A major goal in structural biology and biophysics is to rationally design biomolecules that have specific characteristics at the atomic level. There have been significant advances in the design of proteins that fold into predetermined three-dimensional conformations.[1] However, biomolecular structures also undergo dynamic excursions about their native conformation and transiently access conformational substates that play critical roles in folding, catalysis, recognition, and signal transduction.[1–5] The rational design of such dynamics is a formidable challenge given the broad energy landscape that has to be considered and the complex spatiotemporal dependence of dynamics on sequence and structure, particularly for highly flexible molecules such as RNA.[6,7]
We recently showed, using NMR spectroscopy,[8,9] that the transactivation response element (TAR) RNA from HIV-I[10] codes for a broad dynamic structure ensemble and that various ligands bind distinct conformers within the ensemble by conformational selection. In unbound TAR, the two helices are on average highly bent relative to one another (ca. 28°) and undergo large-amplitude rigid-body global motions about a flexible pivot point consisting of spacer bulge residues C24 and U25 and the junctional A22-U40 base pair, which does not form the expected Watson–Crick hydrogen bond alignment (Figure 1 a).[8,9,11] Binding of the ligand argininamide (ARG), which is widely used as a mimic of the TAR cognate viral protein, the transactivator Tat, results in coaxial stacking of helices, a stable A22-U40 canonical base pair, a U23·A27-U38 base triple, and an increase in the local flexibility of bulge residues C24 and U25, which adopt a looped-out conformation (Figure 1 a).[8–10,12]
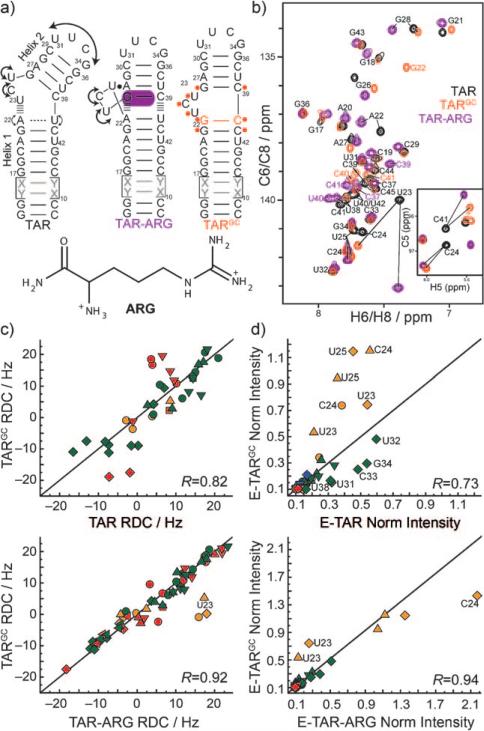
Figure 1.
Biasing the TAR dynamic structure ensemble toward an ARG-bound state. a) Design of the TARGC sequence based on the structural dynamics of the TAR and TAR-ARG complexes. ‘X-Y’ base pairs denote alternating A-U/U-A or G-C/C-G base pairs used in the elongation.[8,9] Residues undergoing the largest chemical shift perturbations due to the G-C mutation are highlighted in orange on the TARGC secondary structure. b) Representative 2D CH HSQC spectra of TAR (black), TARGC (orange), and TAR-ARG (purple). Inset: 2D C,H HSQC C5-H5 spectra. c,d) Correlation plots. Red: residues in helix I, green: helix II, orange: in the bulge. R = correlation coefficients. ■ C2-H2, ▼ C8-H8, ▲ C6-H6, &25cf C5-H5, ◆ C1’-H1’, ◀ N1/3-H1/3. c) Correlation plots between RDCs measured in non-elongated TAR, TARGC, and TAR-ARG complexes. d) Correlation plots between normalized resonance intensities measured in EI-TAR, EI-TARGC, and EI-TAR-ARG.
