Abstract
The resection of DNA double-strand breaks (DSBs) to generate ssDNA tails is a pivotal event in the cellular response to these breaks. In the two-step model of resection, primarily elucidated in yeast, initial resection by Mre11/CtIP is followed by extensive resection by two distinct pathways involving Exo1 or BLM/WRN/Dna2. However, resection pathways and their exact contributions in humans in vivo are not as clearly worked out as in yeast. Here, we examined the contribution of Exo1 to DNA end resection in humans in vivo in response to ionizing radiation (IR) and its relationship with other resection pathways (Mre11/CtIP or BLM/WRN). We find that Exo1 plays a predominant role in resection in human cells along with an alternate pathway dependent on WRN. While Mre11 and CtIP stimulate resection in human cells, they are not absolutely required for this process and Exo1 can function in resection even in the absence of Mre11/CtIP. Interestingly, the recruitment of Exo1 to DNA breaks appears to be inhibited by the NHEJ protein Ku80, and the higher level of resection that occurs upon siRNA-mediated depletion of Ku80 is dependent on Exo1. In addition, Exo1 may be regulated by 53BP1 and Brca1, and the restoration of resection in BRCA1-deficient cells upon depletion of 53BP1 is dependent on Exo1. Finally, we find that Exo1-mediated resection facilitates a transition from ATM- to ATR-mediated cell cycle checkpoint signaling. Our results identify Exo1 as a key mediator of DNA end resection and DSB repair and damage signaling decisions in human cells.
Keywords: DNA damage response, DNA double-strand break, DNA end resection, homologous recombination, ATR activation, Exo1
1. Introduction
The resection of DNA double-strand breaks (DSBs) to generate 3’-single-stranded tails is a critical step in the DNA damage response (DDR) [1], and is a prerequisite for both ATR activation and homologous recombination (HR). Based primarily upon yeast studies, a “twostep” model of DSB resection has recently been propounded [2–4]. According to this model, Mre11 [5], in cooperation with CtIP (Sae2 in yeast) [6, 7], carries out initial limited resection at the sites of DSBs. The minimally-resected DNA then serves as a template for extensive resection by two alternate “downstream” pathways involving: 1) the 5’-3’ exonuclease Exo1 alone or 2) the helicases BLM or WRN (Sgs1 in yeast) in conjunction with either Exo1 or the endonuclease Dna2 [8–12]. This model has been further refined by recent biochemical studies using purified yeast or human components showing that Mre11/CtIP also stimulate resection by promoting the association of Exo1 or BLM with DNA ends [13–15]. Homologues of the yeast proteins have conserved functions in end resection in mammalian cells [8, 16, 17]. However, resection pathways and their inter-relationships in humans in vivo are not as clearly worked out as in yeast.
We have previously demonstrated that Exo1 is a key player in DDR, being important for HR [18], cell cycle checkpoint activation [19], as well as apoptosis [20]. Here, we examined the contribution of Exo1 to DNA end resection in humans in vivo in response to ionizing radiation (IR), its relationship with other resection pathways, and its influence on DNA repair and damage signaling events regulated by resection.
2. Materials and Methods
2.1. Cell culture and knockdown of proteins
Clones of wild type (1BR3) fibroblasts [19] with shRNA-mediated knockdown of Exo1 were generated by lipofection with Exo1-shRNA or scrambled-shRNA vectors (Santa Cruz Biotechnology) followed by selection and continued maintenance in puromycin. Additional proteins were depleted using siRNAs (Invitrogen) as described [19] and the efficiency of knockdown for every experiment was verified by Western blotting (see Supplement, Table 1 for siRNAs used and Fig. S1 for knockdown efficiencies). Cells transfected with scrambled siRNA served as controls. To rule out any “off-target” effects of these siRNAs, key experiments were repeated after knockdown with a second set of different siRNAs (see Supplement, Table 2). 1BR3 and HSF cells [19] were maintained in α-MEM medium and ATLD cells [21] in RPMI medium supplemented with 10% fetal calf serum and penicillin/streptomycin. HCC1937 cells [22] were maintained in RPMI1640 medium with 20% fetal calf serum, penicillin/streptomycin, and 200 µg/ml G418. All cells were mycoplasma free.
2.2. Irradiation of cells
Cells were irradiated with gamma rays from a cesium source (JL Shepherd and Associates) at the indicated doses. For laser irradiation, cells were micro-irradiated with a pulsed nitrogen laser (Spectra-Physics; 365nm, 10Hz) with output set at 75% of the maximum, as described [19].
2.3. Live-cell imaging combined with laser micro-irradiation
Briefly, cells were transfected with GFP-Exo1 [18], GFP-RPA [23], GFP-ATM [24], or GFP-ATR [25] constructs, laser micro-irradiated, time-lapse imaged, and fluorescence intensities of micro-irradiated areas relative to non-irradiated areas calculated as described previously [18]. The laser micro-irradiation and live-cell imaging set up used in these experiments has been described previously [26–32]. Briefly, live-cell images were taken with a Carl Zeiss Axiovert 200M microscope (63X oil-immersion objective) coupled to a pulsed nitrogen laser (Spectra-Physics; 365nm, 10Hz) and fluorescence intensities determined using Axiovision software v4.5. Mean value of the fluorescence intensities for each time point was calculated from at least 30 independent measurements.
