Abstract
Cystathionine beta-synthase (CBS), a heme-containing PLP-dependent enzyme, catalyzes the condensation of serine and homocysteine to yield cystathionine. Missense mutations in CBS, the most common cause of homocystinuria, often result in misfolded proteins. Arginine 266, where the pathogenic missense mutation R266K was identified, appears to be involved in the communication between heme and the PLP-containing catalytic center. Here, we assessed the effect of a short affinity tag (6xHis) compared to a bulky fusion partner (glutathione S-transferase - GST) on CBS wild type (WT) and R266K mutant enzyme properties. While WT CBS was successfully expressed either in conjunction with a GST or with a 6xHis tag, the mutant R266K CBS had no activity, did not form native tetramers and did not respond to chemical chaperone treatment when expressed with a GST fusion partner. Interestingly, expression of R266K CBS constructs with a 6xHis tag at either end yielded active enzymes. The purified, predominantly tetrameric, R266K CBS with a C-terminal 6xHis tag had ~ 82% of the activity of a corresponding WT CBS construct. Results from thermal pre-treatment of the enzyme and the denaturation profile of R266K suggests a lower thermal stability of the mutant enzyme compared to WT, presumably due to a disturbed heme environment.
Keywords: Cystathionine beta-synthase, heme, protein expression, affinity chromatography, protein folding, missense mutation
Introduction
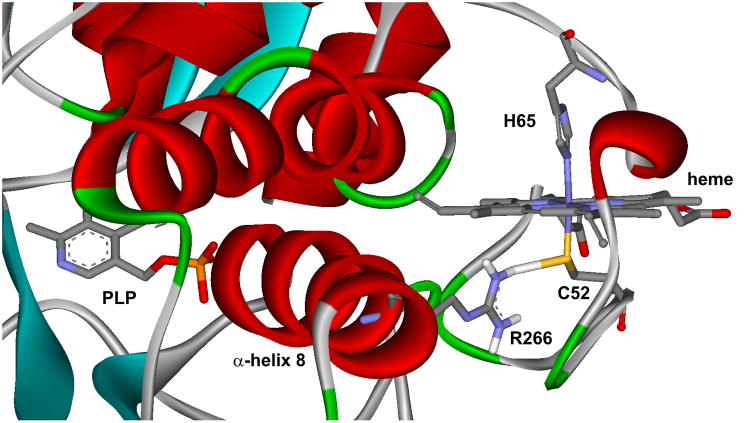
Cystathionine β-synthase (CBS) [EC# 4.2.1.22] lies at the critical branchpoint of sulfur amino acid metabolism, where homocysteine, a toxic metabolite of the methionine cycle, is diverted to the transsulfuration pathway. In this pathway, CBS condenses homocysteine with serine to yield cystathionine, which is then converted to cysteine and ultimately to glutathione. Human CBS has a modular structure and a complex regulatory mechanism (reviewed in [1, 2]). The enzyme contains four identical 63 kDa subunits each consisting of an N-terminal heme-binding region, a central pyridoxal-5′-phosphate (PLP)-containing catalytic core and a C-terminal S-adenosyl-L-methionine (AdoMet)-binding regulatory region with a CBS domain tandem. The interaction of AdoMet with these CBS domains results in up to an 8-fold increase in enzyme activity. A similar activation of CBS can be achieved either by thermal pre-treatment of the enzyme close to its melting temperature, by the introduction of certain point mutations into CBS domains or by a complete removal of the C-terminal region [3, 4]. Removal of the C-terminal regulatory domain is accompanied by a loss of response to AdoMet along with a change in oligomeric status of the full-length homotetramer into a more active homodimer, whose crystal structure was successfully solved [5, 6]. The crystal structure revealed that the heme cofactor is relatively surface exposed and axially coordinated by residues C52 and H65 (Fig. 1). Redox properties of the heme lead to a proposal for its regulatory role providing a potential feedback between the glutathione pool and the transsulfuration pathway [7]. However, the complex pH- and temperature-dependent redox behavior of CBS heme renders this regulatory mechanism unlikely [8–10].
Figure 1. Relationship between heme and active site PLP in human CBS.
Heme cofactor is axially coordinated by residues C52 and H65. Thiolate of C52 is stabilized by hydrogen bonding with R266 residing on the α-helix 8. Disturbances in heme’s coordination, due to e.g. the presence of a pathogenic mutation such as R266K, may be projected by the other end of α-helix 8 into the PLP-containing active site.
CBS-deficient homocystinuria (i.e. classical homocystinuria) [OMIM# 263200] is an inherited autosomal recessive metabolic disorder characterized by elevated plasma homocysteine and methionine levels and by symptoms such as dislocated optic lenses, vascular manifestations, skeletal deformities and mental retardation (reviewed in [11–13]). A deficiency in CBS activity is primarily caused by the presence of missense mutations in the CBS polypeptide. As most of them do not target key residues involved in catalysis, it was proposed that missense mutations induce CBS misfolding [14]. Recently, it was shown that the presence of chemical chaperones or induction of molecular chaperones partially or fully restored the activity and native conformation of several CBS mutants [15–17]. These observations further support the notion that, at least in some cases, enzyme misfolding is the pathogenic mechanism in CBS deficiency. One of the affected residues is R266, where a couple of pathogenic missense mutations, R266K and R266G, were identified [18, 19]. The R266K mutation is highly prevalent among Norwegian patients, who are responsive to therapeutic doses of the PLP precursor, vitamin B6. The R266G mutation was identified in a Japanese patient, who did not respond to vitamin B6 supplementation and suffered from a severe homocystinuric phenotype. The R266 residue seems to stabilize heme’s C52 axial ligand by hydrogen bonding (Fig. 1). Moreover, R266 resides on α-helix 8, whose other end extends to the PLP-containing active site. Therefore, it has been postulated that the R266 residue is critical for communication between heme and the catalytic center containing PLP [20]. Indeed, a spectroscopic study involving three R266 mutants (naturally occurring R266K and R266G and an artificial R266M mutation) showed that the electrostatic interaction between the C52 axial ligand and R266 is important for stabilizing the ferrous heme and its disruption by missense mutation leads to a facile formation of an inactive CBS species missing the C52 heme ligand [20].
