Abstract
Caveolin is an integral membrane protein that is found in high abundance in caveolae. Both the N- and C- termini lie on the same side of the membrane, and the transmembrane domain has been postulated to form an unusual intra-membrane horseshoe configuration. To probe the structure of the transmembrane domain, we have prepared a construct of caveolin-1 that encompasses residues 96-136 (the entire intact transmembrane domain). Caveolin-1(96-136) was over-expressed and isotopically labeled in E.coli, purified to homogeneity, and incorporated into lyso-myristoylphosphatidylglycerol micelles. Circular dichroism and NMR spectroscopy reveal that the transmembrane domain of caveolin-1 is primarily α-helical (57–65%). Furthermore, chemical shift indexing reveals that the transmembrane domain has a helix-break-helix structure which could be critical for the formation of the intra-membrane horseshoe conformation predicted for caveolin-1. The break in the helix spans residues 108 to 110, and alanine scanning mutagenesis was carried out to probe the structural significance of these residues. Our results indicate that mutation of glycine 108 to alanine does not disrupt the structure, but mutation of isoleucine 109 and proline 110 to alanine dramatically alters the helix-break-helix structure. To explore the structural determinants further, additional mutagenesis was performed. Glycine 108 can be substituted with other small side chain amino acids (i.e. alanine), leucine 109 can be substituted with other β-branched amino acids (i.e. valine), and proline 110 cannot be substituted without disrupting the helix-break-helix structure.
Keywords: caveolin, caveolin-1, transmembrane domain, helix-break-helix, NMR spectroscopy, circular dichroism spectroscopy, membrane protein, caveolae, chemical shift indexing
Introduction
Caveolae are 50 to 100 nm wide invaginations found in the plasma membrane [1–3]. Caveolae are particularly abundant in endothelial cells, smooth muscle cells, macrophages, cardiac myocytes, and fibroblasts [2, 3]. These structures are involved in a variety of cellular processes including signal transduction, membrane trafficking, calcium signaling, and lipid recycling [4]. Caveolin is the most important protein found in caveolae, as caveolae structures are lost when the caveolin gene is silenced [5]. Furthermore, misregulation and mutations to caveolin have been implicated in a variety of diseases including cancer, muscular dystrophy, Alzheimer’s, and heart disease [6–10]. In addition to the caveolin protein, caveolae are enriched in cholesterol, and sphingolipids when compared to the bulk plasma membrane [11].
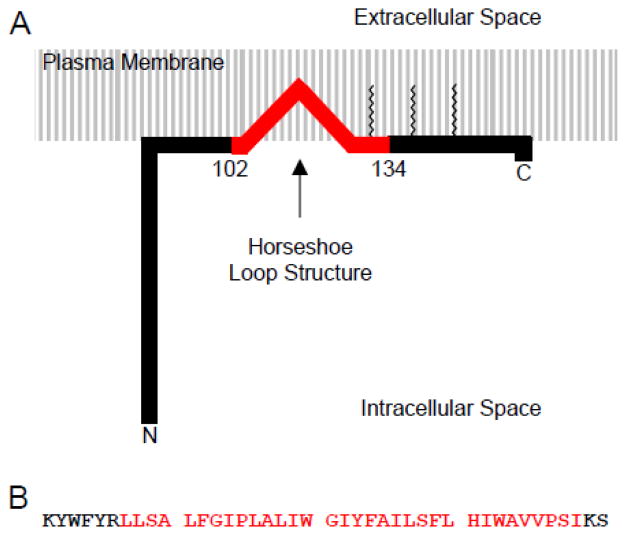
Caveolin is a highly hydrophobic membrane protein that exists in three isoforms (caveolin-1, -2, -3), although caveolin-1 is the most ubiquitous [2]. In vivo studies show that both the N- and C- termini of caveolin are exposed to cytoplasm [12, 13]. Moreover, based on primary sequence analysis, the predicted length of the caveolin transmembrane domain is too long to span the membrane once, but too short to span twice. This data has led to the postulation that the caveolin transmembrane domain adopts a horseshoe conformation in the membrane (Figure 1) [12, 14]. The horseshoe conformation is thought to be critical for the ability of caveolin to curve the plasma membrane to form caveolae by giving the transmembrane domain of caveolin a wedge shape which asymmetrically stretches the inner and outer leaflets of the bilayer. Although this horseshoe conformation is unusual, it has been observed or predicted in other membrane proteins notably NogoC, DP1, and DGKε [15, 16]. Furthermore, in vivo studies have shown that the mutation of proline 110 (found in the transmembrane domain) to alanine, alters the topology and/or orientation of the caveolin protein [17]. However, the presence of the intra-membrane horseshoe conformation has not been verified.
Figure 1.