2.4. Western blotting / antibodies
Western blotting was carried out as described previously after irradiation of cells with 8 Gy of gamma rays [19]. The following primary antibodies were used for blotting and immunofluorescence : RPA, [Abcam] pRPA(S4/8), CtIP, BLM, WRN, ATR, 53BP1, [Bethyl Laboratories], Chk1, pChk1(S317), Chk2, pChk2(T68) [Cell Signaling], Brca1 [Calbiochem], ATM, Mre11 [Genetex], Rad51, Cyclin A [Santa Cruz Biotechnology], actin [Sigma], Exo1 [Thermo Fisher], γH2AX, pHistone-H3(S10) [Upstate Biotechnology], and Ku80 [kind gift from Dr. B.P. Chen]. HRP-conjugated secondary antibodies were procured from Biorad and Alexa488/568-conjugated secondary antibodies were procured from Molecular Probes.
2.5. Immunofluorescence staining for Rad51 or RPA foci
Cells were seeded onto chamber slides (Lab-Tek) and irradiated with 6 Gy of gamma rays. For Rad51 foci, cells were co-immunostained with Rad51 and Cyclin A antibodies, 3 hours later, as described (Tomimatsu et al., 2009) The average number of Rad51 foci for Cyclin A- positive (S/G2) nuclei was determined after scoring at least 100 nuclei. For RPA foci, cells were subjected to in situ extraction (Tomimatsu et al., 2009) 3 hours after irradiation and immunostained with RPA antibody. The percentage of RPA positive cells (cell with 10 or more foci) was determined after scoring at least 100 nuclei. Images were captured using a Leica DH5500B fluorescence microscope (40X objective lens) coupled to a Leica DFC340 FX camera using Leica Application Suite v3 acquisition software.
2.6. Metaphase chromosome preparations
Cells were irradiated with 2 Gy of gamma rays and metaphase chromosome spreads scored for asymmetrical exchanges as described [33]. Aberrations were quantified by analyzing at least 100 metaphase spreads.
2.7. G2/M checkpoint assay
The G2/M checkpoint was evaluated by quantifying histone H3 phosphorylation by flow cytometry as described [19].
2.8. Statistical analyses
Statistical significance was determined by a two-tailed t test using GraphPad Prism software (*, P< 0.05; **, P< 0.01; ***, P< 0.001). Error bars represent standard error of the mean for all plots.
3. Results and Discussion
3.1. Exo1 plays a major role in DSB resection in human cells in response to IR
To determine the contribution of Exo1 to DSB resection in human cells compared to Mre11/CtIP or BLM/WRN, we utilized several different assays to quantify DSB resection in vivo. First, we utilized a novel assay to quantify overall DSB resection kinetics in real time. This was carried out by live-cell imaging of recruitment of the 32-kDa subunit of the ssDNA-binding complex, replication protein A (henceforth referred to as RPA) [23] at DSBs induced focally by laser micro-irradiation [18]. The live-cell imaging set up used in these experiments has been extensively used to study the dynamics of a large number of DDR proteins at DSBs [26–32]. To validate this assay, we first established that GFP-RPA recruitment to laser-induced breaks reflects physiologically-relevant DNA end resection: 1) We confirmed that RPA accumulation occurs only in S/G2 cells and not in G1 cells where extensive resection and HR would be actively inhibited [34] (Fig. S1d, S2). 2) CDK activity in S/G2 cells triggers DNA end resection and CDK inhibitors have been previously demonstrated to attenuate resection and RPA foci formation [17, 35]. Therefore, we also confirmed that GFP-RPA recruitment is inhibited by the CDK inhibitor AZD5438 [36], demonstrating that GFP-RPA accumulation kinetics in our livecell assay reflects the generation of ssDNA due to DNA end resection (Fig. S2).
After validating the live-cell end-resection assay, we examined the accumulation of GFP-RPA at laser micro-irradiation sites in wild type 1BR3 cells. Maximum accumulation was observed by approximately 45 minutes (Fig. 1A). Knockdown of Mre11or CtIP (Fig. S1a) resulted in slower recruitment of GFP-RPA, but peak levels were still attained by approximately 120 minutes. These results indicate that while Mre11 and CtIP stimulate DSB resection in human cells, resection can still proceed to completion in their absence, albeit at a slower rate. Ablation of Exo1 resulted in a severe resection defect such that peak resection levels were not reached by even 120 minutes confirming that Exo1 plays a major role in end resection in human cells. Attenuation of the other pathway, by knockdown of either of the two helicases involved (BLM or WRN), also resulted in resection defects. Knockdown of WRN caused a more severe defect compared to BLM, indicating that WRN may play a prominent role in this pathway in humans in vivo, in accord with a recent Xenopus study [12]. Combined knockdown of both WRN and Exo1 resulted in a further decrease in resection, confirming that these two “downstream” players function in alternate pathways for extensive DSB resection in human cells, similar to that seen for their yeast counterparts [9, 11].