As pathogenic missense mutations introduce perturbations in enzyme folding and structure, often accompanied by a decrease in enzyme activity, many CBS mutants represent a difficult target for enzyme purification and characterization. Thus, most of the data supporting the positive effect of chemical chaperones on mutant CBS folding, assembly and catalytic activity were obtained by studying the overexpressed CBS in crude extracts [15, 17, 21]. We showed that eight CBS mutants, following expression in the presence of a chemical chaperone in the growth medium, were amenable to purification to homogeneity and thus allowed for detailed characterization [16]. We used our established system for protein expression and purification using glutathione-S-transferase (GST)-tagged CBS constructs and a two-column purification procedure including the capturing Glutathione Sepharose affinity and the polishing DEAE Sepharose anion exchange chromatography steps [16, 22, 23]. The direct purification of CBS from mammalian tissues is complicated by its tendency to aggregate and its susceptibility to proteolysis [24]. As GST has been reported to enhance the production and in some cases the solubility of its fusion partners [25, 26], the selection of a GST-based system for CBS expression represented an ideal choice. The GST-CBS expression system has many advantages including high purity after a single affinity step and a final protein sequence with only a single extra glycine residue at the N-terminus following the removal of the GST partner. The shared disadvantage of the GST-based constructs is the requirement for protease cleavage of the fusion partner and its subsequent removal by an additional chromatography step. Moreover, some proteins or their mutant forms may form insoluble aggregates after they are cleaved from the GST partner. In addition, GST forms a homodimer where each subunit contains four solvent exposed cysteines and may also interfere with the folding of its fused partner [26, 27]. These facts may impact the folding and assembly of CBS polypeptides, particularly the CBS mutants, and may result in a final conformation induced by GST and not solely by the studied mutation. Recently, a wild type CBS containing a short affinity (6xHis) tag has been successfully purified demonstrating the cost-effectiveness and efficiency of various CBS constructs with an N-terminal 6xHis tag for purification and subsequent kinetic studies of wild type and possibly mutants of CBS [28].
In this study, we have observed that the GST partner interfered with folding of the R266K CBS mutant polypeptide. To study this problem in more detail, we prepared three different CBS constructs carrying a 6xHis tag (Fig. 2). The 6xHis tag is significantly smaller than GST, thus the effect of the affinity tag on wild type or mutant CBS polypeptide folding and assembly or biochemical activity is minimal [26, 28]. We compared the 6xHis-tagged CBS constructs of wild type and the R266K mutant to our established GST-CBS construct in terms of protein expression, CBS native conformation and activity. We also tested whether chemical chaperones have any effect on CBS properties. Subsequently, we purified the R266K CBS to homogeneity and characterized it.
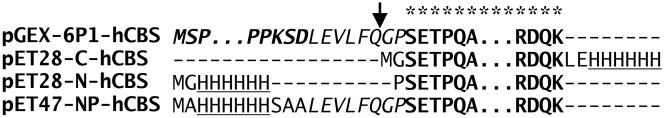
Figure 2. Protein sequence alignment of the CBS constructs.
Bold sequence marked by asterisks designates CBS sequence that is conserved among the constructs. The pGEX-6P1-hCBS yields fusion protein with the GST (semibold italic) and the HRV3C protease recognition site (italic) at the N-terminus of CBS. The pET28-C-hCBS and pET28-N-hCBS constructs carry permanent 6xHis tag (underlined) at the C-terminus and the N-terminus of CBS, respectively. The pET47-NP-hCBS construct yields CBS enzyme with 6xHis tag and HRV3C protease recognition site at the N-terminus of CBS. The GST partner or 6xHis tag can be removed after cleavage with HRV3C protease (designated by an arrow) yielding native CBS sequence with only one extra amino acid (glycine) at the N-terminus from the pGEX-6P1-hCBS and the pET47-NP-hCBS constructs. Dashes represent gaps in protein alignment, while dots stand for the remaining sequence of either GST or CBS.
Materials and Methods
Chemicals
Unless stated otherwise, all chemicals were purchased from Sigma or Fisher Scientific. L-[14C(U)]-serine was obtained from PerkinElmer Life Sciences.
Preparation of CBS constructs
We used our established GST-CBS construct pGEX-6P1-hCBS as a template for PCR amplification of the human CBS coding sequence and also as a reference for expression studies and comparison with a variety of CBS constructs carrying the 6xHis tag ([22]; Fig. 2). In order to create the 6xHis-tagged constructs, we inserted CBS into two commercially available vectors, pET-28a(+) and pET-47b(+) (Novagen) to obtain three differently tagged CBS constructs: the first one with a permanent C-terminal 6xHis tag (pET28-C-hCBS), a second one with a permanent N-terminal 6xHis tag (pET28-N-hCBS) and a third one with a removable N-terminal 6xHis tag followed by the PreScission protease recognition site (pET47-NP-hCBS). The last construct yields an identical CBS polypeptide, after cleaving off the 6xHis tag, as the pGEX-6P1-hCBS construct after the proteolytic removal of the GST.
For the preparation of the pET28-C-hCBS construct, CBS coding sequence was PCR-amplified using a forward primer containing an NcoI site (5′-ctagCCATGGgttctgagaccccccagg) and a reverse primer with an XhoI site, which did not include the STOP codon (5′-ctagCTCGAGcttctggtcccgctcctg). After cleavage with NcoI-HF and XhoI (NEB Biolabs), the CBS fragment was cut out from a 1% agarose gel and cleaned up using the QIAquick gel extraction kit (Qiagen). Subsequently, the CBS fragment was ligated with NcoI-XhoI linearized pET-28a(+) vector.
The preparation of pET28-N-hCBS followed the same strategy as mentioned in the previous paragraph except for the PCR primers. The forward primer contained an NcoI site and an additional sequence for the 6xHis tag (5′-ctagCCATGGgtcatcatcatcatcaccacccctctgagaccc) and the reverse primer possessed an XhoI site along with a STOP codon (5′-ctagCTCGAGtcacttctggtcccgctcctggg).
For the preparation of the pET47-NP-hCBS construct, first the original vector was modified in order to allow for cloning into the ApaI and XhoI sites. The internal ApaI site at position 1405 was abolished and subsequently the SanDI site in the PreScission protease recognition site was mutated into an ApaI site both using the QuikChange site directed mutagenesis kit (Agilent). The CBS coding sequence was PCR-amplified using a forward primer with an ApaI site (5′-ccagGGGCCCtctgag) and reverse primer with an XhoI site (5′-gccgCTCGAGtcgactc). After cleavage with ApaI and XhoI (NEB Biolabs), the CBS fragment was cut out from a 1% agarose gel and cleaned up using the QIAquick gel extraction kit (Qiagen). Subsequently, the CBS fragment was ligated into an ApaI-XhoI linearized modified pET-47b(+) vector.
All constructs were transformed into E. coli XL1-Blue cells (Agilent) and their authenticity was confirmed by DNA sequencing. The R266K missense mutation was subsequently introduced into the wild type CBS constructs by the QuikChange site directed mutagenesis kit (Agilent) using forward (5′-cacgggcattgccAGGaagctgaaggag) and reverse (5′-ctccttcagcttCCTggcaatgcccgtg) oligonucleotides. The presence of the desired mutation was confirmed by DNA sequencing. Verified plasmids for wild type as well as R266K mutant CBS were finally transformed into E. coli Rosetta2 (DE3) expression host cells (Novagen).