We redesigned the TAR sequence to bias its dynamic structure ensemble towards the ARG-bound state without losing its ability to bind ARG. Our approach was to introduce mutations that do not disrupt ARG binding but that promote interactions uniquely observed in the TAR-ARG state. We replaced the flexible A22-U40 base pair, which is a confluence point for local and global dynamics and is not involved in any sequence-specific contacts with ARG, with a stronger hydrogen-bond-forming G-C base pair (Figure 1 a). The goal of this modification was to promote a canonical G-C base-pair and co-axial helical stacking due to improved G26-C39/G22-C40 stacking of about 1.2 kcalmol–1.[13]
We used NMR spectroscopy to site-specifically characterize the structural dynamics and ligand-binding properties of the TAR mutant (TARGC) in which A22-U40 is replaced with G22-C40 (Figure 1 a). The spectrum of the imino region of TARGC immediately revealed a signal that could unambiguously be assigned to G22 (Supporting Information, Figure S1), confirming that unlike A22-U40 in unbound TAR, G22-C40 forms the expected Watson–Crick hydrogen bond alignment in TARGC. The A-U to G-C mutation also resulted in large changes in 2D C,H HSQC spectra of TAR, indicating that the mutation affects structural dynamics across a variety of sites (Figure 1 b; Supporting Information, S1). Interestingly, the largest differences were observed for many residues that experience large chemical shift perturbations on binding to ARG, including the three bulge residues and the G26-C39 base pair above the bulge. A closer analysis revealed that many of the perturbations induced by the G-C mutation were along directions similar to those observed with ARG binding. For example, the downfield U23 carbon chemical shift perturbations are consistent with unstacking and coaxial stacking of helices.[14]
To further investigate the structural dynamics of TARGC, we measured residual dipolar couplings (RDCs)[15,16] between directly bonded C–H and N–H bonds and compared values with those previously reported for TAR[17] and TAR-ARG[12] (Figure 1 c; Supporting Information, Table S1). RDCs probe the orientation dynamics of bond vector relative to a molecule-fixed order tensor frame and are sensitive to internal motions occurring over a broad range of timescales (up to milliseconds). The RDCs measured at various sites in TARGC differed significantly from those measured in TAR but were in strikingly good agreement with those measured in TAR-ARG (Figure 1 c). Thus, the G-C mutation significantly alters the TAR structural dynamics and specifically biases it, on a site-by-site basis, towards the ARG-bound state.
To quantitatively characterize the structural dynamics of TARGC, we used domain elongation[8,9] (Supporting Information, Figure S2) to broaden the timescale sensitivity of spin relaxation measurements and to reduce couplings between internal motions from overall reorientation, which is key for quantitatively interpreting RDCs. Ignoring chemical exchange, the resonance intensities measured in 2D C,H HSQC spectra of elongated RNAs report the net dynamics of a given site relative to the applied magnetic field occurring at picosecond–nanosecond timescales. In unbound domain I elongated TAR (EI-TAR), elevated intensities were observed for domain II owing to collective interhelical motions, and A22, U40, C24, and U25 owing to elevated local mobility (Supporting Information, Figure S3).[9] Binding of ARG arrests interhelical motions and local fluctuations involving A22 and U40, but leads to a dramatic increase in the dynamics of C24 and U25, which adopt a looped-out conformation.[9] Although the EI-TARGC resonance intensities differ significantly from those of EI-TAR (R = 0.73), they were in very good agreement with those of EI-TAR-ARG (R = 0.94) (Figure 1 d; Supporting Information, S3); the only exception was U23, which is more flexible in TARGC. Thus, whilst U23 adopts a looped-out conformation in TARGC, it does not form the U23·A27-U38 base triple observed in TAR-ARG. Therefore, the G-C mutation biases the local and global picosecond–nanosecond dynamical properties of TAR towards the ARG-bound state.