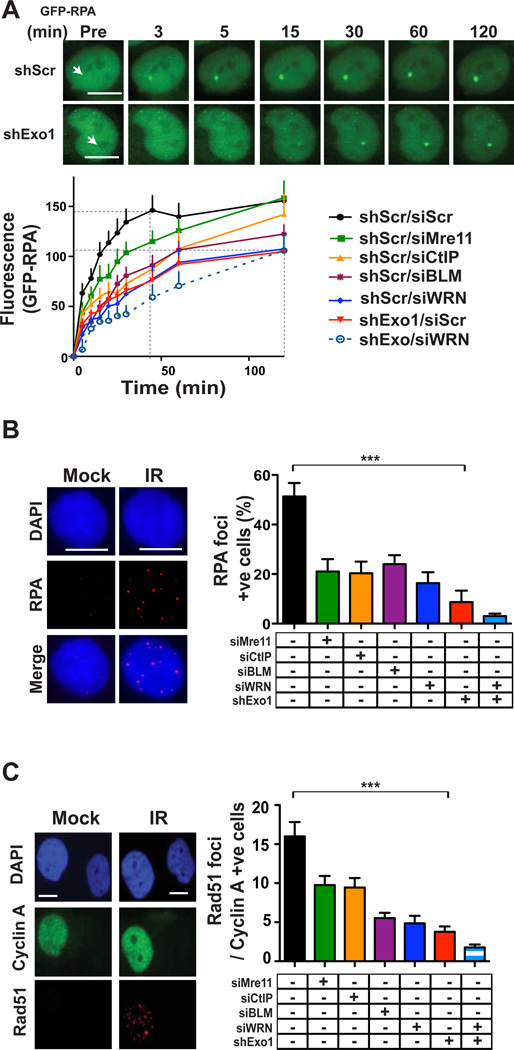
Figure 1. Exo1 plays a major role in DSB resection and HR in human cells.
(A) Time-lapse images of accumulation of GFP-RPA (green) at DSBs induced by laser micro-irradiation (arrows) of wild type 1BR3 cells with or without Exo1 knockdown. Scale bars, 10 µm. Plot shows kinetics of recruitment of GFP-RPA in 1BR3 cells with knockdown of Mre11, CtIP, BLM, WRN or Exo1. Cells transfected with scrambled siRNA (siScr) and/or with a scrambled shRNA vector (shScr) serve as controls. (B) Representative image of gamma-irradiated 1BR3 cells immunnostained with anti-RPA antibody (red) after 3 hours. Nuclei are stained with DAPI (blue). Percentages of RPA-positive cells (10 or more foci) are plotted for 1BR3 cells with depletion of Mre11, CtIP, BLM. WRN, or Exo1. (C) Representative image of gamma-irradiated 1BR3 cells co-immunofluorescence stained with anti-Cyclin A antibody (green) and anti-Rad51 antibody (red) after 3 hours. Average numbers of Rad51 foci for Cyclin A-positive (S/G2) nuclei at 3 hours post-irradiation are plotted for 1BR3 cells with depletion of Mre11, CtIP, BLM, WRN, or Exo1.
We further validated the major conclusions from the GFP-RPA accumulation experiment by directly quantifying the generation of ssDNA by staining for 5-bromo-2-deoxyuridine (BrdU) foci under nondenaturing conditions [37] (Fig. S3). In gamma ray-irradiated 1BR3 cells, the time taken to reach peak numbers of BrdU/ssDNA foci (Fig. S3) was similar to that needed for maximum accumulation of GFP-RPA in live-cell experiments (Fig. 1a), further validating the live-cell end-resection assay. Knockdown of Mre11 or CtIP reduced, but did not eliminate, BrdU/ssDNA foci similar to that reported before for CtIP knockdown [37]. Exo1 knockdown, on the other hand, largely eliminated BrdU/ssDNA foci in these cells (Fig. S3). These results, which are in accord with results from the live-cell assay, confirm that Exo1 plays a predominant role in end resection in human cells.
In order to re-confirm the major role that Exo1 plays in resection using a different assay, we quantified the induction of RPA foci in response to gamma rays. RPA foci were reduced, but not absent, upon Mre11 or CtIP knockdown while foci formation was maximally attenuated upon Exo1 knockdown (Fig. 1B; Fig. S1a) To corroborate results obtained with Mre11 knockdown, we assessed RPA foci formation in two primary Mre11-deficient ATLD cell lines [21] and found that IR-induced RPA foci formation was reduced, but not absent, in these cells compared to primary human skin fibroblasts (HSF), similar to that reported before for a different ATLD line [38] (Fig. S4). Upon quantifying the induction of Rad51 foci in gamma-irradiated S/G2 cells (cyclin A-positive [39]), we found maximum attenuation of Rad51 foci with Exo1 knockdown (Fig. 1C). To reconfirm these results and to rule out any “off-target” effect of the siRNAs used in these experiments (Supplement; Table 1), we quantified RPA and Rad51 foci after knockdown of Mre11, CtIP, BLM, WRN, or Exo1 with a different set of siRNAs (Supplement; Table 2) and found similar effects on the induction RPA and Rad51 foci (Fig. S5). Combined knockdown of both Exo1 and WRN reduced RPA and Rad51 foci numbers further to nearbackground levels, indicating that Exo1 functions in resection in vivo along with an alternate pathway involving WRN (Fig. 1B C: Fig. S1a).