(A) Cartoon of caveolin-1 topology. Red denotes the transmembrane domain. Zigzag lines denotes palmitoylation. (B) Sequence of Cav196-136.
In the present study, we reconstituted the entire intact transmembrane domain (residues 96-136) of caveolin-1 into LMPG micelles to probe its structure using NMR and circular dichroism spectroscopy. The transmembrane domain of caveolin-1 is highly helical and displays a helix-break-helix structure. Three amino acids G108, I109, and P110 are critical for this motif. These results strongly suggest that transmembrane domain by itself is responsible for the formation of the intra-membrane horseshoe conformation.
Material and Methods
Overexpression and purification of Cav196-136
Cloning, expression, and purification of uniformly 15N- labeled Cav196-136 was performed as described in Diefenderfer et al [18]. For compatibility with cyanogen bromide cleavage, the single methionine at position 111, which is not strictly conserved, was mutated to leucine based on sequence homology to caveolin-2 and caveolin-3. C133 a site of cysteine palmitoylation was mutated to serine to avoid unwanted and biologically irrelevant disulfide bonding. Previous studies have shown that caveolin-1 still retains the ability to fold correctly and traffic to the plasma membrane when C133 is mutated to serine [19–21]. Expression of uniformly 15N and 13C-labeled Cav196-136, was done according to the procedure of Marley et al [22]. Specific amino acid labeling was performed as described in Truhlar et al with slight modification [23]. The cell growth was done on a 1 L scale using M9 minimal media supplemented with 100 mg of the desired 15N amino acid and 500 mg each of the other 19 14N amino acids. Expression was induced at an OD600 of 0.6 using 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside) and grown for 6– 8 hours at 37°C.
Sample Preparation
NMR samples were prepared by adding 500 μL of buffer (100 mM LMPG, 100 mM NaCl, 20 mM phosphate pH 7, and 10% D2O) to a tube containing 2.5 mg of lyophilized Cav196-136. The sample was vortexed vigorously for 2 minutes followed by immersion in a hot water bath (95°C) for one minute. At this point a clear solution was obtained with a minimal amount of precipitate. Finally, the sample was filtered through a 0.2 μm regenerated cellulose spin filter. The final protein concentration was approximately 1 mM. For circular dichroism studies the NMR sample was diluted 20-fold with buffer.
Circular Dichroism spectroscopy
All circular dichroism experiments were performed using a JASCO circular dichroism spectrophotometer (Easton, MD). The experiments were carried out at 25°C. Spectra were obtained from 260 to 190 nm using 16 accumulations. A background spectrum containing no caveolin protein was subtracted from the spectra containing protein. In all cases a 0.1 mm path length cuvette was used.
Two and Three dimensional NMR spectroscopy
All NMR spectra were acquired at 37°C using the a 600 MHz Bruker Avance II spectrometer (Billerica, MA) equipped with a cryoprobe. For analysis and backbone assignments, the following TROSY-based pulse sequences were employed: HSQC, HNCA, and HN(CO)CA. The spectra were processed using NMRPipe and Sparky [24, 25].
Alanine scanning mutagenesis
All mutant constructs were prepared using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA).
Results
Circular Dichroism
Figure 2 shows the circular dichroism spectrum of Cav196-136. There are two minima at 208 and 222 nm which is indicative of α-helical properties. Analysis of the spectrum using the CDSSTR and K2D algorithms (Dichroweb) indicates that Cav196-136 is approximately 57% α-helical [26, 27]. Mutation of proline 110 to alanine results in a circular dichroism spectrum that is remarkably similar to that of the wild-type. Therefore, mutation of proline 110 to alanine does not dramatically alter the secondary structure of Cav196-136.
Figure 2.
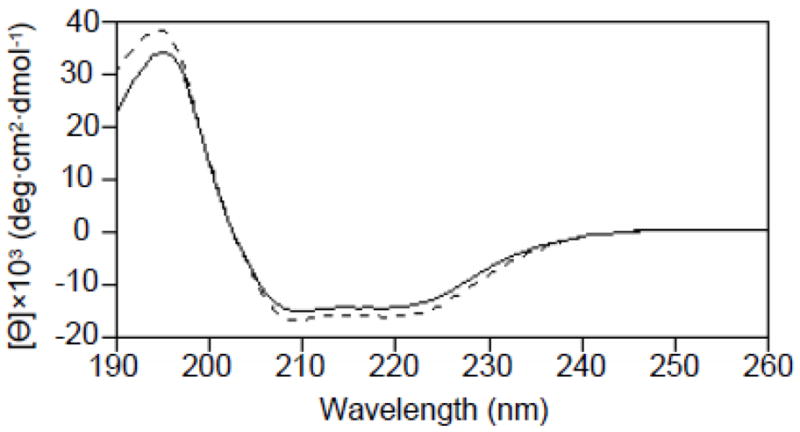
Circular dichroism spectra of Cav196-136 (solid line) and Cav196-136 P110A (dashed line). Minima at 208 nm and 222 nm are indicative of a highly α-helical structure.