Preparation of crude extracts
Crude extracts for initial screening and for a screening of chemical chaperones were prepared following the previously published procedure [16]. Briefly, bacterial cells with the desired construct were grown (30 °C, 275 rpm) in 30 ml of LB medium supplemented with 0.001% thiamine-HCl, 0.0025% pyridoxine-HCl, 0.1 mM ferric chloride, 0.3 mM δ-aminolevulinic acid, 100 μg/ml ampicillin (for GST-based constructs) or 30 μg/ml kanamycin (for 6xHis-tagged constructs) and one of the chemical chaperones at the selected final concentration, if applicable. When the cell density reached A600 ~0.8, CBS expression was induced by adding IPTG to a final concentration of 1 mM. After induction, the cells continued to grow overnight and were then harvested by centrifugation at 9000×g for 7 min at 4°C. The cell pellets were washed with 1x PBS and either processed immediately or kept at −80°C before processing. The crude extracts (i.e. soluble fractions) were prepared according to the initial steps of our CBS purification procedure described elsewhere [16, 22].
Purification of CBS enzymes with the C-terminal 6xHis tag
The cells were grown similarly as for a small scale study described above in six 2.8 l baffled Fernbach flask containing 1 liter of appropriately supplemented LB medium. The cell pellets were resuspended in lysis buffer (50 mM sodium phosphate pH 7.4, 300 mM NaCl, 0.1 mM PLP and Protease inhibitor cocktail VII (A.G. Scientific)) at a 1:5 (w/v) ratio using a homogenizer. Resuspended cells were treated with 2 mg/ml lysozyme for 1 hour at 4°C prior to sonication (8 cycles of 2 min at 50% duty cycle; Misonix S-3000, Qsonica). The cell lysate was centrifuged at 58000×g for 30 min at 4°C. The supernatant was loaded on a 50 ml TALON column (Clontech) equilibrated in 50 mM sodium phosphate pH 7.4 and 300 mM NaCl. The column was subsequently washed with at least 5 column volumes of wash buffer (50 mM sodium phosphate pH 7.4, 300 mM NaCl, 10 mM imidazole) and the bound protein eluted with 200 mM imidazole in the wash buffer. Subsequently, the fraction was desalted on a Sephadex G-25 (GE Healthcare) column (3.2 cm I.D. × 22 cm) and the buffer was exchanged with the DEAE loading buffer (15 mM potassium phosphate pH 7.2, 1 mM EDTA, 1 mM DTT, 10% ethylene glycol). The desalted sample was loaded onto a DEAE Sepharose (GE Healthcare) column (2.5 cm I.D. × 8.0 cm) and washed with 2 column volumes of the DEAE loading buffer followed by 5 column volumes of the DEAE wash buffer (65 mM potassium phosphate pH 7.2, 1 mM EDTA, 1 mM DTT, 10% ethylene glycol). The protein was eluted with 300 mM potassium phosphate in the DEAE loading/wash buffer. The CBS was formulated into a final buffer (20 mM HEPES pH 7.4, 1 mM TCEP, 0.01% Tween 20) on a Sephadex G-25 column and subsequently concentrated using an ultrafiltration device (Amicon) equipped with a YM-100 (Millipore) membrane. Finally, the enzyme was aliquoted, flash-frozen in liquid nitrogen and stored at −80°C.
Protein gel electrophoresis and Western blot analysis
Protein concentrations were determined by the Bradford method (Thermo Pierce) using bovine serum albumin (BSA) as a standard according to the manufacturer’s recommendations. Denatured proteins were separated by SDS-PAGE using a 9% separating gel with a 4% stacking gel. Native samples were separated in 4–15% polyacrylamide gradient precast gels (Mini-PROTEAN TGX, Bio-Rad). For visualization, the denatured gels were stained with Simple Blue (Invitrogen). Western blot analysis of crude cell lysates under denaturing or native conditions was performed as described previously [16].
CBS activity assay
The CBS activity in the classical reaction was determined by a previously described radioisotope assay using [14C(U)] L-serine as the labeled substrate [29]. CBS enzyme (420 ng) was assayed in a 100 μl reaction mixture for 30 min at 37 °C. The reaction mixture contained 100 mM Tris–HCl pH 8.6, 10 mM L-serine, 0.3 μCi L-[14C(U)]-serine and 0.5 mg/ml BSA. The extent of saturation of the enzyme with PLP was tested by running the activity assay in the presence or absence of 0.2 mM PLP. The reaction was performed in the presence or absence of AdoMet in a final concentration of 0.3 mM. The reaction mixture with enzyme was incubated at 37°C for 5 min and the reaction was initiated by addition of L-homocysteine to a final concentration of 10 mM.
The thermal pre-treatment of the enzyme prior the the CBS activity assay was performed as described before [16]. Briefly, the purified enzyme was diluted to a final concentration of 0.1 mg/ml in Tris-buffered saline pH 8.6, 100 μM PLP. The enzyme solutions (50 μl) were heated in 200-μl thin-walled PCR tubes in a Mastercycler gradient PCR thermal cycler (Eppendorf). The temperature was raised from 37°C to 60°C in 0.5°C-increments with a 1 min incubation at each temperature. Aliquots (12 μl) were collected into separate tubes at designated temperatures and kept on ice until the last aliquot was taken. The CBS activity was subsequently determined as described above.
Thermal denaturation
To characterize the thermal stability of the purified enzymes, the protein absorption spectrum was monitored between 25°C and 90°C every 2.5°C after 2 min of equilibration at each temperature. A full spectrum was recorded on an Agilent diode array model 8453 UV-visible spectrophotometer equipped with a Peltier temperature controller. One ml of a protein sample (0.2 mg/ml) was prepared by diluting the stock protein with the appropriate amount of buffer (1x TBS) and then placed in a quartz cuvette with a 1-cm path length. A micro stir bar (100 rpm) was put into the cuvette to reduce the thermal gradient of the sample.
Results
Construction and initial evaluation of CBS constructs
Three different constructs for CBS, carrying a short affinity (6xHis) tag, were prepared and tested (Fig. 2). They were compared to each other and to our established GST-based expression/purification system for CBS in order to test the hypothesis that a bulky fusion partner (GST) may interfere with the folding of this CBS mutant polypeptide and to investigate the usefulness of the 6xHis-based expression/purification system for studying the CBS mutants. The pET28-C-hCBS construct yields an enzyme bearing a permanent 6xHis tag at the C-terminus, while the pET28-N-hCBS and the pET47-NP-hCBS constructs yield enzymes with a permanent or a cleavable 6xHis tag at the N-terminus, respectively (Fig. 2). Moreover, the expressed CBS from the pET47-NP-hCBS construct resulted in an identical amino acid sequence, after cleaving off the 6xHis tag with HRV3C protease, as the CBS enzyme from the pGEX-6P1-hCBS construct after proteolytic removal of the GST partner. This approach allowed for a direct comparison of the effect of a bulky fusion partner (GST) and a short affinity tag (6xHis).