Interestingly, Exo1 knockdown could further reduce RPA and Rad51 foci in Mre11- or CtIP-depleted cells, indicating that Exo1 can also function independently of these two proteins in resection (Fig. S6). Therefore, to examine if Exo1 could be recruited to DSBs independently of Mre11/CtIP, we quantified the accumulation of GFP-tagged Exo1 at DSBs induced focally by laser micro-irradiation of 1BR3 cells [18]. We first confirmed that the recruitment of Exo1 to laser-induced breaks was physiologically relevant by showing that significant Exo1 accumulation occurred only in those phases of the cell cycle (S/G2) where extensive resection would be permissible [34] (Fig. S1d, S2). Exo1 recruitment to DSBs was unaffected by the knockdown of Mre11and CtIP (Fig. S7). Taken together, these results indicate that Exo1 plays a major role in resection in humans and can function independently in resection even without initial limited resection by Mre11/CtIP.
3.2. Ku inhibits Exo1-mediated DSB resection and homologous recombination in human cells
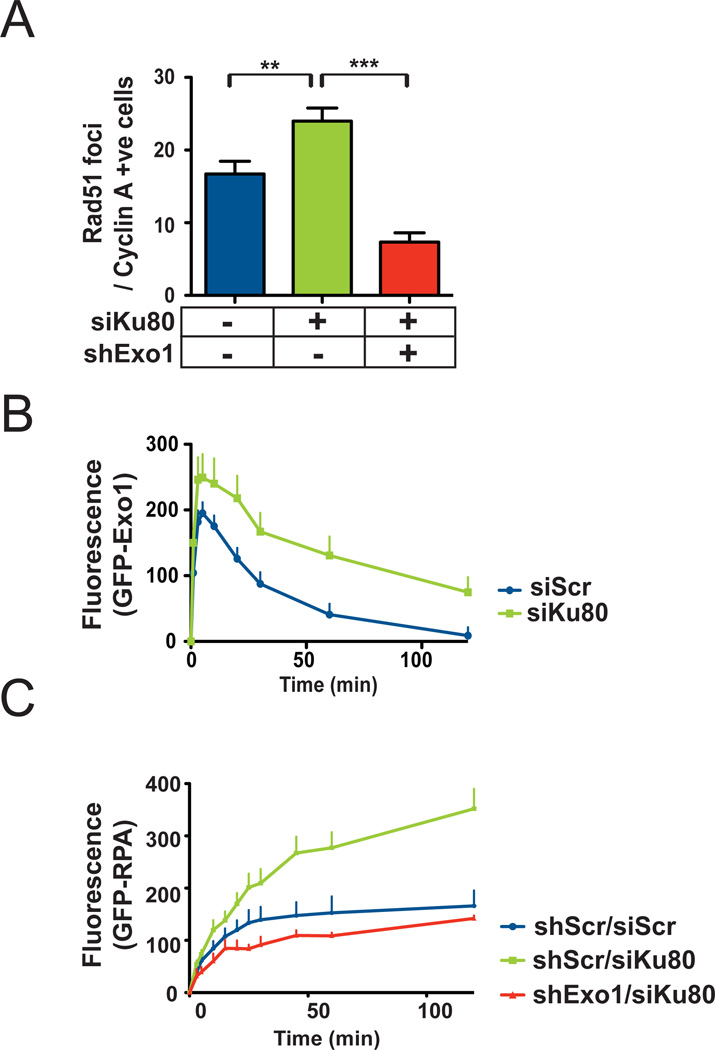
Recent genetic studies in yeast suggest that the NHEJ protein Ku might antagonize long-range resection by Exo1, especially in the absence of Mre11 or Sae2 [40, 41]. We, therefore, wanted to investigate whether Ku might act as a general inhibitor of Exo1-mediated resection in human cells. We quantified Rad51 foci in gamma-irradiated 1BR3 cells with siRNA-mediated knock down of Ku80 (Fig. 2A; S1b). We found that Ku80 knockdown resulted in an increase in Rad51 foci upon irradiation and this increase could be negated by additional knockdown of Exo1 indicating that increased resection upon Ku80 depletion is mediated by Exo1. To rule out any “off-target” effect of the Ku80 siRNA used in this experiment (Supplement; Table 1), we quantified Rad51 foci after knockdown of Ku80 with a different siRNA (Supplement; Table 2) and found a similar increase in Rad51 foci upon Ku80 depletion (Fig. S8). We wanted to examine whether Ku80 loss might promote resection by enhancing Exo1 recruitment to DNA breaks. We quantified the accumulation of GFP-tagged Exo1 at DSBs induced focally by laser micro-irradiation of 1BR3 cells [18] (Fig. S1d, S2) and found that Ku80 knockdown increased the recruitment and retention of GFP-Exo1 at DNA damage sites (Fig. 2B). A corresponding increase in GFP-RPA recruitment was also seen upon Ku80 knockdown and this increase could be negated by additional knockdown of Exo1 (Fig. 2C). Taken together, these results suggest that Ku80 exerts an inhibitory effect on DSB resection by preventing Exo1 from accessing DNA ends.
Figure 2. Ku inhibits Exo1-mediated DSB resection and homologous recombination in human cells.
(A) Average numbers of Rad51 foci for S/G2 nuclei at 3 hours post-irradiation are plotted for wild 1BR3 cells with knockdown of Ku80 or both Ku80 and Exo1. (B) Kinetics of recruitment and dissociation of GFP-Exo1 at DSBs induced by laser micro-irradiation of 1BR3 cells with or without knockdown of Ku80. (C) Kinetics of recruitment of GFP-RPA at DSBs induced by laser micro-irradiation of wild type 1BR3 cells or cells with knockdown of Ku80 or both Ku80 and Exo1.