Peak assignments
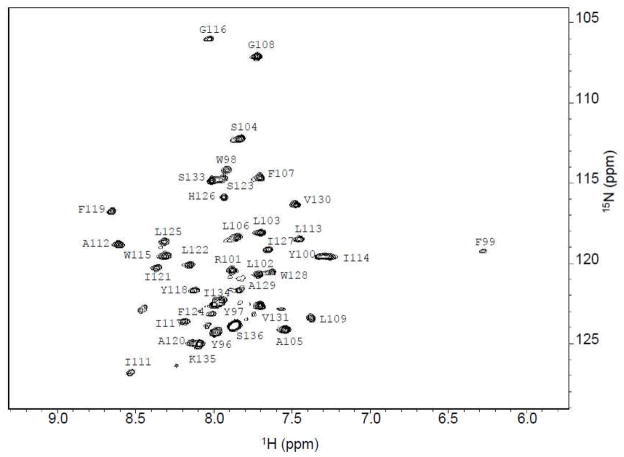
Uniformly labeled Cav196-136 (13C, 15N ) in LMPG micelles gave the highest quality NMR spectra [28]. To assign the backbone resonances the following two and three dimensional TROSY-based experiments were performed: HSQC, HNCA, and HN(CO)CA. To aid in backbone assignments, specific amino acid labeling was employed (Phe, Ala, Leu, Ile, and Val). The assigned 1H, 15N-TROSY spectrum is shown in Figure 3.
Figure 3.
1H-15N TROSY-HSQC spectrum of Cav196-136. The spectrum was acquired with 256 complex points in t1 (15N) and 2048 complex points in t2 (1H).
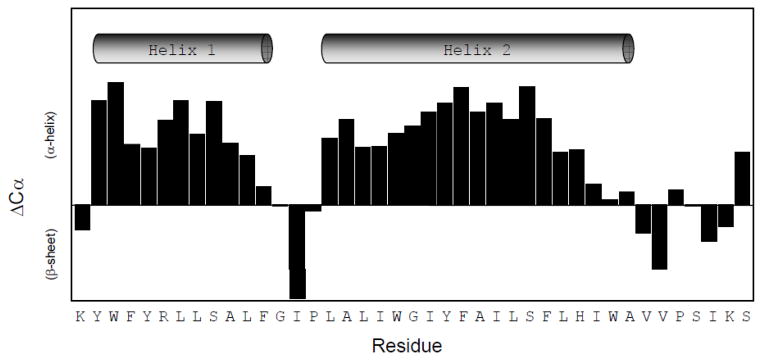
Chemical shift indexing
Figure 4 shows the chemical shift indexing of Cav196-136. Residues 97-107 and residues 111–129 have consistently positive ΔCα’s which is indicative of α-helical structure [29]. Residues 108–110 which lie between these regions do not have positive ΔCα’s. This indicates breakage between the two α-helices. Residues 130-136 do not display consistent ΔCα’s indicating, most likely, a disordered conformation at the end of the transmembrane domain. Therefore, the transmembrane domain of caveolin is displaying a helix-break-helix structure. Based on chemical shift indexing, 65% of Cav196-136 is α-helical which agrees well with the circular dichroism data which predicted 57% α-helicity.
Figure 4.
Chemical shift indexing of Cav196-136. Cav196-136 displays a helix-break-helix structure.