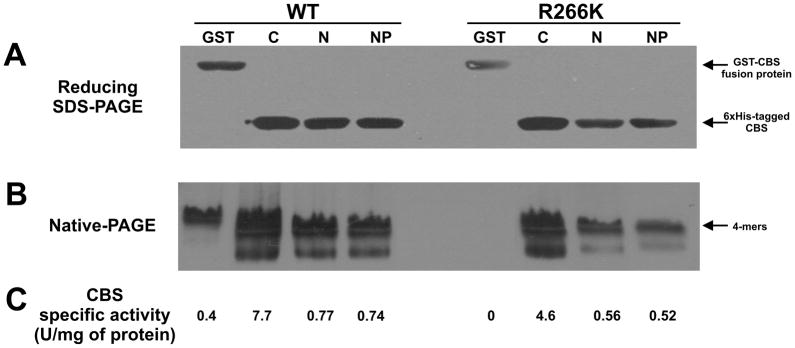
After the introduction of the R266K mutation by site directed mutagenesis and the transformation of the verified constructs into expression host cells, we performed initial expression of the 6xHis-tagged constructs along with our established GST-based one (Fig. 3). We successfully expressed wild type as well as the R266K mutant CBS proteins from all four constructs based on denaturing SDS-PAGE Western blot analysis of the crude extracts (Fig. 3A). The expression from the constructs with the C-terminal 6xHis tag resulted in somewhat higher yields compared to the other constructs for both wild type and R266K CBS. Interestingly, the native gel-Western blot analysis showed the presence of CBS tetramers [16, 23, 30] for all constructs except for the GST-based construct of R266K CBS (Fig. 3B). This construct completely lacked the native tetramers which fully corresponded with this construct having no detectable CBS activity (Fig. 3C). The relative amounts of detectable CBS tetramers were substantially higher for the pET28-C-hCBS constructs of wild type as well as R266K CBS compared to the other ones, which also translated to significantly higher CBS specific activities of these two constructs (Fig. 3B, 3C). Direct comparison of similar 6xHis-tagged constructs of wild type and mutant CBS showed that R266K CBS has approximately 65% of the wild type activity. This result suggests that the short affinity tag located at the C- or the N-terminus does not interfere with the folding, assembly and activity of the mutant in sharp contrast to the GST-CBS mutant construct, which yielded an inactive unassembled enzyme.
Figure 3. Expression, native conformation and activity of various CBS constructs of wild type and the R266K mutant.
The SDS-PAGE-Western blot (A), native-PAGE-Western blot (B) and CBS activity (C) of the CBS constructs of wild type and the R266K mutant. The crude extracts (soluble fraction, 30 μg) were separated either in a 10% Tris-HCl gel using reducing denaturing conditions (A) or in a 4–15% Tris-HCl gel under native conditions (B), transferred to a polyvinylidene difluoride membrane and probed with monoclonal anti-CBS antibody (Abnova). Arrows designate the GST-CBS fusion protein, 6xHis-tagged CBS protein and native tetramers, respectively.
Chemical chaperone screening
In our previous work, we have successfully used small chemical chaperones, such as dimethylsulfoxide (DMSO), trimethylamine-N-oxide (TMAO) or ethanol, to promote folding and assembly of several mutant CBS enzymes expressed using the GST-based system [16]. For that reason, we were interested in whether the inclusion of the chemical chaperone in the growth medium would enhance the folding and ultimately activity of the GST-based as well as the 6xHis-tagged mutant CBS constructs. The presence of a chemical chaperone in the bacterial culture expressing the GST-CBS R266K construct did not improve the folding or activity of the enzyme (Fig. S1A). Western blot of a native gel showed the total absence of CBS tetramers in lieu of inactive aggregates. Similarly, chemical chaperones did not enhance the mutant CBS folding and activity when expressed with a 6xHis tag (Fig. S1B). Even though there was an apparent abundance of CBS tetramers, when DMSO was included in growth medium of the C-terminally 6xHis-tagged construct, compared to a non-chaperoned sample, the specific activity was not affected. The similar results were obtained when the R266K CBS construct with an N-terminal 6xHis tag was screened in the presence of the chemical chaperones (data not shown). These results suggest that the inclusion of a chemical chaperone in the growth medium cannot rescue the inactive GST-CBS R266K construct and has no significant impact on the 6xHis-tagged constructs.
Purification of the R266K mutant enzyme
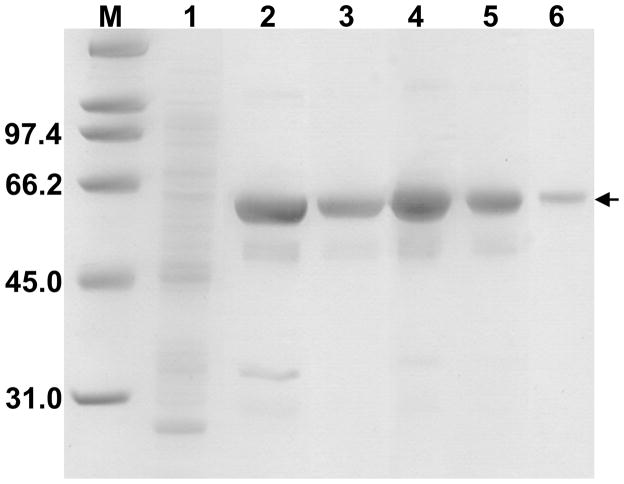
As we were unsuccessful with the expression of an assembled tetrameric R266K CBS mutant, the presence of an active tetrameric 6xHis-tagged CBS mutant (Fig. 3) encouraged us to purify this mutant to homogeneity. We selected the CBS construct with a permanent C-terminal 6xHis tag, whose crude extracts in initial screening and the screening of chemical chaperones showed the highest amount of CBS protein as well as active tetramer and specific activity (Fig. 3). The purification of the C-6xHis-tagged R266K CBS resulted in ~ 80% purity after a single TALON chromatography step (Fig. 4). After an additional DEAE chromatography step, the purity of the enzyme increased to ≥ 90% (Fig. 4) and in total a 15-fold increase in specific activity compared to the crude extract was achieved (data not shown). The saturation of the enzyme with heme cofactor was expressed as a ratio of absorbance at 430 nm (Soret peak) to 280 nm (total protein peak). The A430/280 of R266K CBS, equal to 0.91, was slightly lower compared to the wild-type CBS construct prepared using the same purification procedure (1.01; Fig. S2); however, within the range of variability for both wild-type and mutant CBS (0.9–1.2) [16]. Overall yield from the two column purification of R266K CBS yielded about 5.5 mg per liter of culture, which is approximately half of the yield for a similar wild type CBS construct (10.2 mg per liter of culture). These results suggest that the 6xHis tag construct represents an efficient way for high level expression and purification of wild type and mutant CBS.
Figure 4. Two-step purification of R266K CBS with C-terminal 6xHis tag.
Each protein purification fraction was separated on a 9% SDS–PAGE gel and stained using Simply Blue Safe stain (Invitrogen). Lanes: M, Molecular weight marker (Biorad); 1, crude extract (10 μg); 2, Cobalt TALON column elution (10 μg); 3, DEAE Sepharose elution (4.5 μg); 4–6, purified R266K CBS (10 μg, 5 μg and 1 μg, respectively). The arrow designates the CBS subunit.
