Abstract
Background
Alcohol consumption is one of the top-10 risks for the worldwide burden of disease and an established cause of head and neck cancer as well as cancer at other sites. Acetaldehyde, the major metabolite of ethanol, reacts with DNA to produce adducts, which are critical in the carcinogenic process and can serve as biomarkers of exposure and possibly of disease risk. Acetaldehyde associated with alcohol consumption is considered “carcinogenic to humans”. We have previously developed the technology to quantify acetaldehyde-DNA adducts in human tissues, but there are no studies in the literature defining the formation and removal of acetaldehyde-DNA adducts in people who consumed alcohol.
Methods
We investigated levels of N2-ethylidene-dGuo, the major DNA adduct of acetaldehyde, in DNA from human oral cells at several time points after consumption of increasing alcohol doses. Ten healthy non-smokers were dosed once a week for three weeks. Mouthwash samples were collected before and at several time points after the dose. N2-Ethylidene-dGuo was measured as its NaBH3CN reduction product N2-ethyl-dGuo by LC-ESI-MS/MS.
Results
N2-ethylidene-dGuo levels increased as much as 100-fold from baseline within 4h after each dose for all subjects and in a dose responsive manner (p = 0.001).
Conclusion
These results demonstrate an effect of alcohol on oral cell DNA adduct formation, strongly supporting the key role of acetaldehyde in head and neck cancer caused by alcohol drinking.
Impact
Our results provide some of the first conclusive evidence linking exposure to a lifestyle carcinogen and kinetics of DNA adduct formation in humans.
Keywords: DNA adducts, alcohol, oral cavity
Introduction
Many epidemiological studies have established the relationship between alcohol consumption and different types of cancers including those of the upper aerodigestive tract (1,2). An understanding of mechanisms of alcohol-induced cancer is critical for developing rational approaches to cancer prevention. A variety of mechanisms may contribute to alcohol-mediated carcinogenesis, including the effect of ethanol in increasing solubility of carcinogens, the production of toxic reactive oxygen species, the perturbation of methyl transfer and other enzymatic systems or the abnormal metabolism of vitamin A and its derivative retinoic acid (3). However there is convincing evidence that acetaldehyde, the major metabolite of ethanol and a DNA-reactive compound, is at least partially responsible for the carcinogenic effects of alcohol. Therefore, acetaldehyde associated with alcohol consumption has been classified recently as “carcinogenic to humans” by the International Agency for Research on Cancer (4).
Acetaldehyde reacts with DNA bases to produce adducts which are critical in the carcinogenic process because they can cause miscoding resulting in mutated genes and loss of normal growth control mechanisms. DNA adducts can serve as biomarkers of carcinogen exposure and potentially of cancer risk. The major DNA adduct of acetaldehyde is N2-ethylidenedeoxyguanosine (N2-ethylidene-dGuo) (5). This adduct can be analyzed as N2-ethyldeoxyguanosine (N2-ethyl-dGuo) after treatment of the DNA with NaBH3CN (6) or can undergo in vivo reduction to N2-ethyl-dGuo which has been found in the DNA of both ethanol treated mice and human alcoholics (7–9). Several other adducts as well as DNA-DNA cross-links are also formed in the reactions of acetaldehyde with DNA (10). The impact of heavy drinking and aldehyde dehydrogenase polymorphisms on levels of acetaldehyde-DNA adducts have been reported (9,11), suggesting a potential use of N2-ethyl-dGuo as a biomarker for understanding alcohol carcinogenesis. However, no studies have reported the relationship between alcohol consumption and the kinetics and extent of N2-ethylidene-dGuo formation in humans.
Although ethanol is mainly metabolized in the liver, the concentration of acetaldehyde in saliva after ingesting ethanol is much higher than in the blood, due to oral microflora metabolism of ethanol as well as the acetaldehyde content of alcoholic beverages. Bacteria present in the normal oral flora such as Streptococcus salivarius and Neisseria contribute to the production and accumulation of acetaldehyde from ethanol in the oral cavity (12–14). Therefore, oral cell DNA appeared to be an appropriate target for investigating levels of N2-ethylidene-dGuo after alcohol consumption. We quantified N2-ethylidene-dGuo in oral cell DNA at intervals up to 120 h after consumption of increasing amounts of ethanol by 10 healthy subjects. There are little if any data in the literature relating exposure to an environmental or lifestyle carcinogen such as ethanol and the kinetics and extent of DNA adduct formation in humans.