Alanine scanning mutagenesis
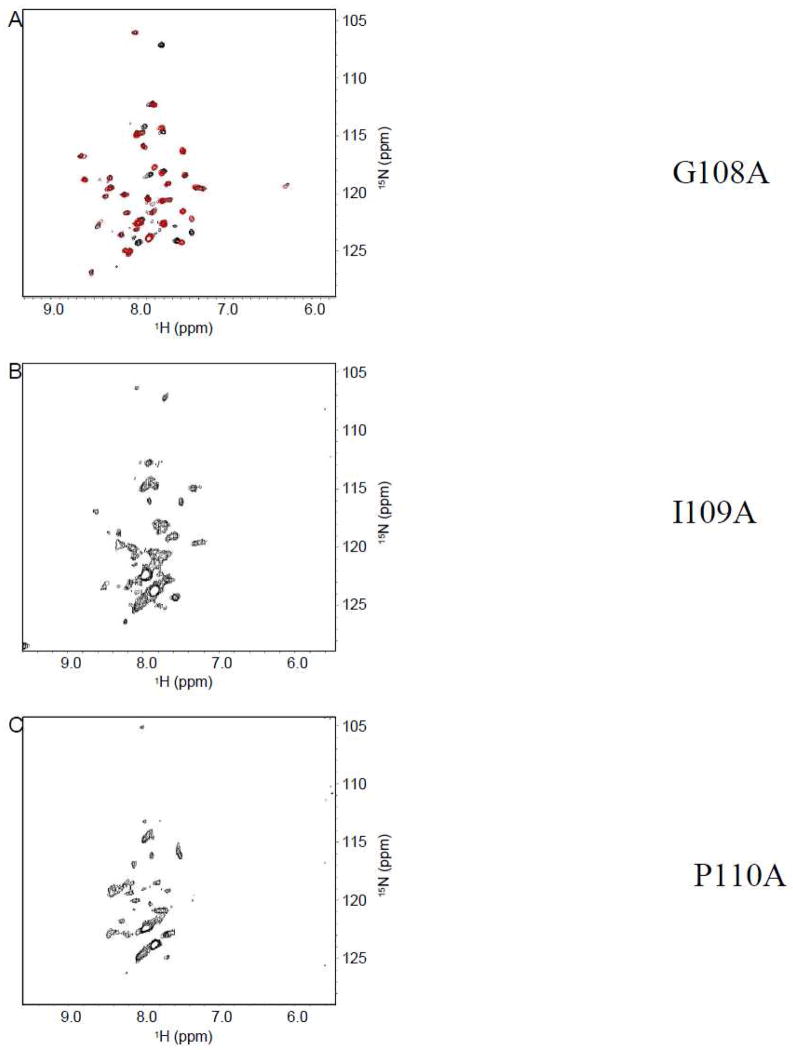
Chemical shift indexing reveals that the break encompasses G108, I109, and P110. To probe whether or not these specific residues were critical for the observed helix-break-helix structure, alanine scanning mutagenesis was employed by mutating G108, I109, and P110 individually to alanine. Alanine was chosen because it has very “neutral” characteristics (small chemically inert nonpolar side chain and conformational freedom that is similar to most amino acids). Figure 5A shows a comparison of the G108A spectrum to that of wild-type. Although the resonances neighboring G108 displayed small shifts, the bulk of the resonances remained the same. Therefore, the global structure of Cav196-136 was retained when glycine 108 was mutated to alanine. Figure 5B and 5C show the I109A and P110A mutations, respectively. Clearly these two spectra are significantly different from that of the wild-type. Specifically, the chemical shift dispersion of the spectra is greatly reduced, and the number of distinct residues has diminished. This indicates that there was a dramatic change in the structure when these mutations are made. Therefore, I109 and P110 appear to be critical for stabilizing the helix-break-helix motif.
Figure 5.
TROSY-HSQC of Cav196-136. (A) Red denotes G108A mutant and black denotes wild-type. (B) I109A mutant. (C) P110A mutant.
Additional Mutagenesis
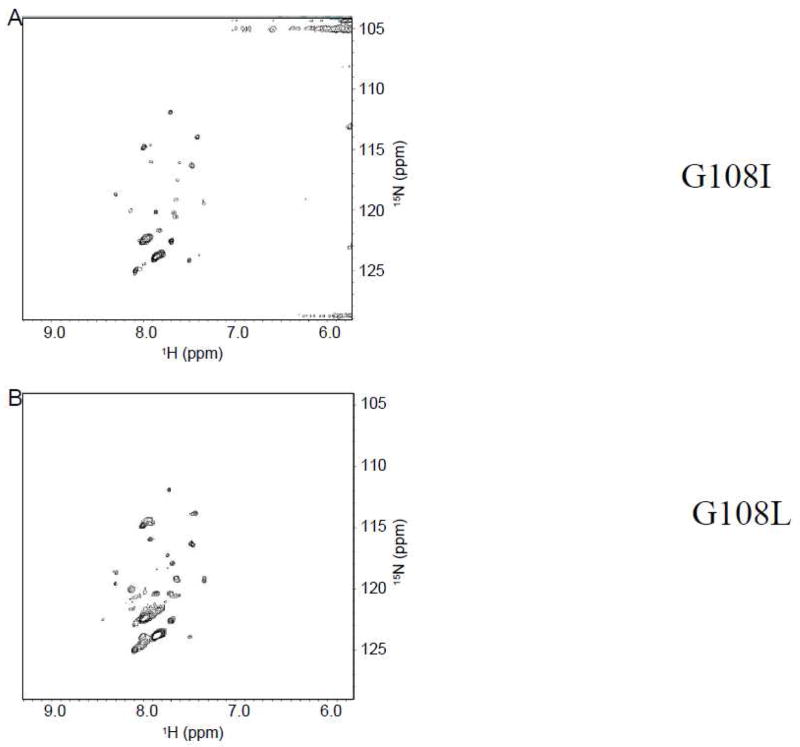
Based on the alanine scanning mutagenesis result, glycine 108 was mutated to isoleucine and leucine to probe whether position 108 can tolerate amino acids with bulky side chains. Figure 6A and 6B show that both G108I and G108L have spectra that are significantly different from that of the wild-type. Therefore, position 108 can tolerate small side chain amino acids such as glycine and alanine, but not bulky side chain amino acids such as leucine or isoleucine.