Enzymatic activity and effect of increasing temperature on R266K CBS
We compared the purified wild type and the R266K mutant CBS enzymes to each other in terms of their response to (i) an additional PLP, (ii) a CBS allosteric activator AdoMet and (iii) an increased temperature pre-treatment of the enzyme prior to the activity assay. Table 1 shows that activity of R266K CBS in the absence of an additional PLP and AdoMet was essentially equal to that of wild type. However, a mutant enzyme seemed to be less saturated with the PLP cofactor than a wild type as the addition of the PLP to the assay increased its activity ~1.5 times. On the other hand, the R266K CBS response to AdoMet stimulation was significantly lower (2-fold increase) compared to that of wild type (4-fold increase) when assayed simultaneously (Table 1). Comparison of wild type CBS enzymes purified using either 6xHis tag or the GST fusion partner showed that they are practically identical in their specific activity, PLP and AdoMet response (Table 1) suggesting that the presence of the additional sequence at the C terminus does not affect catalytic properties of the enzyme.
Table 1. Activity, PLP and AdoMet response of purified R266K CBS mutant compared to wild type.
The purified enzymes were assayed for CBS catalytic activity in the classical reaction. The saturation of the purified enzyme with PLP cofactor was tested by running the activity assay in the presence or absence of 200 μM PLP. The AdoMet response was determined by assaying the enzyme in the presence or absence of 300 μM AdoMet. The − and + signs denote the absence and presence, respectively, of PLP or AdoMet in the reactions. Values represent average values with S.E.M. from at least three independent assays.
| PLP (200 μM) | AdoMet (300 μM) | CBS Specific Activity (U/mg of protein)
|
||
|---|---|---|---|---|
| R266K CBS | WT CBS | WT CBS (pGEX)* | ||
|
| ||||
| − | − | 130 ± 10 | 128 ± 6 | 128 ± 15 |
| − | + | 299 ± 35 | 444 ± 22 | 470 ± 34 |
| + | − | 191 ± 17 | 137 ± 5 | 148 ± 21 |
| + | + | 386 ± 26 | 507 ± 34 | 530 ± 45 |
For comparative purposes the specific activities for CBS expressed from pGEX-6P1-hCBS were taken from Majtan et al. [16].
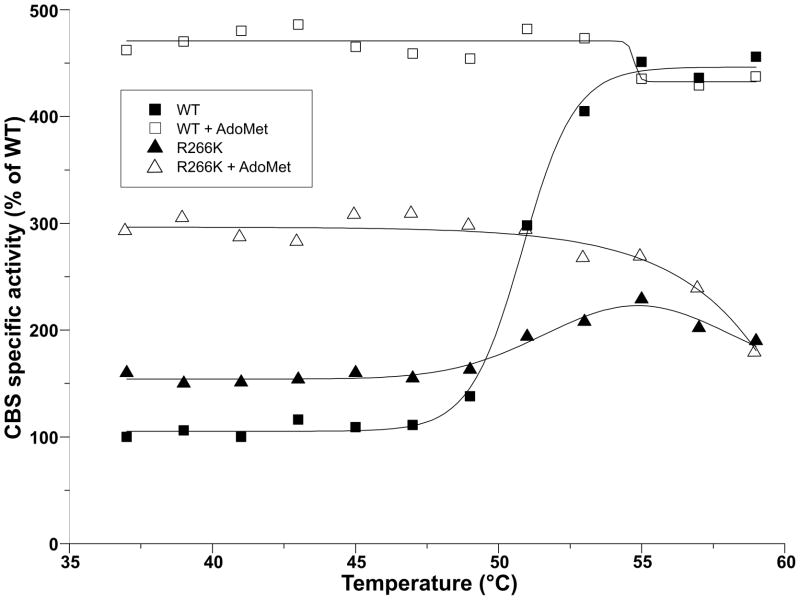
The temperature pre-treatment of R266K CBS showed an interesting feature compared to the wild type enzyme (Fig. 5). Thermal pre-treatment at 53°C of human CBS, close to its melting point, is known to stimulate its activity and thus mimic the effect of AdoMet [3, 16]. The activity of thermally pre-treated R266K CBS never reached the activity of an AdoMet-stimulated enzyme as the wild type enzyme did. Moreover, after reaching 55°C the mutant enzyme started to lose activity. Compared to a profile of the wild type CBS, it seemed that the increased temperature caused a premature destabilization of mutant enzymes resulting in its decreasing activity.
Figure 5. The effect of thermal pre-treatment of wild type and R266K CBS on their specific activity in the absence and presence of AdoMet.
Filled and open squares designate wild type CBS in the absence and presence of AdoMet, respectively. Filled and open triangles represent R266K CBS in the absence and presence of AdoMet, respectively. CBS activities are expressed in percent relative to the activity of wild type CBS at 37°C in the absence of AdoMet representing 100% (128 ± 0.4 U/mg of protein).
Thermal denaturation
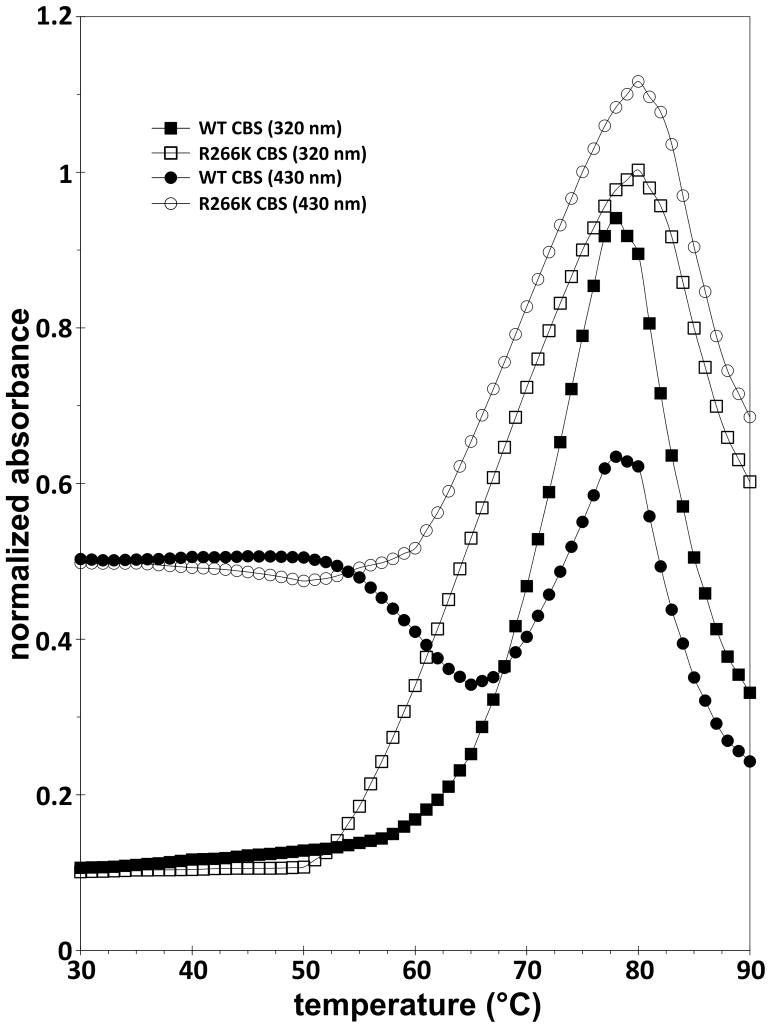
Fig. 6 shows the thermal denaturation profiles of R266K and wild type CBS. We compared the thermal profiles of the enzymes recorded at two different wavelengths: absorbance at 320 nm is mainly due to the light scattering of the thermally unfolded and aggregated protein and absorbance at 430 nm corresponds to an absorption maximum of CBS (Soret peak). The R266K CBS mutant started to form aggregates at a lower temperature than the wild type (Tonset = 52°C, Tm = 66°C at 320 nm for the mutant compared to Tonset = 58°C, Tm = 68°C at 320 nm for the wild type enzyme). Monitoring the intensity of the CBS Soret peak also revealed a significant difference between R266K CBS compared to that of wild type. For wild type CBS, an increase in heme absorbance at 430 nm is preceded by an initial decrease, whereas for R266K only an increase in heme absorbance with temperature was detected (Fig. 6). The presented data suggest that the R266K mutant is thermally less stable than wild type, presumably due to changes in the heme environment.