Materials and Methods
Subjects
Ten healthy volunteers, 5 men and 5 women, were recruited from University of Minnesota students and staff. All subjects were social drinkers accustomed to consuming alcoholic beverages regularly in moderation. This criterion was defined as having consumed at least one drink a week and 3 drinks over a 4 h period at least once in the month preceding the study. All subjects were more than 21 years old, non-smokers, in good physical and mental health, with no unstable medical conditions and no history of alcohol abuse based on a medical history questionnaire. Exclusion criteria were as follows: Asian ethnicity, periodontal disease or other oral lesions that might affect drug absorption, chronic use of any drug that could interact with alcohol, and insulin-dependent diabetes. For women, pregnancy and current breastfeeding were two additional exclusion criteria. Subjects were asked to refrain from using any recreational drug and from ingesting any ethanol-containing product other than the dose provided for the study, for the entire 4 week duration of the study.
Study design
The study was approved by the University of Minnesota Human Research Protection Programs Institutional Review Board. Subjects were enrolled after signing a consent form and evaluation of the eligibility criteria. Medical history and alcohol drinking history, both in the past 12 months and lifetime, were obtained through a questionnaire. An open-label study design, in which each subject served as his/her own control, was used. Subjects participated in an initial meeting in which they were provided with information about the study, and a mouthwash sample was collected. Starting from this meeting participants were asked to refrain from using any mouthwash containing alcohol and from drinking any alcohol other than that to be administered in the study. One week later the subjects came to the clinic for the administration of the first alcohol dose. The meeting was set at 8 a.m. and subjects were asked to come to the clinic after having had a light breakfast (cereal, milk and coffee) which was to be consumed at least 90 min before the dose. Subjects were asked to provide a mouthwash sample before drinking the alcohol dose. Vodka, 100 proof, was mixed with tonic water and Rose lime juice (all purchased at a local liquor store). The amount of the drink served to each subject was calculated based on body weight and gender (15) in order to reach a blood alcohol level of 0.03%. The drinking was paced so that subjects had one sip every 5 min over a 20 min period. Systemic alcohol concentration was monitored using a breath alcohol analyzer 30 min after the last sip (16). Mouthwash samples were collected 2, 4 and 6 h after completion of the dosing period. Subjects returned to the clinic between 8 and 10 a.m. one, two and five days after the dose to provide a mouthwash sample for each time point. They were asked to refrain from drinking and eating between completion of the alcohol dose and the 4h sample collection. Between the 4 h and 6 h samples, the subjects had a light meal. One week later, the subjects came back to the clinic for the second alcohol dose. The session started at 8 a.m. They were asked to provide a mouthwash sample before drinking. The alcoholic beverage was prepared as in the first session, but with a target blood alcohol level of 0.05%. Subjects had one sip every 5 min over a 30 min period. Samples were collected following the protocol described for the first dosing session. One week later, they came back to the clinic for the third and final alcohol dose. The session started at 8 a.m. They were asked to provide a mouthwash sample before drinking. The alcoholic beverage was prepared as in the first dosing meeting, with a target blood alcohol level of 0.07%. They had one sip every 5 min over a 40 min period. Samples were collected following the protocol described for the first dosing session, with addition of one sample collected 7 days after the last dose.
Mouthwash sample collection
Oral cell samples were collected by mouthwash using a commercially available sterile 0.9% saline solution (Medline, Mundelein IL) following a protocol reported in the literature (17). Subjects were asked to brush their teeth with a standard toothbrush and toothpaste. Forty-five min later, they were given 15 mL of the saline solution in a sterile mono-dose tube, and they were asked to swish vigorously 30 times, and then spit back into a 50-mL centrifuge tube. Mouthwash samples were immediately frozen and stored until DNA isolation.