Figure 6.
TROSY-HSQC of Cav196-136. (A) G108I mutant. (B) G108L mutant.
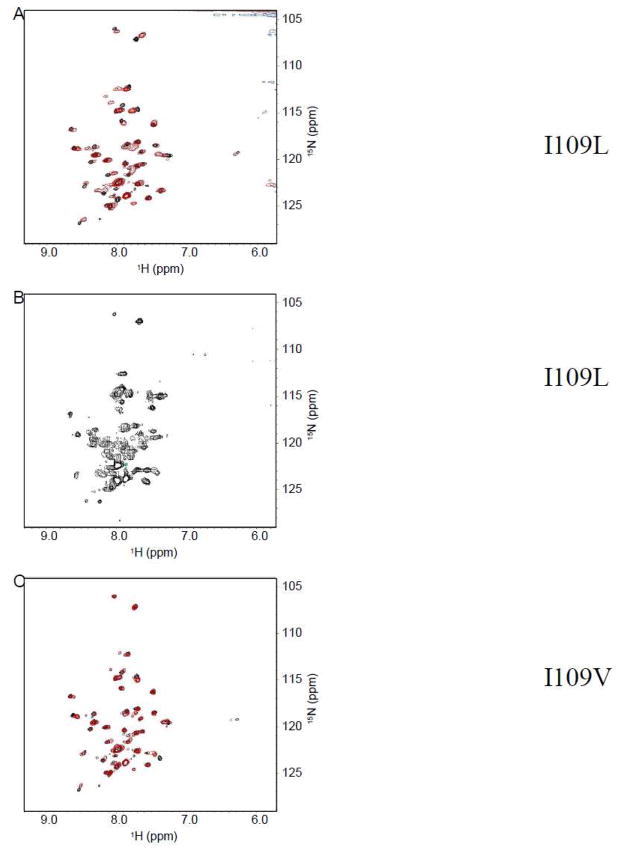
Next, isoleucine at position 109 was mutated to leucine. Leucine and isoleucine have very similar side chains aside from the fact that in leucine, the methyl branching occurs off of the gamma carbon while it occurs off of the beta carbon in isoleucine. Figure 7A shows the spectrum of I109L. Overall the spectrum looks similar to that of wild-type. Extra peaks were observed, and the resolution of the spectrum was mildly diminished when compared to that of the wild-type. Figure 7B shows the spectrum of I109L after one week. In contrast, this spectrum is dramatically different from that of the wild-type. This suggests that position 109 may require a beta-branched amino acid for structure stability. To probe this further, isoleucine 109 was mutated to valine which is another beta branched amino acid. Figure 7C shows that the I109V spectrum overlays well with that of the wild-type, but as expected, small changes in the resonances around position 109 were observed. Therefore a beta-branched amino acid at position 109 is critical for the helix-break-helix structure.
Figure 7.
TROSY-HSQC of Cav196-136. (A) Overlay of I109L mutant (red) with wild-type (black). (B) I109L after 1 week. (C) Overlay of I109V mutant (red) with wild-type (black).
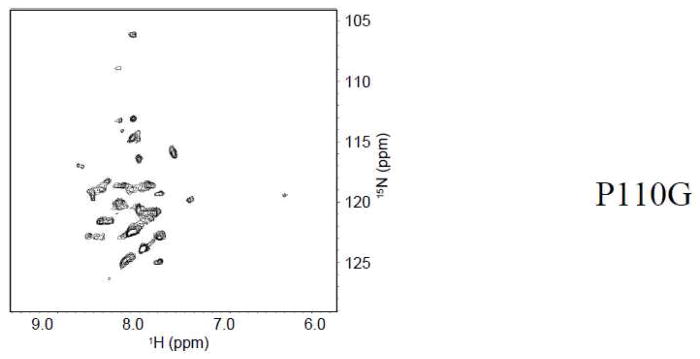
Finally, the proline at position 110 was mutated to glycine. Glycine was chosen because it is the amino acid with the most conformational freedom. This would give position 110 the ability to adopt a tight turn geometry that may be necessary to stabilize the helix-break-helix motif. Figure 8 shows the spectrum of P110G. Although glycine has the most conformational freedom of all twenty standard amino acids, the spectrum of P110G was significantly different from that of wild-type. Therefore, proline at position 110 is critical for the helix-break-helix structure.
Figure 8.
TROSY-HSQC of Cav196-136 P110G.