3.3. Exo1 is a regulated by the interplay between 53BP1 and Brca1
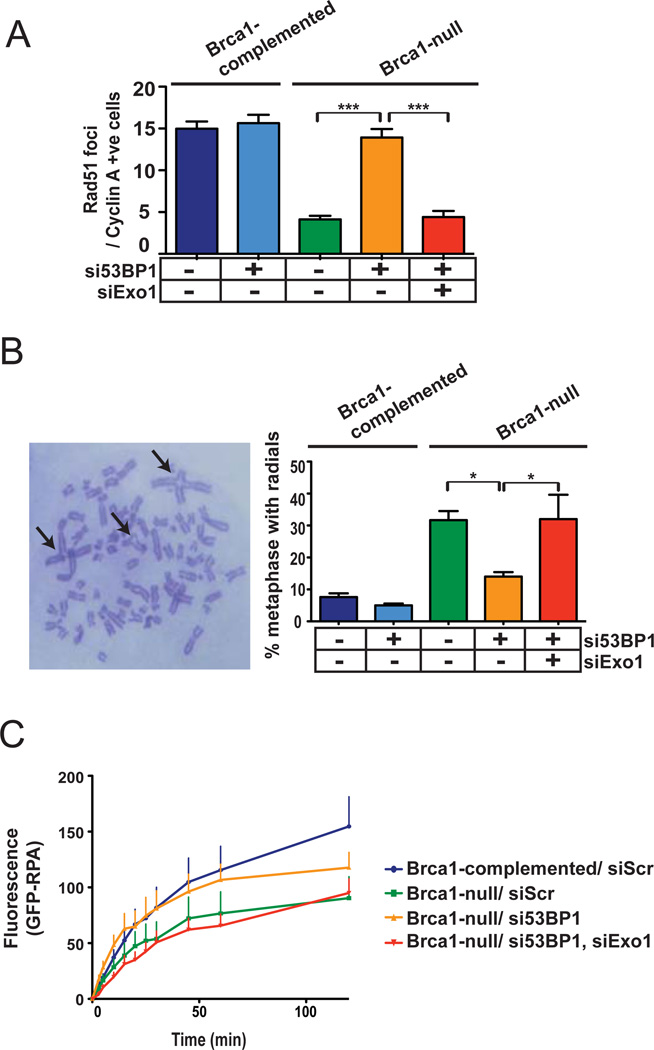
A very important NHEJ to HR switching model was recently formulated involving 53BP1, a “mediator” protein that accumulates at DSBs and Brca1, a HR protein that is mutated in a subset of familial breast and ovarian cancers; reviewed in [34]. According to this model, the accumulation of 53BP1 at DSBs promotes NHEJ while suppressing HR, presumably by inhibiting resection. However, Brca1 can trigger a switch from NHEJ to HR by displacing 53BP1 from DNA ends, thereby facilitating resection [42, 43]. Thus, according to these reports, the HR defect of Brca1-mutant mouse cells can be corrected by additional loss of 53BP1 [42, 43]. In accord with these reports, we found that 53BP1 knockdown in Brca1-deficient HCC1937 breast cancer cells [22] corrected the HR defect of these cells. Specifically, the reduced numbers of IR-induced Rad51 foci (Fig. 3A; S1c) and the increased numbers of radial chromosome structures [43] (Fig. 3B) in Brca1-null cells could be reversed by knockdown of 53BP1. To rule out any “off-target” effect of the 53BP1 siRNA used in this experiment (Supplement; Table 1), we quantified Rad51 foci after knockdown of 53BP1 with a different siRNA (Supplement; Table 2) and found a similar increase in Rad51 foci upon 53BP1 depletion (Fig. S9). These results confirm that the interplay between Brca1 and 53BP1, elucidated in cells from transgenic mice [42, 43], also occurs in human breast cancer cells. Importantly, the rescue of the HR defect could be negated by additional knockdown of Exo1 indicating that 53BP1 inhibits Exo1 in these cells (Fig. 3A, B). Finally, we found that GFP-RPA recruitment, as measured by live-cell imaging, was significantly attenuated in Brca1-null cells compared to Brca1-complemented cells (Fig. 3C). Strikingly, 53BP1 knockdown partially restored RPA recruitment in Brca1-null cells and this rescue was negated by additional knockdown of Exo1. Taken together, these results indicate that Exo1-mediated DNA resection is a pivotal event in repair pathway choice that is controlled by the interplay between Brca1 and 53BP1.
Figure 3. Exo1 is regulated by the interplay between 53BP1 and Brca1.
(A) Average numbers of Rad51 foci for S/G2 nuclei at 3 hours post-irradiation are plotted for Brca1-deficient HCC1937 cells or Brca1-complemented cells with siRNA-mediated knockdown of 53BP1 or both 53BP1 and Exo1. (B) Percentages of metaphases with radial chromosomes (arrows in representative metaphase) are plotted for gamma-irradiated HCC1937 cells with knockdown of 53BP1 or both 53BP1 and Exo1. (C) Kinetics of recruitment of GFP-RPA at DSBs induced by laser micro-irradiation of Brca1-complemented or Brca1-null HCC1937 cells with knockdown of 53BP1 or both 53BP1 and Exo1.