Figure 6. Thermal denaturation profiles of wild type and the R266K CBS enzymes.
Absorbance was recorded as a function of temperature at two different wavelengths. The absorbance at 320 nm (squares) is mainly due to light scattering of the thermally unfolded and aggregated protein. The absorbance at 430 nm (circles) corresponds to an absorption maximum of CBS (Soret peak). Filled symbols designate wild type CBS, while open symbols correspond to the R266K CBS mutant.
Discussion
The successful purification of proteins of interest to homogeneity is the first critical step in their detailed characterization. The purification of mutant forms of proteins has its own challenges due to the effect of the mutation on their native properties. Recently, we have reported a successful purification and characterization of wild type and eight mutant CBS enzymes [16, 22]. The CBS enzymes were expressed as fusion proteins with a GST partner at the N-terminus, which after cleavage with the HRV3C (also known as PreScission) protease and subsequent chromatographic separation of the GST yielded a ≥90% pure CBS polypeptide with only a single extra amino acid residue, glycine, at the N-terminus of CBS compared to the native sequence. We have used the same expression/purification system to prepare CBS enzyme containing cobalt protoporphyrin instead of heme in order to unravel the role of the heme cofactor in CBS enzymes from higher eukaryotes [23]. However, several mutant forms of CBS including R266K eluded our efforts of purification and characterization using this procedure. Our data presented here shows the GST-tagged R266K CBS mutant forms inactive aggregates, which precluded its purification. To our surprise, Singh et al. [20] reported a successful purification and spectroscopic characterization of an R266K CBS mutant using a GST-based system. However, compared to our GST-CBS construct the spacer between the GST fusion partner and CBS was substantially longer and after cleavage with thrombin protease, the final CBS carried an extra 11 amino acid residues. These opposing results, using similar expression systems, suggested that although our construct yields a CBS polypeptide as similar to the native CBS sequence as possible, which works for wild type as well as many CBS mutants, the presence of GST likely interferes with the folding and/or proper assembly of the CBS polypeptide for some mutants, such as R266K. The extra 11 residues in the construct of Singh et al. [20] most likely prevented undesired interactions between GST and CBS. The potential disadvantage of an extended spacer between GST and CBS is an alteration of some of the enzymes properties. Janosik et al. [3] reported a significantly lowered (~9-fold) Km of CBS, with 23 extra residues at the N-terminus, for homocysteine. Altogether, the data suggests that there was a need for a new construct, which would allow for efficient expression and purification of both wild type and mutant CBS. Such construct(s) should avoid the use of a bulky fusion partner such as GST, which may interfere with the structural properties of CBS polypeptides, and at the same time should yield the CBS sequence as close to its native sequence as possible to avoid alterations in the biochemical properties of the purified CBS enzymes.
We prepared three CBS constructs carrying a short affinity (6xHis) tag and evaluated their efficacy for the production of pure wild-type and mutant CBS enzymes. Generally, the small size of the 6xHis tag does not interfere with the folding, assembly or biochemical properties of the partner protein [26]. Recently, a new cost-effective and efficient expression/purification system for CBS based on a short affinity (6xHis) tag has been reported [28]. The authors reported that the CBS construct bearing the C-terminal 6xHis tag was insoluble probably due to a destabilization of the CBS oligomeric structure, which resulted in aggregation and precipitation. In our hands, the C-terminally 6xHis-tagged CBS construct yielded a soluble CBS polypeptide. Moreover and in stark contrast to the previously reported study [28], our construct yielded more active tetrameric CBS enzyme in crude extract than any other 6xHis-tagged or GST-based constructs tested herein. The plausible explanation of such different results may possibly involve a failure to verify the DNA sequence or a problem with expression and/or purification of Belew’s et al. construct [28].
The presence of chemical chaperones such as ethanol, TMAO or DMSO during CBS expression was recently shown to facilitate proper folding and to rescue the activity of several CBS mutants [15–17]. We have also shown that the effect of chemical chaperones may be indirect by increasing the steady-state levels of molecular chaperones, such as DnaJ, a microbial analog of HSP40 [16]. The lowering of the temperature from 37°C to 18°C during CBS expression had a similar effect as the presence of chemical chaperones on recovery of native tetramers and activity of several CBS mutants [21]. This data suggests that the deficiency in CBS activity may be in some instances caused by misfolding that may be alleviated by folding under permissive conditions. The R266K CBS mutant was found to be responsive to a chaperone treatment by inclusion of betaine or glycerol in the growth medium or to the lowering of the temperature during bacterial growth and CBS expression [17, 21]. In this study we demonstrated that the presence of ethanol, TMAO or DMSO as chemical chaperones did not promote a significant increase in R266K CBS activity when expressed from a GST-based or 6xHis-based construct. However, we detected an increased amount of native tetramer for the C-terminally 6xHis-tagged R266K CBS, which indicates that chemical chaperones promoted proper folding and/or tetramer assembly of this protein. The lack of correlation between the increased amount of native tetramer and no significant changes in the specific activity of R266K may be explained by a topology of this particular mutation. According to the crystal structure of the truncated form of human CBS, the R266 is a residue buried in the structure of the CBS globule [5, 6]. Kozich et al. [21] showed that several buried mutations, particularly G307S, lacked the correlation between the amount of native tetramers and specific activity of the CBS mutant. On the other hand, increased activity of several CBS mutants containing a solvent-exposed mutation mirrored the increased amount of tetramers suggesting that solvent-exposed mutations are more likely to respond to interventions aimed at correcting their impact on CBS polypeptide folding.