Discussion
Caveolin is the most important protein in caveolae, and it plays a dual role. It is critical both structurally for facilitating the highly-curved nature of caveolae, and for interacting with a myriad of proteins in various signal transduction pathways. Based on hydropathy analysis, the transmembrane domain of caveolin is predicted to be 33 residues in length. Therefore, the transmembrane domain of caveolin is too long to span the membrane once, but too short to span twice. In addition, there are no loop segments within the transmembrane domain that would facilitate a polytopic topology. Furthermore, in vivo studies have shown that both the N- and C- termini of caveolin face the cytoplasm [12]. These two pieces of data have led to the postulation that the transmembrane domain of caveolin forms an unusual horseshoe or loop conformation in the membrane. Moreover, the intra-membrane horseshoe conformation is thought to facilitate the high-degree of membrane curvature found in caveolae. However, there is no direct experimental data to support the postulation that the transmembrane domain is indeed adopting a horseshoe conformation.
Based on hydropathy analysis, the transmembrane domain of caveolin-1 is predicted to encompass residues 102–134. To allow for a margin of error in transmembrane prediction, additional residues are added to the N- and C- termini so that the final construct contains residues 96–136. Next, the transmembrane domain is reconstituted into LMPG micelles. Circular dichroism spectroscopy reveals that the secondary structure in the transmembrane domain is predominately α-helical (57%).
The HSQC spectrum of the caveolin-1 transmembrane domain (Figure 3) shows relatively sharp resonances and a high degree of chemical shift dispersion which is indicative of a well-structured protein. This shows that the transmembrane domain alone has a stable conformation. Chemical shift indexing (Figure 4) reveals that the transmembrane domain has four distinct regions: There is an α-helix from residues 97–107 (helix 1), a break from residues 108-110, another α-helix from residues 111-129 (helix 2), and an unstructured region from residues 130-136. This data clearly shows that the transmembrane domain of caveolin is displaying a helix-break-helix structure. This result is remarkable because if the transmembrane domain forms a horseshoe conformation, it is expected that a break in the helix would be necessary for the polypeptide to make a tight turn and return to the membrane surface. Therefore, a helix-turn-helix structure would be consistent with the caveolin transmembrane domain forming a wedge shape in the membrane. Importantly this wedge shape could induce the high degree of membrane curvature observed in caveolae by asymmetrically stretching the inner and outer leaflets of the bilayer. This represents, to our knowledge, the first piece of direct experimental structural data on the intact transmembrane domain of caveolin-1. Furthermore, our data strongly supports the premise that the transmembrane domain of caveolin-1 alone is primarily responsible for the horseshoe conformation. Based on the chemical shift index analysis, the transmembrane domain is approximately 65% helical which agrees well with the circular dichroism data which shows 57% α-helicity.
The break between helix 1 and helix 2 encompasses three amino acid residues, G108, I109, and P110. Next, alanine scanning mutagenesis is performed for each of the three residues to deduce the roles of these amino acids in the transmembrane domain structure. Comparison of the mutant G108A spectrum to that of the wild-type spectrum shows that mutation of G108 to alanine did not significantly alter the structure of the caveolin transmembrane domain (Figure 5A). However, the mutation of G108 to leucine or isoleucine dramatically altered the HSQC spectrum when compared to that of the wild-type (Figure 6). This reveals that position 108 is somewhat restricted sterically, and can not accommodate amino acids with large side chains.
Isoleucine resides at position 109. When isoleucine is mutated to alanine, the HSQC spectrum is altered significantly (Figure 5B). This indicates that there is a dramatic change in the structure of the transmembrane domain. Analysis of the I109L mutant shows a spectrum that is mildly altered, but over time (1 week) the structure is lost (Figure 7A & B). This reveals that the substitution of isoleucine for leucine produces a structure that is only meta-stable. However, this meta-stability can not be contributed to changes in side chain bulkiness as leucine and isoleucine are very similar. Mutation of I109 to valine shows a spectrum that is virtually identical to that of the wild-type revealing that valine is able to preserve the structure (Figure 7C). Both isoleucine and valine are β-branched amino acids. β-branched amino acids are more conformationally restricted than the other amino acids except for proline. Therefore at this position a conformationally restricted amino acid is needed to stabilize the structure.
Position 110 contains proline which is the most conformationally restricted of all of the amino acids. It is not surprising that the proline would be required for a helix-break-helix structure as it is often found in places where the polypeptide makes a tight turn. Mutation of P110 to alanine, again, results in a loss of structure showing that it is critical at this position (Figure 5C). Clearly, proline provides the necessary rigidity to support the helix-break-helix structure. Glycine, like proline, is also commonly found in places where the polypeptide makes tight turns. Glycine is able to adopt different conformations easily because of its conformational freedom. Mutation of P110 to glycine showed that it was not a suitable substitute for proline (Figure 8). Therefore, at position 110, proline appears to be absolutely critical. In summary, all three amino acids found in the break, for different reasons, play an important role in stabilizing the helix-break-helix structure.