3.4. DSB resection by Exo1 facilitates a transition from ATM- to ATR-mediated DNA damage signaling
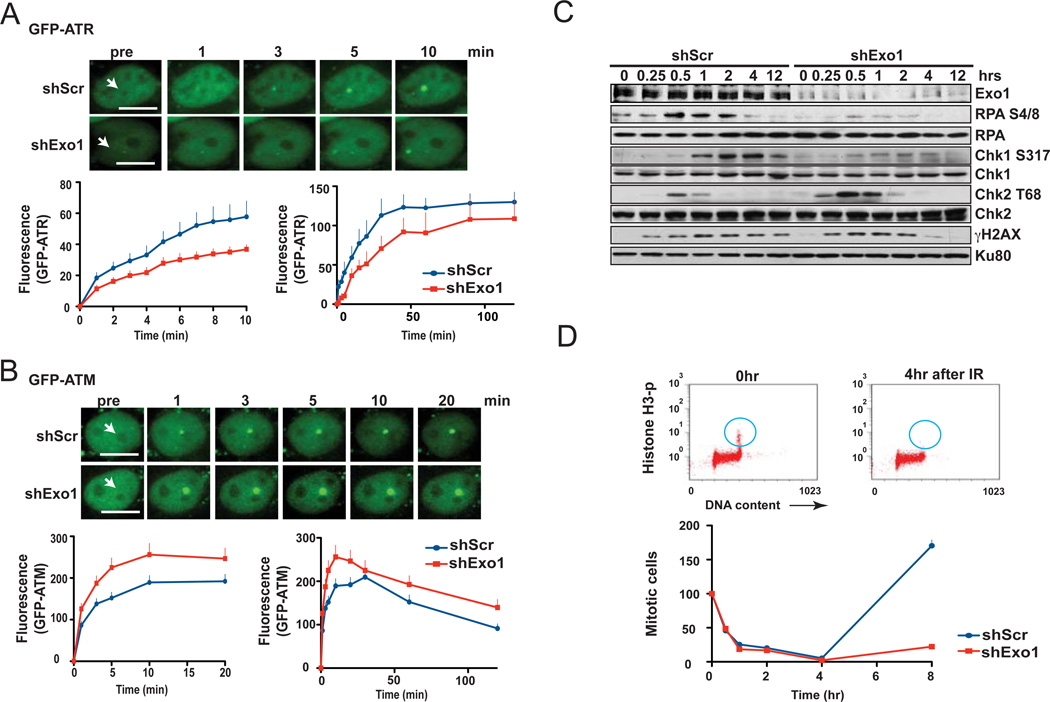
In addition to a switch from NHEJ to HR, DSB resection is expected to facilitate a transition from ATM- to ATR-mediated signaling, as the generation of 3’-ssDNA is crucial for ATR activation and is reported to disfavor ATM activation in vitro [44]. To investigate a possible role for Exo1 in such a transition in vivo, we examined the kinetics of GFP-ATM and GFP-ATR recruitment by live-cell imaging of 1BR3 cells. Interestingly, Exo1 knockdown resulted in delayed recruitment of ATR (Fig. 4A; S1d) and augmented recruitment of ATM (Fig. 4B; S1d) to the sites of laser-induced DNA damage. In a complementary experiment, we found that ectopic expression of Exo1 in 1BR3 cells significantly augmented the recruitment of GFPATR and attenuated the recruitment of GFP-ATM to the sites of laser-induced DSBs (Fig. S10). As Chk1 and Chk2 are signature substrates for ATR and ATM, respectively [1], we validated the live-cell imaging results by examining the kinetics of Chk1 and Chk2 phosphorylation upon gamma irradiation by Western blotting (Fig. 4C). Exo1-deficient human cells showed markedly impaired DSB resection as evident from the reduced phosphorylation of RPA at serines 4/8, a surrogate marker for DSB resection [7]. Chk1 phosphorylation was attenuated upon Exo1 knockdown, clearly indicating that, in the absence of Exo1, residual resection is insufficient to stimulate robust ATR signaling. In contrast, Chk2 phosphorylation was increased upon Exo1 knockdown, indicating that attenuated resection might augment ATM activation. Using histone H3 phosphorylation to distinguish between mitotic and G2 cells [19], we found that Exo1- depleted cells displayed a more prolonged gamma ray-induced G2/M block, presumably due to the enhanced Chk2 phosphorylation in these cells and in keeping with the role of Chk2 in maintaining the G2/M block at 8 hours post-irradiation [45] (Fig. 4D). It is clear from these results that end resection by Exo1 augments DNA damage-signaling by ATR and concurrently curtails signaling by ATM. We previously postulated that phosphorylation of Exo1 by ATM might restrain the resection activity of Exo1 [18]. Given the inhibitory effect of resection on ATM activation, it is logical to assume that ATM might inhibit Exo1 to ensure that its own activation and retention at breaks is not prematurely terminated due to DSB to ssDNA conversion.
Figure 4. DSB resection by Exo1 facilitates a transition from ATM- to ATR-mediated DNA damage signaling.
Plots and representative images show recruitment of (A) GFP-ATR or (B) GFP-ATM to the sites of DSBs induced by laser micro-irradiation (arrows) of wild type 1BR3 cells with or without Exo1 knockdown. Scale bars, 10 µm. (C) 1BR3 cells with or without Exo1 knockdown were irradiated with gamma rays and phosphorylation of DDR proteins was assayed at the indicated times by Western blotting with phospho-specific antibodies. (D) Early G2/M block in irradiated 1BR3 cells, with or without Exo1 knockdown, was assayed by dual-parameter flow cytometry (representative distributions are shown). Percent cells in M-phase (normalized to mock-irradiated controls) are plotted against post-irradiation time points - the G2/M checkpoint manifests as a decrease in mitotic cells (blue circles) at 4 hr post-irradiation.