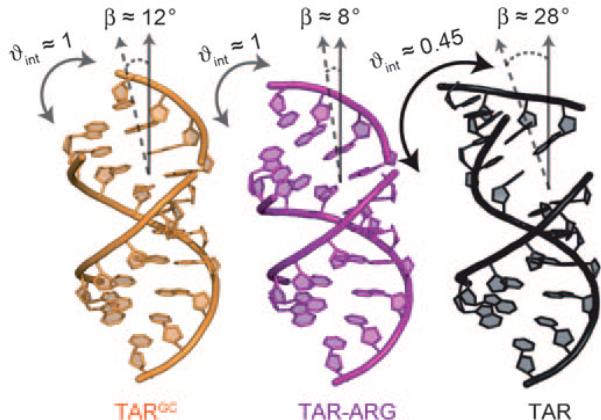
To characterize the TARGC dynamic structure ensemble over longer timescales, we measured RDCs in EI-TARGC and subjected values measured in each helical domain to a model-free order tensor analysis (Supporting Information, Table S2), as described previously for EI-TAR and EI-TAR-ARG.[8] In this analysis, RDCs and an idealized A-form helix geometry are used to determine five order tensor elements that describe ordering of each helix relative to the internal elongated helix axis. An excellent RDC fit to the idealized A-form geometry was obtained for each helix (Supporting Information, Table S2), confirming that they adopt a standard canonical geometry as reported previously for TAR.[8] The analysis yielded a TARGC global interhelical conformation in which the helices undergo very limited interhelical motions (ϕint = ϕshort/ϕelongated ≈ 1.02 ± 0.1 and ranges between 0 and 1 for maximum and minimum motions) about a nearly coaxially stacked (bend angle ca. 12 ± 7°) conformation (Figure 2). This conformation differs considerably from the previously reported[8] highly bent (ca. 28 ± 3°) and flexible (θint ≈ 0.45 ± 0.1) EI-TAR conformation, but it is in very good agreement with the coaxial (ca. 8 ± 4°) and globally rigid (θint ≈ 1.09 ± 0.1) TAR-ARG complex (Figure 2). The EI-TARGC bulge RDCs are near zero, which is consistent with a looped-out highly flexible conformation (Supporting Information, Table S1).
Figure 2.
Interhelical structural dynamics of EI-TAR, EI-TARG22C40, and EI-TAR-ARG from order tensor analysis of RDCs measured in helix I elongated constructs. There are no constraints on rotations around the elongated axis.
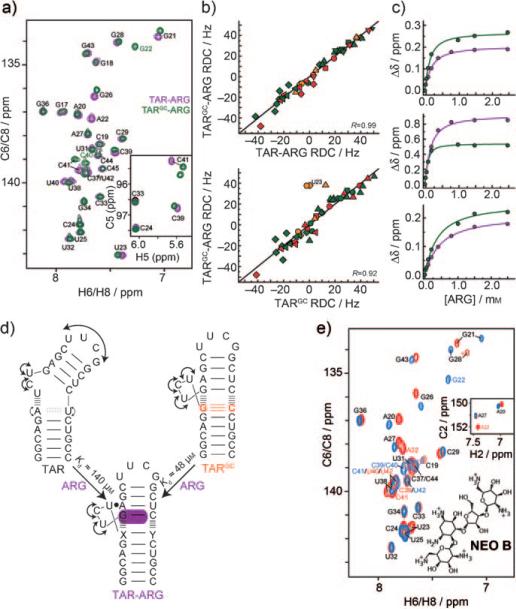
Our results suggest that despite having reduced and distinct structural dynamics, TARGC can dynamically sample the TAR ARG-bound state (Figure 2). We therefore examined whether TARGC can bind ARG. Strikingly, the NMR spectra (Figure 3 a; Supporting Information, S4) and also RDCs (Figure 3 b) measured in TARGC bound to ARG were in excellent agreement with counterparts measured in the TAR-ARG complex. Thus, despite large differences in their unbound structural dynamics, TAR and TARGC converge to a common ARG-bound conformation. The RDCs reveal that ARG binding only induces minor conformational changes in TARGC, which are centered on residue U23, and which most likely involve formation of the U23·A27-U38 base triple (Figure 3 c). ARG binds TARGC with slightly higher (threefold) affinity as compared to TAR, which is consistent with it having a lower free-energy cost associated with the RNA conformational change in the pre-organized TARGC. Additional kinetic experiments will be required to determine how the mutation impacts the rates of complex formation and dissociation.[18] As an inverse experiment, we compared the binding of the aminoglycoside neomycin B (NEOB), which binds a more bent (ca. 30°) TAR conformation[19,20] that is likely less accessible to TARGC. Unlike ARG, NEOB induces entirely different chemical shift perturbations in TAR and TARGC (Figure 3 e). This result is consistent with the expectation that NEOB is less effective at binding/stabilizing the bent conformation observed with TAR when interacting with TARGC and that instead it binds an alternate structure, possibly using a different NEOB conformation.[21]
Figure 3.