As discussed earlier, the spectrum of P110A is of poor quality (increased line broadening) when compared to that of the wild-type. However, the circular dichroism spectrum of P110A (Figure 2) is very similar to that of the wild-type, indicating that this mutation does not result in a global alteration of the secondary structure (α-helicity). Furthermore, gel filtration analysis (data not shown) of the P110A mutant when compared to the wild-type reveals that large aggregates are not forming. However, the data does not preclude the formation of low order (dimers or trimers) aggregates. Consequently, the poor quality of the P110A mutant spectrum could be due to significantly lower reorientation rates caused by small scale sample aggregation. Another possibility is that the mutation inhibits the association of the transmembrane domain with the micelles. However this is not probable based on in vivo data which shows that the caveolin-1 P110A mutant traffics to and inserts into the plasma membrane albeit with an altered orientation [30]. We postulate that the mutation of P110A significantly alters the tertiary structure of the transmembrane domain which in turn causes small scale sample aggregation. Therefore, if we make the assumption that the break between helix 1 and helix 2 is a turn region that supports the horseshoe conformation, the mutation of P110 to alanine likely causes a loss of the horseshoe conformation.
Lastly, our results also agree with a topological model of caveolin based solely on sequence analysis which predicted that the turn of the polypeptide chain occurred at residues GIPM (108-111) [14]. However, our results reveal that only GIP (108-110) is involved in the break. Overall our results provide much needed experimental data on the structure of the transmembrane domain of caveolin, and will help to clarify and support existing caveolin models.
Highlights.
Caveolin-1 transmembrane domain displays a helix-break-helix structure.
Transmembrane domain of caveolin-1 is highly helical.
Residues 108-110 (G, I, P) are critical amino acids for the break between helices.
Acknowledgments
We thank to Professor Fang Tian and Dr. Xingsheng Wang (Penn State Hershey College of Medicine) for useful discussion about NMR and data processing. We thank Penn State Hershey College of Medicine for NMR spectrometer use. We thank Sarah Plucinsky and Niall Buckley for laboratory assistance.
Abbreviations and symbols
- LMPG
lyso-myristoylphosphatidylglycerol
- NMR
nuclear magnetic resonance spectroscopy
- CD
circular dichroism spectroscopy
- CSI
chemical shift indexing
- HSQC
heteronuclear single quantum coherence
- TROSY
transverse relaxation optimized spectroscopy
- Cav196-136
caveolin-1(96-136)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stan RV. Structure of caveolae. Biochimica Et Biophysica Acta. 2005;1746:334–348. doi: 10.1016/j.bbamcr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 2.Williams TM, Lisanti MP. The Caveolin genes: from cell biology to medicine. Annals of Medicine. 2004;36:584–595. doi: 10.1080/07853890410018899. [DOI] [PubMed] [Google Scholar]
- 3.Krajewska WM, Maslowska I. Caveolins: structure and function in signal transduction. Cell Mol Biol Lett. 2004;9:195–220. [PubMed] [Google Scholar]
- 4.Spisni E, Tomasi V, Cestaro A, Tosatto SC. Structural insights into the function of human caveolin 1. Biochem Biophys Res Commun. 2005;338:1383–1390. doi: 10.1016/j.bbrc.2005.10.099. [DOI] [PubMed] [Google Scholar]
- 5.Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, Schedl A, Haller H, Kurzchalia TV. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 6.Bonuccelli G, Casimiro MC, Sotgia F, Wang C, Liu M, Katiyar S, Zhou J, Dew E, Capozza F, Daumer KM, Minetti C, Milliman JN, Alpy F, Rio MC, Tomasetto C, Mercier I, Flomenberg N, Frank PG, Pestell RG, Lisanti MP. Caveolin-1 (P132L), a common breast cancer mutation, confers mammary cell invasiveness and defines a novel stem cell/metastasis-associated gene signature. Am J Pathol. 2009;174:1650–1662. doi: 10.2353/ajpath.2009.080648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shatz M, Liscovitch M. Caveolin-1: A tumor-promoting role in human cancer. International Journal of Radiation Biology. 2008;84:177–189. doi: 10.1080/09553000701745293. [DOI] [PubMed] [Google Scholar]
- 8.Gaudreault SB, Dea D, Poirier J. Increased caveolin-1 expression in Alzheimer's disease brain. Neurobiol Aging. 2004;25:753–9. doi: 10.1016/j.neurobiolaging.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Ahn M, Kim H, Matsumoto Y, Shin T. Increased expression of caveolin-1 and-2 in the hearts of Lewis rats with experimental autoimmune myocarditis. Autoimmunity. 2006;39:489–495. doi: 10.1080/08916930600929321. [DOI] [PubMed] [Google Scholar]