The R266 is not just a buried residue within the CBS structure, but also an important residue in the second coordination sphere of the axial heme ligand C52. Singh et al. [20] showed that the electrostatic interaction between R266 and C52 is critical for the stabilization of the ferrous heme and its disruption leads to the irreversible formation of an inactive ligand-switched species. The relevance of their finding is somewhat questionable since it is not known whether CBS heme undergoes redox changes in vivo and thus whether there is any heme-based allosteric redox regulation of CBS activity. Here we showed that the R266K mutation affects (i) the enzyme saturation with a PLP cofactor, (ii) the response to the CBS allosteric activator AdoMet and (iii) the thermal stability compared to wild type. The modest increase in CBS mutant activity after addition of exogenous PLP correlates well with the results reported by Singh et al. [20]. The extent of CBS response to AdoMet varies greatly and partially correlates with the folding, assembly of tetramers and particularly with the relative position of the catalytic core to its C-terminal regulatory region. In addition, the thermal pre-treatment of CBS prior to an activity assay mimics the enzyme activation with AdoMet [3, 16]. Our data showed that the response of the R266K CBS mutant to AdoMet is significantly lower compared to wild type. Moreover, thermal pre-treatment resulted in only a modest increase in activity and then began to decrease in activity after 55°C possibly due to premature aggregation.
Indeed, the R266K mutant formed aggregates at a lower temperature than wild type suggesting its decreased thermal stability. The differences at 430 nm, monitoring the Soret peak of the CBS heme, between the wild type and mutant spectra may be plausibly explained by a premature aggregation of the mutant enzyme. The decrease of Soret peak intensity for wild type enzyme with increasing temperature is most likely due to the loss of the heme ligand, which converts the heme from a low-spin into a high-spin species [31, 32]. The subsequent increase in 430 nm absorbance with increasing temperature is most likely due to protein aggregation, which increases the baseline absorbance. In R266K, the loss of the heme ligand was either not present or masked by a more facile aggregation. We assume that the R266K heme environment, where the pathogenic mutation already affects the stability of the heme thiolate ligand, is more sensitive to heme ligand loss with increasing temperature than wild type, which in turn immediately triggers protein aggregation.
Our previous studies showed that heme is essential for the expression of an active human CBS and that the saturation of enzyme with heme cofactor directly correlates with an amount of bound PLP and thus with CBS catalytic activity [33–35]. Destabilization of CBS heme’s second coordination sphere by the R266K missense mutation may be communicated to the PLP-containing active site of CBS via α-helix 8 and possibly resulting in premature irreversible loss of PLP cofactor in the partially unfolded enzyme by the thermal pre-treatment. Such a link would further confirm the importance of an intact connection between heme and PLP in human CBS. Detailed spectroscopic characterization of the R266K heme environment will be a subject of another study.
Supplementary Material
Highlights.
Nature and position of affinity tag is critical for mutant CBS folding and assembly
C-terminal His-tagged WT and R266K CBS has the highest activity among all constructs
The heme in R266K mutant is more susceptible to ligand switch than WT
The R266K mutation most likely destabilizes the thiolate ligand of the heme
Acknowledgments
We would like to thank Aaron T. Smith (University of Wisconsin, Madison) for helpful comments regarding the CBS heme and Sarah Venezia (University of Colorado, Denver) for critical reading of the manuscript. This work was supported by Postdoctoral Fellowship 0920079G from the American Heart Association (to TM), by National Institutes of Health Grant HL065217, by American Heart Association Grant In-Aid 09GRNT2110159 and by a grant from the Jerome Lejeune Foundation (to JPK).
Abbreviations
- δ-ALA
δ-aminolevulinic acid
- AdoMet
S-adenosyl-L-methionine
- BSA
bovine serum albumin
- CBS
cystathionine β-synthase
- GST
glutathione S-transferase
- IPTG
isopropyl-β-D-1-thiogalactopyranoside
- PLP
pyridoxal-5′-phosphate
- SEM
standard error of the mean
- WT
wild type
Footnotes
Supporting material containing figures (i) from the screening of chemical chaperons of the GST-fused and the C-terminally 6xHis-tagged R266K CBS constructs and (ii) the UV-visible spectrum of the purified R266K CBS compared to that of wild type is available free of charge online on the journal’s website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miles EW, Kraus JP. Cystathionine beta-synthase: structure, function, regulation, and location of homocystinuria-causing mutations. J Biol Chem. 2004;279:29871–29874. doi: 10.1074/jbc.R400005200. [DOI] [PubMed] [Google Scholar]
- 2.Singh S, Madzelan P, Banerjee R. Properties of an unusual heme cofactor in PLP-dependent cystathionine beta-synthase. Nat Prod Rep. 2007;24:631–639. doi: 10.1039/b604182p. [DOI] [PubMed] [Google Scholar]
- 3.Janosik M, Kery V, Gaustadnes M, Maclean KN, Kraus JP. Regulation of human cystathionine beta-synthase by S-adenosyl-L-methionine: evidence for two catalytically active conformations involving an autoinhibitory domain in the C-terminal region. Biochemistry. 2001;40:10625–10633. doi: 10.1021/bi010711p. [DOI] [PubMed] [Google Scholar]
- 4.Maclean KN, Gaustadnes M, Oliveriusova J, Janosik M, Kraus E, Kozich V, Kery V, Skovby F, Rudiger N, Ingerslev J, Stabler SP, Allen RH, Kraus JP. High homocysteine and thrombosis without connective tissue disorders are associated with a novel class of cystathionine beta-synthase (CBS) mutations. Hum Mutat. 2002;19:641–655. doi: 10.1002/humu.10089. [DOI] [PubMed] [Google Scholar]
- 5.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Structure of human cystathionine beta-synthase: a unique pyridoxal 5′-phosphate-dependent heme protein. Embo J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R. Human cystathionine beta-synthase is a heme sensor protein. Evidence that the redox sensor is heme and not the vicinal cysteines in the CXXC motif seen in the crystal structure of the truncated enzyme. Biochemistry. 2002;41:10454–10461. doi: 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee R, Zou CG. Redox regulation and reaction mechanism of human cystathionine-beta-synthase: a PLP-dependent hemesensor protein. Arch Biochem Biophys. 2005;433:144–156. doi: 10.1016/j.abb.2004.08.037. [DOI] [PubMed] [Google Scholar]
- 8.Pazicni S, Lukat-Rodgers GS, Oliveriusova J, Rees KA, Parks RB, Clark RW, Rodgers KR, Kraus JP, Burstyn JN. The redox behavior of the heme in cystathionine beta-synthase is sensitive to pH. Biochemistry. 2004;43:14684–14695. doi: 10.1021/bi0488496. [DOI] [PubMed] [Google Scholar]