TAR and TARGC converge to a common ARG-bound conformation. a) Representative 2D C,H HSQC spectra of TAR-ARG (green) and TARGC-ARG (purple). Inset: 2D HSQC C5-H5 spectra. b) Correlation plots between RDCs measured in TARGC, TARGC-ARG, and TAR-ARG. For symbol and color key, see Figure 1 c. c) Representative ARG titration curves for TAR (purple) and TARGC (green) as a function of total ARG concentration. Individual Kd values: top: G22 (TARGC) 96.6 ± 16.1 μm, A22 (TAR) 141 ± 13.2 μm; middle: U23 (TARGC) 31.3 ± 3.24 μm, U23 (TAR) 116 ± 16.1 μm; bottom: G28 (TARGC) 311 ± 65.1 μm, G28 (TAR) 414 ± 10.3 μm. Global Kd values: TARGC 47.7 ± 9.3 μm, TAR 140 ± 9.0 μm. d) TAR and TARGC adopt distinct unbound dynamic structure ensembles that converge to a common ARG-bound state. e) Representative 2D C,H HSQC spectra of TAR-NEOB (red) and TARGC-NEOB (blue) with the chemical structure of NEOB shown.
In summary, the dynamics of unbound RNA can be used to tune ligand binding affinities without altering the ligand-bound RNA conformation. Our results also suggest that the sequence of Watson–Crick base pairs flanking interhelical junctions are important determinants of local and global RNA dynamics and that these effects can be predicted in part based on simple thermodynamic considerations. This, together with sequence-independent topological constraints imposed by length and asymmetry of junctions,[19,22] provide a framework for rationally designing RNA structures with specific dynamic and functional properties.
Supplementary Material
Footnotes
The authors acknowledge the Michigan Economic Development Cooperation and the Michigan Technology Tri-Corridor for the support of the purchase of a 600 MHz spectrometer. This work was supported by the NIH (R01 AI066975-01) and the National Science Foundation Graduate Research Fellowship Program (ACS).
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.201000814.
References
- 1.Das R, Baker D. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin AJ, Kay LE. Nat. Chem. Biol. 2009;5:808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 3.Henzler-Wildman K, Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 4.Boehr DD, Nussinov R, Wright PE. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. Angew. Chem. 2007;119:1232–1240. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 6.Al-Hashimi HM, Walter NG. Curr. Opin. Struct. Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fürtig B, Buck J, Manoharan V, Bermel W, Jäschke A, Wenter P, Pitsch S, Schwalbe H. Biopolymers. 2007;86:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 10.Puglisi JD, Tan R, Calnan BJ, Frankel AD, Williamson JR. Science. 1992;257:76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- 11.Aboul-ela F, Karn J, Varani G. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pitt SW, Majumdar A, Serganov A, Patel DJ, Al-Hashimi HM. J. Mol. Biol. 2004;338:7–16. doi: 10.1016/j.jmb.2004.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner DH, Sugimoto N, Freier SM. Annu. Rev. Biophys. Biophys. Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- 14.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. J. Mol. Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tjandra N, Bax A. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 16.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. Proc. Natl. Acad. Sci. USA. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Hashimi HM, Gossera Y, Gorina A, Hua W, Majumdara A, Patel DJ. J. Mol. Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 18.Jucker FM, Rhillips RM, McCallum SA, Pardi A. Biochemistry. 2003;42:2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- 19.Bailor MH, Sun X, Al-Hashimi HM. Science. 2010;327:202–206. doi: 10.1126/science.1181085. [DOI] [PubMed] [Google Scholar]
- 20.Faber C, Sticht H, Schweimer K, Rosch P. J. Biol. Chem. 2000;275:20660–20666. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 21.Namanja AT, Wang XJ, Xu B, Mercedes-Camacho AY, Wilson BD, Wilson KA, Etzkorn FA, Peng JW. J. Am. Chem. Soc. 2010;132:5607–5609. doi: 10.1021/ja9096779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu VB, Lipfert J, Bai Y, Pande VS, Doniach S, Herschlag D. NAR. 2009;15:2195–2192. doi: 10.1261/rna.1747509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.