Abstract
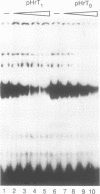
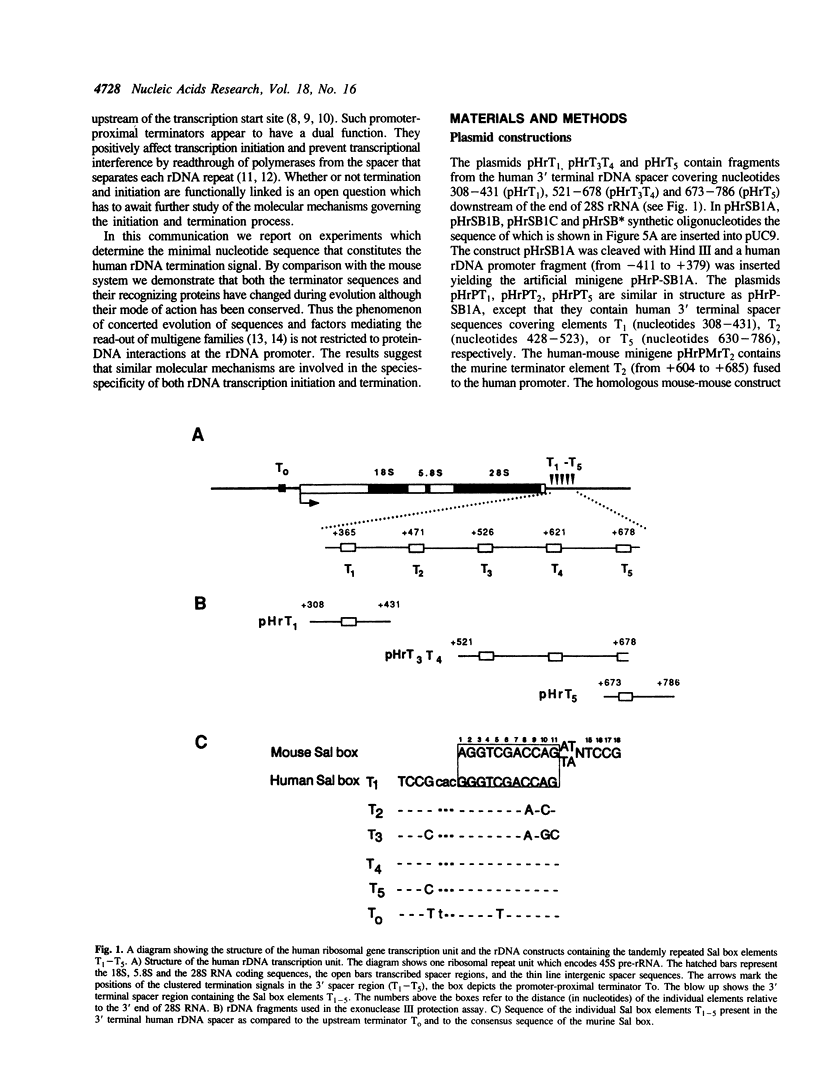
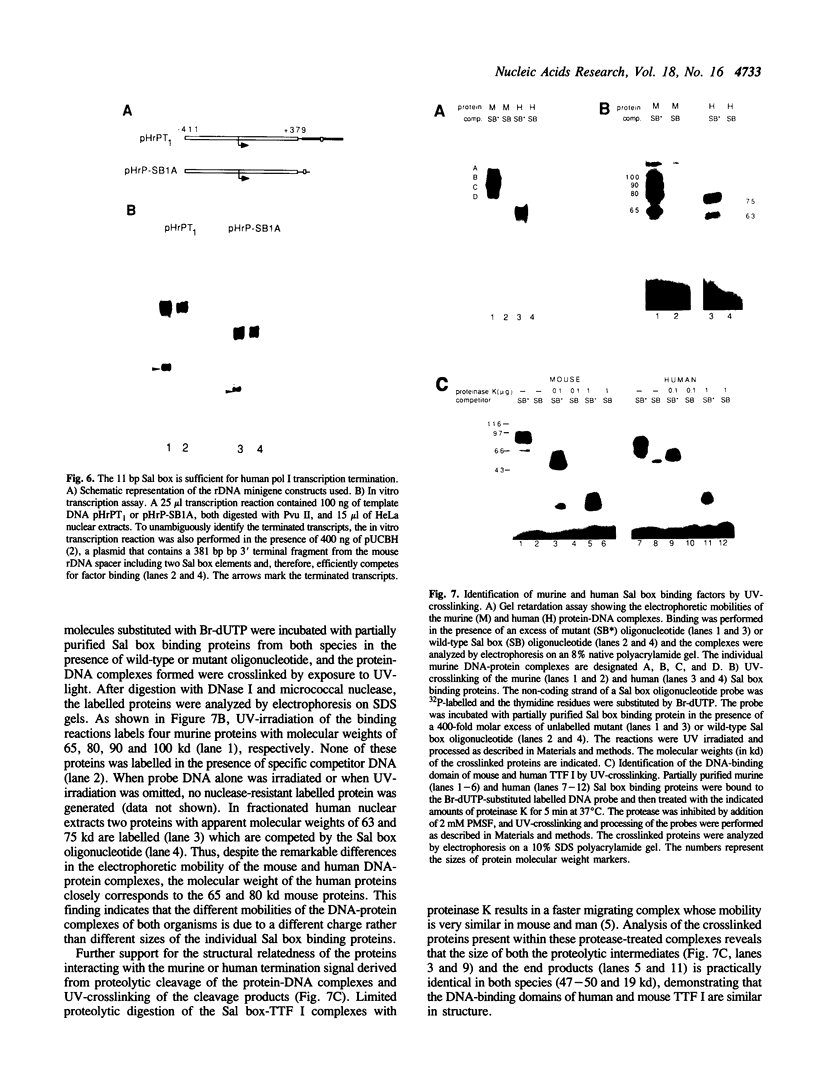
Previously we have shown that a repetitive 18 bp sequence motif, the Sal box (AGGTCGACCAGA/TT/ANTCCG), present in the 3' terminal spacer of mouse rDNA constitutes a termination signal for RNA polymerase I (pol I). Similar sequence elements which are functionally analogous to the murine terminator are present in the spacer of human rDNA. However, the human termination signal is shorter encompassing only 11 bp (GGGTCGACCAG) which correspond to the proximal part of the mouse sequence. Two out of the five human Sal box elements are functionally inactive due to natural point mutations which damage factor binding. A similar sequence motif with a 10 of 11 base identity with the downstream terminators is located upstream of the human transcription initiation site. The upstream element interacts with the same factor(s) as the downstream terminators and is also capable to stop elongating human RNA polymerase I. Despite the human and mouse factors exert different electrophoretic mobilities in gel retardation assays, UV-crosslinking and proteolytic clipping experiments indicate that both the sizes and the tertiary structure of the Sal box binding proteins of both species are very similar. When bound to DNA, both the human and the mouse factor terminate transcription of pol I from the heterologous species. The results implicate that changes in signal sequences necessary for termination have been accompanied by compensatory changes in the DNA binding domain of the protein(s) interacting with the termination signal. In contrast, the protein-protein interactions between the termination factor and the transcribing RNA polymerase I appear to have been conserved during evolution.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartsch I., Schoneberg C., Grummt I. Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribsomal genes. Mol Cell Biol. 1987 Jul;7(7):2521–2529. doi: 10.1128/mcb.7.7.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch I., Schoneberg C., Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol. 1988 Sep;8(9):3891–3897. doi: 10.1128/mcb.8.9.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Promoter occlusion during ribosomal RNA transcription. Cell. 1988 Sep 23;54(7):985–992. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- Clos J., Buttgereit D., Grummt I. A purified transcription factor (TIF-IB) binds to essential sequences of the mouse rDNA promoter. Proc Natl Acad Sci U S A. 1986 Feb;83(3):604–608. doi: 10.1073/pnas.83.3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. A., Flavell R. B. Molecular coevolution: DNA divergence and the maintenance of function. Cell. 1984 Oct;38(3):622–623. doi: 10.1016/0092-8674(84)90255-1. