to improve detection of rare DNA targets in stool for colon cancer screening [11]. In the present study, we designed two probes for each parasite's 18S rRNA gene in order to capture nucleic acid during DNA extraction. The extracts were then amplified in multiplex using our Cryptosporidium and Giardia PCR assay. The result is an integrated protocol that maximizes sensitivity of detection of Cryptosporidium and Giardia.
We first modified our previously-reported Giardia A/B subtype-specific PCR assay [7] into a common Giardia assay through redesign of the forward primer, reverse primer, and internal probe (Table 1). This pan-Giardia assay exhibited comparable performance as the parent assay using stool samples spiked with Giardia cysts (Human isolate H-3, Waterborne, Inc., New Orleans, La). Specifically, 105 Giardia cysts spiked into parasite-free stool was subjected to DNA extraction using the QIAamp DNA Stool Mini kit followed by qPCR using either the subtype-specific or the common Giardia protocol, and these assays yielded similar CT (29.8 ± 1.0, P = NS). This Giardia PCR was then combined with our Cryptosporidium PCR [8] into a multiplex assay. With proper PCR master mix and cycling conditions (25 μl volume containing iQ Multiplex Powermix (Bio-Rad, Hercules, CA), 7.5 pmol of each Scorpion probe, 30 pmol each primer, amplified on a Bio-Rad iCycler under the following cycling conditions: 95 °C for 10 min; 40 cycles of 95 °C for 15 s, 51 °C for 15 s and 72 °C for 20 s; 75 °C for 10 min) we were able to achieve comparable sensitivity with the multiplex assay versus the singleplex counterparts. For example, 105 Cryptosporidium (Iowa isolate) or Giardia cysts spiked into parasite-free stool exhibited similar CT (28.8 ± 1.0, P = NS) whether amplified by singleplex or multiplex. Additionally, when a dilution series of 105, 104, 103 Cryptosporidium and Giardia cysts were co-spiked into stool, DNA extracted, and PCR performed in duplicate with the multiplex and singleplex assay, the multiplex CT were not statistically different than the singleplex CT (P = NS, n = 12). We also tested the specificity of the assay by using rotavirus, sapovirus, astrovirus, norovirus, Enterohemorrhagic E. coli, Enteroaggregative E. coli, Enteropathogenic E. coli, Enterotoxigenic E. coli, Campylobacter, Vibrio, Salmonella, Shigella, Entamoeba histolytica, and E. dispar and all were PCR negative with the assay (data not shown).
Table 1.
Primer and probe sequences.
| Oligonucleotide | Sequence (5’-3’) | Position (Accession) |
|---|---|---|
| Giardia forward primer | CGGTCGATCCTGCCGGA | 5-21 (AF199446) |
| Giardia internal probe (linked to forward primer) | GCCATGCATGCCCG-FAM | 46-59 (AF199446) |
| Giardia reverse primer | AGGACAACGGTTGCACCCC | 86-104 (AF199446) |
| Giardia capture probe 1 | GCTAGCCGGACACCGCTGGCAACa | 119-141 (AF199446) |
| Giardia capture probe 2 | ATCATCCTGTTTCACCCGTCa | 647-666 (AF199446) |
| Cryptosporidium forward primer | GGTTGTATTTATTAGATAAAGAAC | 198-221 (AF093491) |
| Cryptosporidium internal probe (linked to reverse primer) | HEX-GTGACATATCATTCAAGTTTCTGAC | 267-291 (AF093491) |
| Cryptosporidium reverse primer | AGACGGTAGGGTATTGGCCT | 303-322 (AF093491) |
| Cryptosporidium capture probe 1 | GAGCCATTCGCAGTTTAACCGa | 78-99 (AF093491) |
| Cryptosporidium capture probe 2 | ACAAGTATCAATTGGAGGGCAa | 515-535 (AF093491) |
Biotin-triethylene glycol-9 carbon spacer added at 3’ end of capture oligonucleotides.
We then developed the DNA capture protocol. We started with our existing Giardia capture probe and added another capture probe to capture the opposing strand. We then designed two new Cryptosporidium capture probes that would target the human pathogenic Cryptosporidium species (C. hominis, C. parvum, C. meleagridis, C. suis, C. felis, C canis). Target DNA captured by biotinylated probes (biotin-triethyleneglycol-9 carbon spacer at 3’ end) was magnetically eluted away from the stool extracts using streptavidin-coated magnetic beads. In brief, stool was first lysed with Qiagen ASL buffer, beat for 3 minutes with 212-300 μm glass beads, boiled for 5 minutes, centrifuged twice at 20,000×g, treated with 2.5 μg RNAse, chloroform extracted, ethanol precipitated, and reconstituted in 100μl Qiagen PCR buffer with 4 mM MgCl2 and 125 pmol of each capture oligonucleotide. The mixture was denatured at 95°C for 5 min followed by annealing at 70°C and hybridization at 53°C for 5 min. Captured DNA was incubated on a rotator at room temperature with 200μl streptavidin coated Dynabeads M-280 (Invitrogen, Carlsbad, CA) in 131 μl Binding Solution. Beads were magnetically separated, washed twice in the included Wash Solution (Invitrogen, Carlsbad, CA), once in Tris 10mM pH 8.0, and eluted by heat at 80°C for 20 min in 30 μL of Tris 10mM/10mM EDTA pH 8.0.