- 9.Pazicni S, Cherney MM, Lukat-Rodgers GS, Oliveriusova J, Rodgers KR, Kraus JP, Burstyn JN. The heme of cystathionine beta-synthase likely undergoes a thermally induced redox-mediated ligand switch. Biochemistry. 2005;44:16785–16795. doi: 10.1021/bi051305z. [DOI] [PubMed] [Google Scholar]
- 10.Cherney MM, Pazicni S, Frank N, Marvin KA, Kraus JP, Burstyn JN. Ferrous human cystathionine beta-synthase loses activity during enzyme assay due to a ligand switch process. Biochemistry. 2007;46:13199–13210. doi: 10.1021/bi701159y. [DOI] [PubMed] [Google Scholar]
- 11.Mudd SH, Levy HL, Kraus JP. Disorders of transsulfuration. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler K, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 2007–2056. [Google Scholar]
- 12.Kraus JP, Kozich V. Cystathionine b-Synthase and Its Deficiency. In: Carmel R, Jacobsen DW, editors. Homocysteine in Health and Disease. Cambridge University Press; Cambridge: 2001. pp. 223–243. [Google Scholar]
- 13.Mudd SH. Hypermethioninemias of genetic and non-genetic origin: A review. Am J Med Genet C Semin Med Genet. 2011;157:3–32. doi: 10.1002/ajmg.c.30293. [DOI] [PubMed] [Google Scholar]
- 14.Janosik M, Oliveriusova J, Janosikova B, Sokolova J, Kraus E, Kraus JP, Kozich V. Impaired heme binding and aggregation of mutant cystathionine beta-synthase subunits in homocystinuria. Am J Hum Genet. 2001;68:1506–1513. doi: 10.1086/320597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh LR, Chen X, Kozich V, Kruger WD. Chemical chaperone rescue of mutant human cystathionine beta-synthase. Mol Genet Metab. 2007;91:335–342. doi: 10.1016/j.ymgme.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Majtan T, Liu L, Carpenter JF, Kraus JP. Rescue of cystathionine beta-synthase (CBS) mutants with chemical chaperones: purification and characterization of eight CBS mutant enzymes. The Journal of biological chemistry. 2010;285:15866–15873. doi: 10.1074/jbc.M110.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kopecka J, Krijt J, Rakova K, Kozich V. Restoring assembly and activity of cystathionine beta-synthase mutants by ligands and chemical chaperones. J Inherit Metab Dis. 2011;34:39–48. doi: 10.1007/s10545-010-9087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim CE, Gallagher PM, Guttormsen AB, Refsum H, Ueland PM, Ose L, Folling I, Whitehead AS, Tsai MY, Kruger WD. Functional modeling of vitamin responsiveness in yeast: a common pyridoxine-responsive cystathionine b-synthase mutation in homocystinuria. Hum Mol Genet. 1997;6:2213–2221. doi: 10.1093/hmg/6.13.2213. [DOI] [PubMed] [Google Scholar]
- 19.Katsushima F, Oliveriusova J, Sakamoto O, Ohura T, Kondo Y, Iinuma K, Kraus E, Stouracova R, Kraus JP. Expression study of mutant cystathionine beta-synthase found in Japanese patients with homocystinuria. Mol Genet Metab. 2006;87:323–328. doi: 10.1016/j.ymgme.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Singh S, Madzelan P, Stasser J, Weeks CL, Becker D, Spiro TG, Penner-Hahn J, Banerjee R. Modulation of the heme electronic structure and cystathionine beta-synthase activity by second coordination sphere ligands: The role of heme ligand switching in redox regulation. J Inorg Biochem. 2009;103:689–697. doi: 10.1016/j.jinorgbio.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozich V, Sokolova J, Klatovska V, Krijt J, Janosik M, Jelinek K, Kraus JP. Cystathionine beta-synthase mutations: effect of mutation topology on folding and activity. Hum Mutat. 2010;31:809–819. doi: 10.1002/humu.21273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank N, Kent JO, Meier M, Kraus JP. Purification and characterization of the wild type and truncated human cystathionine beta-synthase enzymes expressed in E. coli. Arch Biochem Biophys. 2008;470:64–72. doi: 10.1016/j.abb.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majtan T, Freeman KM, Smith AT, Burstyn JN, Kraus JP. Purification and characterization of cystathionine beta-synthase bearing a cobalt protoporphyrin. Arch Biochem Biophys. 2011;508:25–30. doi: 10.1016/j.abb.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraus JP, Rosenberg LE. Cystathionine beta-synthase from human liver: improved purification scheme and additional characterization of the enzyme in crude and pure form. Arch Biochem Biophys. 1983;222:44–52. doi: 10.1016/0003-9861(83)90500-3. [DOI] [PubMed] [Google Scholar]
- 25.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 26.Walls D, Loughran ST. Tagging recombinant proteins to enhance solubility and aid purification. Methods in molecular biology (Clifton, NJ. 2011;681:151–175. doi: 10.1007/978-1-60761-913-0_9. [DOI] [PubMed] [Google Scholar]
- 27.Waugh DS. Making the most of affinity tags. Trends Biotechnol. 2005;23:316–320. doi: 10.1016/j.tibtech.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Belew MS, Quazi FI, Willmore WG, Aitken SM. Kinetic characterization of recombinant human cystathionine beta-synthase purified from E. coli. Protein Expr Purif. 2009;64:139–145. doi: 10.1016/j.pep.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Kraus JP. Cystathionine beta-synthase (human) Methods Enzymol. 1987;143:388–394. doi: 10.1016/0076-6879(87)43068-1. [DOI] [PubMed] [Google Scholar]
- 30.Frank N, Kery V, Maclean KN, Kraus JP. Solvent-accessible cysteines in human cystathionine beta-synthase: crucial role of cysteine 431 in S-adenosyl-L-methionine binding. Biochemistry. 2006;45:11021–11029. doi: 10.1021/bi060737m. [DOI] [PubMed] [Google Scholar]
- 31.Taoka S, Green EL, Loehr TM, Banerjee R. Mercuric chloride-induced spin or ligation state changes in ferric or ferrous human cystathionine beta-synthase inhibit enzyme activity. J Inorg Biochem. 2001;87:253–259. doi: 10.1016/s0162-0134(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 32.Ojha S, Wu J, LoBrutto R, Banerjee R. Effects of heme ligand mutations including a pathogenic variant, H65R, on the properties of human cystathionine beta-synthase. Biochemistry. 2002;41:4649–4654. doi: 10.1021/bi011827o. [DOI] [PubMed] [Google Scholar]
- 33.Kery V, Bukovska G, Kraus JP. Transsulfuration depends on heme in addition to pyridoxal 5′-phosphate. Cystathionine beta-synthase is a heme protein. J Biol Chem. 1994;269:25283–25288. [PubMed] [Google Scholar]
- 34.Kery V, Poneleit L, Meyer JD, Manning MC, Kraus JP. Binding of pyridoxal 5′-phosphate to the heme protein human cystathionine beta-synthase. Biochemistry. 1999;38:2716–2724. doi: 10.1021/bi981808n. [DOI] [PubMed] [Google Scholar]
- 35.Majtan T, Singh LR, Wang L, Kruger WD, Kraus JP. Active cystathionine beta-synthase can be expressed in heme-free systems in the presence of metal-substituted porphyrins or a chemical chaperone. The Journal of biological chemistry. 2008;283:34588–34595. doi: 10.1074/jbc.M805928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.