3.5. Concluding remarks
Exo1 is a member of the Rad2 family of nucleases and possesses 5’ to 3’ dsDNA exonuclease and 5’–flap endonuclease activities and functions in a number of important cellular pathways including DNA repair, recombination, replication, and telomere integrity [46]. It is clear from this study that Exo1 plays a predominant role in resection in human cells in response to IR along with an alternate pathway dependent on WRN. We also show that resection is attenuated (but not abrogated) in the absence of Mre11 or CtIP and that Exo1 can carry out resection in the absence of Mre11/CtIP in human cells.
In the “two-step” model of DSB resection, largely derived from yeast studies, the upstream and downstream components act sequentially, with upstream components and initial (limited) resection facilitating the second step (extensive resection) [2–4]. This model was further refined by in vitro observations showing that the upstream components Mre11/Sae2 facilitate downstream processes undertaken by Exo1 or Dna2/Sgs1 though this stimulation was accomplished without Mre11 nuclease activity [13–15]. Therefore Mre11/Sae2 need not generate ssDNA in order to stimulate resection but, rather, may do so in other way(s) that are not fully understood such as stabilization of resection proteins at DNA ends. In Mre11 or Sae2 mutant yeast, resection is slower but not absent and depends on the presence of Exo1 and Sgs1 [8–11]. In the absence of either of the two downstream components in yeast, there is little effect on resection in vivo, but when both are absent then resection is limited to a short region close to break site; this limited resection is Mre11/Sae2 dependent. However, there is some complementarity between these upstream and downstream pathways since yeast cells deficient in Mre11 or Sae2 can still resect DSBs while cells deficient in all three pathways show no resection at all [8–11].
In this manuscript we present data showing that Exo1 can resect IR-induced DSBs without Mre11/CtIP and may function in this process along with an alternate pathway represented by WRN. We show that, in human cells, resection is severely affected by the depletion of Exo1 or WRN but not as much by the depletion of Mre11 or CtIP. However, codepletion of Exo1 and WRN results in a more severe resection defect compared to ablation of either protein alone. Interestingly, co-depletion of both WRN and BLM also results in a more severe resection defect compared to single knockdowns (data not shown), indicating that both BLM and WRN may function as helicases in the second resection pathway in human cells. Consistent with our observations of a more limited role of Mre11 or CtIP in IR-induced resection, conditional deletion of Mre11 in mouse cells attenuates (but does not abrogate) IRinduced RPA and Rad51 foci [16], IR-induced RPA foci are reduced (but not absent) in Mre11-deficient ATLD cells [38], and knockdown of CtIP in human cells reduces (but does not abolish) IR-induced RPA [17] or BrdU/ssDNA [37] foci. Interestingly, it was recently reported that CtIP interacts with Exo1 and actually restrains its exonucleolytic activity [47]. Therefore, it is possible that unbridled Exo1 activity in CtIP knockdown cells might actually compensate for the loss of a CtIP-dependent early step.
It is possible that Mre11/CtIP and the Exo1 pathway might turn out to be complementary with each other and suited for different classes of DSB lesions in human cells which includes clean ends, difficult to repair complex base damage, and DNA-protein cross-links. Endonucleolytic cleavage of 5’ termini by Mre11/CtIP may be essential for resection of DNA ends that are blocked by a covalently-attached protein [48, 49]. Thus, Mre11 and CtIP may be mandatory for resection of DSBs generated by camptothecin (which generates DNA ends linked to topoisomerase I), as reported before [7, 8]. On the other hand, the dependency on Mre11/CtIP may be reduced for IR-induced breaks as these would not necessitate removal of an end-blocking protein. This is borne out by our results and by the reduction (but not absence) of RPA or BrdU/ssDNA foci in irradiated mouse and human cells deficient in Mre11 or CtIP [16, 17, 37, 38]
Our results indicate that the recruitment of Exo1 to DNA breaks might be inhibited by the NHEJ protein Ku80 and the higher level of resection that occurs upon siRNA-mediated depletion of Ku80 is largely dependent on Exo1. Moreover, restoration of resection in BRCA1-deficient cells upon depletion of 53BP1 is also dependent on Exo1. Taken together, our data indicate that resection by Exo1 is modulated by the Ku heterodimer, and by 53BP1 and Brca1 which are components of a recently elucidated NHEJ to HR switching mechanism [34]. Finally, we demonstrate that lower level of resection in the absence of Exo1 correlates with lower level of ATR activation and higher level of ATM activation, consistent with the idea that resection promotes ATR activity and blocks ATM activity [44]. In sum, our results indicate that resection by Exo1 influences key process occurring in human cells in response to breaks – DNA repair by HR versus NHEJ and cell cycle checkpoint signaling by ATR versus ATM. It would be important, in the future, to understand whether CDK-dependent phosphorylation events regulate the function of Exo1 in resection as they do for Sae2 [50] and Dna2 [51].