Upon spiking serial dilutions of Giardia and Cryptosporidium cysts into parasite-free stool samples, the capture DNA extraction method followed by multiplex PCR (Table 2) improved the lower limit of detection by ~1-2 logs versus conventional QIAamp DNA Stool Mini kit DNA extraction and 2-4 logs versus ELISA (ProSpecTGiardia/Cryptosporidium Microplate Assay, Remel, Lenexa, KS). We then utilized the multiplex capture PCR method on 12 clinical samples from HIV patients with diarrhea in Tanzania. Informed consent was obtained from all participants and the human experimentation guidelines of the US Department of Health and Human Services, the University of Virginia, and the Kilimanjaro Christian Medical Centre Research Ethics Committee were followed. The 12 samples were chosen because they had tested positive by both ELISA and qPCR and we had sufficient material to perform the supplemental DNA capture extraction protocol. After capture/multiplex qPCR 10 of 12 samples exhibited a significant improvement in Ct, with an overall improvement from 34.3 ± 3.2 to 22.6 ± 2.3 for Cryptosporidium (Table 1, P < 0.05). PCR products were sent for sequencing and all confirmed amplification of the appropriate Giardia and Cryptosporidium products.
Table 2.
Limit of detection with qPCR
| Cryptosporidium oocysts | Conventional DNA extraction and qPCR | Capture DNA extraction and qPCR | ELISA ODa |
|---|---|---|---|
| 105 | 28.8 ± 0.04 | 17.0 ± 0.01 | 0.250 (+) |
| 104 | 32.3 ± 0.01 | 17.2 ± 0.12 | 0.067 (-) |
| 103 | 38.6 ± 0.62 | 18.0 ± 0.03 | 0.057 (-) |
| 102 | nd | 18.2 ± 0.04 | 0.059 (-) |
| 101 | nd | 20.6 ± 0.02 | 0.065 (-) |
| 0 | nd | nd | 0.067 (-) |
| Giardia cysts | |||
| 105 | 24.1 ± 0.04 | 26.4 ± 0.08 | 3.037 (+)a |
| 104 | 27.3 ± 0.09 | 26.6 ± 0.04 | 1.698 (+) |
| 103 | 29.9 ± 0.06 | 26.9 ± 0.08 | 0.205 (+) |
| 102 | 31.7 ± 0.16 | 27.5 ± 0.01 | 0.067 (-) |
| 101 | nd | 28.5 ± 0.02 | 0.063 (-) |
| 0 | nd | nd | 0.059 (-) |
For ProSpecT Giardia and Cryptosporidium ELISA tests, a positive result is defined as sample OD450-negative control OD450 ≥ 0.050. nd = not detected.
Several features of the DNA capture protocol are worth noting. First, during development we found that capturing both DNA strands was advantageous. Secondly, we observed that some DNA purification is necessary up-front prior to capture (as opposed to capture in crude stool samples), presumably to allow the oligonucleotides to find their template. As such one can also perform the capture method on the Qiagen extracts, and this method also yielded substantial improvement in CT (of 8.4 and 5.4 cycles for Giardia and Cryptosporidium, respectively, data not shown). It is notable that the CT appeared to lose quantitation with the capture method (only spanning ~ 3 CT across 5 log of DNA template), suggesting that the beads can become saturated with captured DNA. Thus we would propose using this method in order to improve limit of detection, but would not infer quantity when positive. This capture method requires about six hours total time for extraction, thus is laborious, but can be considered when sensitivity is of utmost importance such as when initial tests are negative, in archived or fixed specimens where nucleic acid may be damaged or degraded, or on environmental specimens.