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982 Sep 9;299(5879):111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Financsek I., Mizumoto K., Mishima Y., Muramatsu M. Human ribosomal RNA gene: nucleotide sequence of the transcription initiation region and comparison of three mammalian genes. Proc Natl Acad Sci U S A. 1982 May;79(10):3092–3096. doi: 10.1073/pnas.79.10.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I., Kuhn A., Bartsch I., Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986 Dec 26;47(6):901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Rosenbauer H., Niedermeyer I., Maier U., Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986 Jun 20;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Grummt I., Roth E., Paule M. R. Ribosomal RNA transcription in vitro is species specific. Nature. 1982 Mar 11;296(5853):173–174. doi: 10.1038/296173a0. [DOI] [PubMed] [Google Scholar]
- Grummt I. Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc Natl Acad Sci U S A. 1981 Feb;78(2):727–731. doi: 10.1073/pnas.78.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. L., Ryan K., Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev. 1989 Feb;3(2):212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Kermekchiev M. B., Grummt I. Natural point mutations within rat rDNA transcription terminator elements reveal the functional importance of single bases for factor binding and termination. Nucleic Acids Res. 1987 May 26;15(10):4131–4143. doi: 10.1093/nar/15.10.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Bartsch I., Grummt I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990 Apr 5;344(6266):559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989 Feb;3(2):224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Normann A., Bartsch I., Grummt I. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 1988 May;7(5):1497–1502. doi: 10.1002/j.1460-2075.1988.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Volpe A., Simeone A., D'Esposito M., Scotto L., Fidanza V., de Falco A., Boncinelli E. Molecular analysis of the heterogeneity region of the human ribosomal spacer. J Mol Biol. 1985 May 25;183(2):213–223. doi: 10.1016/0022-2836(85)90214-1. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Proudfoot N. J., Platt T. RNA 3'-end formation. Genes Dev. 1989 Dec;3(12B):2218–2222. doi: 10.1101/gad.3.12b.2218. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Parker K. A., Bond U. Analysis of pre-rRNAs in heat-shocked HeLa cells allows identification of the upstream termination site of human polymerase I transcription. Mol Cell Biol. 1989 Jun;9(6):2500–2512. doi: 10.1128/mcb.9.6.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]