- 10.Weiss N, Couchoux H, Legrand C, Berthier C, Allard B, Jacquemond V. Expression of the muscular dystrophy-associated caveolin-3(P104L) mutant in adult mouse skeletal muscle specifically alters the Ca(2+) channel function of the dihydropyridine receptor. Pflugers Arch. 2008;457:361–75. doi: 10.1007/s00424-008-0528-z. [DOI] [PubMed] [Google Scholar]
- 11.Ortegren U, Karlsson M, Blazic N, Blomqvist M, Nystrom FH, Gustavsson J, Fredman P, Stralfors P. Lipids and glycosphingolipids in caveolae and surrounding plasma membrane of primary rat adipocytes. Eur J Biochem. 2004;271:2028–2036. doi: 10.1111/j.1432-1033.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 12.Monier S, Parton RG, Vogel F, Behlke J, Henske A, Kurzchalia TV. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupree P, Parton RG, Raposo G, Kurzchalia TV, Simons K. Caveolae and sorting in the trans-Golgi network of epithelial cells. EMBO J. 1993;12:1597–1605. doi: 10.1002/j.1460-2075.1993.tb05804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parton RG, Hanzal-Bayer M, Hancock JF. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J Cell Sci. 2006;119:787–796. doi: 10.1242/jcs.02853. [DOI] [PubMed] [Google Scholar]
- 15.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 16.Decaffmeyer M, Shulga YV, Dicu AO, Thomas A, Truant R, Topham MK, Brasseur R, Epand RM. Determination of the topology of the hydrophobic segment of mammalian diacylglycerol kinase epsilon in a cell membrane and its relationship to predictions from modeling. J Mol Biol. 2008;383:797–809. doi: 10.1016/j.jmb.2008.08.076. [DOI] [PubMed] [Google Scholar]
- 17.Aoki S, Thomas A, Decaffmeyer M, Brasseur R, Epand RM. The role of proline in the membrane re-entrant helix of caveolin-1. J Biol Chem. 2010;285:33371–33380. doi: 10.1074/jbc.M110.153569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diefenderfer C, Lee J, Mlyanarski S, Guo Y, Glover KJ. Reliable expression and purification of highly insoluble transmembrane domains. Anal Biochem. 2009;384:274–8. doi: 10.1016/j.ab.2008.09.038. [DOI] [PubMed] [Google Scholar]
- 19.Uittenbogaard A, Smart EJ. Palmitoylation of caveolin-1 is required for cholesterol binding, chaperone complex formation, and rapid transport of cholesterol to caveolae. J Biol Chem. 2000;275:25595–25599. doi: 10.1074/jbc.M003401200. [DOI] [PubMed] [Google Scholar]
- 20.Dietzen DJ, Hastings WR, Lublin DM. Caveolin is palmitoylated on multiple cysteine residues. Palmitoylation is not necessary for localization of caveolin to caveolae. J Biol Chem. 1995;270:6838–6842. doi: 10.1074/jbc.270.12.6838. [DOI] [PubMed] [Google Scholar]
- 21.Sotgia F, Razani B, Bonuccelli G, Schubert W, Battista M, Lee H, Capozza F, Schubert AL, Minetti C, Buckley JT, Lisanti MP. Intracellular retention of glycosylphosphatidyl inositol-linked proteins in caveolin-deficient cells. Molecular and Cellular Biology. 2002;22:3905–3926. doi: 10.1128/MCB.22.11.3905-3926.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins RID A-7811-2009. J Biomol NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 23.Truhlar SM, Cervantes CF, Torpey JW, Kjaergaard M, Komives EA. Rapid mass spectrometric analysis of 15N-Leu incorporation fidelity during preparation of specifically labeled NMR samples. Protein Sci. 2008;17:1636–1639. doi: 10.1110/ps.036418.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Goddard TD, Kneller DG. SPARKY. p. 3. [Google Scholar]
- 26.Whitmore L, Wallace B. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data RID C-5651-2008 RID C-3753–2008. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobley A, Whitmore L, Wallace B. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra RID C-5651-2008 RID C-3753-2008. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 28.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44(Spec No):S24–40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 29.Wishart D, Sykes B. The C-13 Chemical-Shift Index - a Simple Method for the Identification of Protein Secondary Structure using C-13 Chemical-Shift Data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 30.Aoki S, Thomas A, Decaffmeyer M, Brasseur R, Epand RM. The role of proline in the membrane re-entrant helix of caveolin-1. J Biol Chem. 2010 doi: 10.1074/jbc.M110.153569. [DOI] [PMC free article] [PubMed] [Google Scholar]