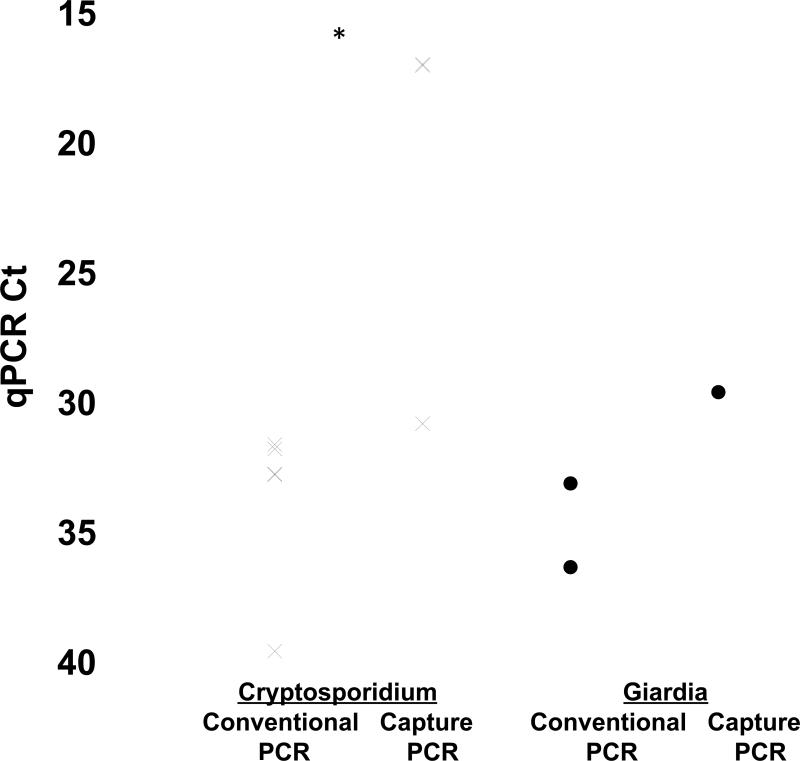
Fig. 1.
Detection of Giardia and Cryptosporidium by DNA capture and multiplex qPCR on clinical specimens. Specimens from patients that were positive for Cryptosporidium and Giardia by both ELISA and qPCR were then subjected to the DNA capture and multiplex qPCR method. qPCR CT shown for both Cryptosporidium and Giardia. *, P < 0.05 comparing CT between conventional and capture extracts.
Acknowledgements
This work was supported by PHS grants AI069598 and AI075396 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hlavsa MC, Watson JC, Beach MJ. Cryptosporidiosis surveillance--United States 1999-2002. MMWR Surveill Summ. 2005;54:1–8. [PubMed] [Google Scholar]
- 2.Hlavsa MC, Watson JC, Beach MJ. Giardiasis surveillance--United States, 1998-2002. MMWR Surveill Summ. 2005;54:9–16. [PubMed] [Google Scholar]
- 3.Addiss DG, Mathews HM, Stewart JM, Wahlquist SP, Williams RM, Finton RJ, et al. Evaluation of a commercially available enzyme-linked immunosorbent assay for Giardia lamblia antigen in stool. J Clin Microbiol. 1991;29:1137–42. doi: 10.1128/jcm.29.6.1137-1142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawitz WG, Faust EC. The probability of detecting intestinal protozoa by successive stool examinations. Am J Trop Med Hyg. 1942;22:131–6. [Google Scholar]
- 5.Ajjampur SS, Rajendran P, Ramani S, Banerjee I, Monica B, Sankaran P, et al. Closing the diarrhoea diagnostic gap in Indian children by the application of molecular techniques. J Med Microbiol. 2008;57:1364–8. doi: 10.1099/jmm.0.2008/003319-0. [DOI] [PubMed] [Google Scholar]
- 6.ten Hove R, Schuurman T, Kooistra M, Moller L, van Lieshout L, Verweij JJ. Detection of diarrhoea-causing protozoa in general practice patients in The Netherlands by multiplex real-time PCR. Clin Microbiol Infect. 2007;13:1001–7. doi: 10.1111/j.1469-0691.2007.01788.x. [DOI] [PubMed] [Google Scholar]
- 7.Ng CT, Gilchrist CA, Lane A, Roy S, Haque R, Houpt ER. Multiplex real-time PCR assay using Scorpion probes and DNA capture for genotype-specific detection of Giardia lamblia on fecal samples. J Clin Microbiol. 2005;43:1256–60. doi: 10.1128/JCM.43.3.1256-1260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroup SE, Roy S, McHele J, Maro V, Ntabaguzi S, Siddique A, et al. Real-time PCR detection and speciation of Cryptosporidium infection using Scorpion probes. J Med Microbiol. 2006;55:1217–22. doi: 10.1099/jmm.0.46678-0. [DOI] [PubMed] [Google Scholar]
- 9.Agency USEP, editor. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. http://www.epa.gov/microbes/1623de05.pdf2005.
- 10.Johnson DW, Pieniazek NJ, Griffin DW, Misener L, Rose JB. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–55. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitney D, Skoletsky J, Moore K, Boynton K, Kann L, Brand R, et al. Enhanced retrieval of DNA from human fecal samples results in improved performance of colorectal cancer screening test. J Mol Diagn. 2004;6:386–95. doi: 10.1016/S1525-1578(10)60536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]