Supplementary Material
Acknowledgements
We thank Prof. Woodring Wright for critical comments on the manuscript. We are grateful to Dr. Cristina Cardoso (Max Delbruck Center for Molecular Medicine, Germany) for the GFP-RPA construct and to Dr. Randal Tibbetts (University of Wisconsin-Madison) for the GFP-ATR construct. We thank Cristel Camacho for generating the DsRed-Exo1 construct. We are grateful to Prof. David Chen for providing access to the laser micro-irradiation facility of UTSW. KK is supported by a Program grant from National Health and Medical Research Council, Australia. SB is supported by grants from the National Institutes of Health (RO1 CA149461), National Aeronautics and Space Administration (NNX10AE08G) and the Cancer Prevention and Research Institute of Texas (RP100644). KD is a recipient of the Barry M. Goldwater Scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair (Amst) 2010;9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, Groocock LM, Cahill D, Hitomi C, Guenther G, Moiani D, Carney JP, Russell P, Tainer JA. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a Cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao S, Toczylowski T, Yan H. Mechanistic analysis of Xenopus EXO1's function in 5'- strand resection at DNA double-strand breaks. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G, Sung P. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huertas P, Jackson SP. Human CtIP mediates Cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolderson E, Tomimatsu N, Richard DJ, Boucher D, Kumar R, Pandita TK, Burma S, Khanna KK. Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient Cells. EMBO Rep. 2009 doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolderson E, Richard DJ, Edelmann W, Khanna KK. Involvement of Exo1b in DNA damage-induced apoptosis. Nucleic Acids Res. 2009;37:3452–3463. doi: 10.1093/nar/gkp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delia D, Piane M, Buscemi G, Savio C, Palmeri S, Lulli P, Carlessi L, Fontanella E, Chessa L. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum Mol Geneta. 2004;13:2155–2163. doi: 10.1093/hmg/ddh221. [DOI] [PubMed] [Google Scholar]
- 22.Tomlinson GE, Chen TT, Stastny VA, Virmani AK, Spillman MA, Tonk V, Blum JL, Schneider NR, Wistuba, Shay JW, Minna JD, Gazdar AF. Characterization of a breast cancer Cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 23.Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol Cell. 2002;10:1355–1365. doi: 10.1016/s1097-2765(02)00729-3. [DOI] [PubMed] [Google Scholar]
- 24.Young DB, Jonnalagadda J, Gatei M, Jans DA, Meyn S, Khanna KK. Identification of domains of ataxia-telangiectasia mutated required for nuclear localization and chromatin association. J Biol Chem. 2005;280:27587–27594. doi: 10.1074/jbc.M411689200. [DOI] [PubMed] [Google Scholar]
- 25.Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, Abraham RT. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y. ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal. 2010;3:rs3. doi: 10.1126/scisignal.2001034. [DOI] [PubMed] [Google Scholar]
- 27.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang SY, Eppink B, Chung YM, Shalev G, Shema E, Shkedy D, Smorodinsky NI, van Vliet N, Kuster B, Mann M, Ciechanover A, Dahm-Daphi J, Kanaar R, Hu MC, Chen DJ, Oren M, Shiloh Y. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richard DJ, Savage K, Bolderson E, Cubeddu L, So S, Ghita M, Chen DJ, White MF, Richard K, Prise KM, Schettino G, Khanna KK. hSSB1 rapidly binds at the sites of DNA double-strand breaks and is required for the efficient recruitment of the MRN complex. Nucleic Acids Res. 2011;39:1692–1702. doi: 10.1093/nar/gkq1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.So S, Davis AJ, Chen DJ. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J Cell Biol. 2009;187:977–990. doi: 10.1083/jcb.200906064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weterings E, Verkaik NS, Keijzers G, Florea BI, Wang SY, Ortega LG, Uematsu N, Chen DJ, van Gent DC. The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol Cell Biol. 2009;29:1134–1142. doi: 10.1128/MCB.00971-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MCellin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, Burma S. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010;70:5457–5464. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowndes NF. The interplay between BRCA1 and 53BP1 influences death, aging, senescence and cancer. DNA Repair (Amst) 2010;9:1112–1116. doi: 10.1016/j.dnarep.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 35.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and Cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 36.Byth KF, Thomas A, Hughes G, Forder C, McGregor A, Geh C, Oakes S, Green C, Walker M, Newcombe N, Green S, Growcott J, Barker A, Wilkinson RW. AZD5438, a potent oral inhibitor of cyclin-dependent kinases 1, 2, and 9, leads to pharmacodynamic changes and potent antitumor effects in human tumor xenografts. Mol Cancer Ther. 2009;8:1856–1866. doi: 10.1158/1535-7163.MCT-08-0836. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foster SS, Balestrini A, Petrini JH. Functional interplay of the Mre11 nuclease and Ku in the response to replication-associated DNA damage. Mol Cell Biol. 2011 doi: 10.1128/MCB.05854-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010 doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient Cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata A, Barton O, Noon AT, Dahm K, Deckbar D, Goodarzi AA, Lobrich M, Jeggo PA. Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G(2)/M checkpoint arrest. Mol Cell Biol. 2010;30:3371–3383. doi: 10.1128/MCB.01644-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran PT, Erdeniz N, Symington LS, Liskay RM. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair (Amst) 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Eid W, Steger M, El-Shemerly M, Ferretti LP, Pena-Diaz J, Konig C, Valtorta E, Sartori AA, Ferrari S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010;11:962–968. doi: 10.1038/embor.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nat Struct Mol Biol. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.