Breath alcohol analysis
Systemic alcohol concentration was monitored using the breath alcohol analyzer Alco Sensor III (Intoximeters, St Louis Mo). After waiting 30 min from the last sip of the alcohol dose, subjects were asked to make a prolonged deep exhalation into the disposable inlet tube of the instrument for at least 6 s. Before each measurement the instrument was calibrated according to the manufacturer’s instructions and a blank was tested measuring the breath alcohol level of a staff member who had not participated in the alcohol dosing.
Chemicals and Enzymes
N2-Ethyl-dGuo and [15N5]N2-ethyl-dGuo were prepared as described(6). Ethanol was obtained from AAPER Alcohol and Chemical Co. (Shelbyville, Ky). Isopropanol was purchased from Acros Organics (Morris Plains, NJ). Puregene DNA purification solutions were obtained from Qiagen (Valencia, CA). Calf thymus DNA was purchased from Worthington Biochemical Corporation (Lakewood, NJ). Alkaline phosphatase (from calf intestine) was obtained from Roche Diagnostics Corporation (Indianapolis, IN). All other chemicals and enzymes were purchased from Sigma-Aldrich (St. Louis, MO).
DNA isolation from oral cells collected with saline mouthwash
DNA was isolated using the DNA purification from oral cell protocol (Qiagen) with several modifications. Briefly, 1 mL of cell lysis solution was added to the oral cell pellet formed after centrifugation of 15 mL of the saline solution used for cell collection. Proteinase K (6 μL) was added and the sample was incubated at room temperature overnight. A solution of RNase A (6 μL of 4 mg/mL) was added and the sample was incubated at room temperature for 2 h. Protein precipitation solution (300 μL) was added to the cell lysate and the mixture was centrifuged to remove proteins. DNA was precipitated from the supernatant by addition of 1.3 mL of isopropanol. After centrifugation the DNA pellet was washed with 1 mL of 70% ethanol in H2O, and then 1 mL of 100 % ethanol. DNA was dried in a stream of N2 and stored at −20 °C until use. Oral cells collected with 15 mL saline mouthwash typically yielded about 20 μg DNA.
Sample enrichment
DNA hydrolysis and sample enrichment and purification were carried out as reported (11). For enzyme hydrolysis, DNA was dissolved in 400 μL of 10 mM Tris/5 mM MgCl2 buffer containing [15N5]N2-ethyl dGuo (50 fmol) and NaBH3CN (30 mg). NaBH3CN treatment converts the major acetaldehyde-DNA adduct, N2-ethylidene-dGuo, to N2-ethyl-dGuo (6). After the pH was adjusted to 7 with 0.1N HCl, the DNA was initially digested overnight at room temperature with 1300 units of DNase I (type II, from bovine pancreas). Then to the resulting mixture were added 1300 additional units of DNase I, 0.07 units of phosphodiesterase I (type II, from Crotalus adamanteus venom), and 750 units of alkaline phosphatase. The mixture was incubated at 37 °C for 70 min and then allowed to stand overnight at room temperature. Enzymes were removed by centrifugation using a centrifree MPS device (MW cutoff of 30,000; Amicon, Beverly, MA). The hydrolysate, after removal of a 10 μL aliquot for dGuo analysis, was desalted and purified using a solid-phase extraction cartridge [Strata-X 33 μm, 30 mg/1 mL (Phenomenex)]. The 70% CH3OH fraction was collected and evaporated to dryness, dissolved in 1 mL of H2O, and purified using a mixed mode, anion exchange reversed phase extraction cartridge (Oasis MAX, 30 mg/cartridge, Waters). Adducts were eluted with 1 mL of 70% CH3OH, and the solution was evaporated to dryness. The residue was dissolved in 20 μL of H2O, and 8 μL aliquots were analyzed by LC-ESI-MS/MS. Samples from each subject were processed separately. Samples from each dose, including baseline and time-points after consumption of the dose, were processed together as a set, resulting in three sets per subject. Buffer blanks containing internal standard were processed as above and analyzed to check the MS instrument baseline and possible contamination. Calf thymus DNA (0.1 mg) with internal standard added as above was used as a positive control to determine inter-day precision and accuracy. Each set of samples was run together with one buffer blank and 3 positive controls.
HPLC-UV analysis
Quantitation of dGuo was carried out with an Agilent 1100 capillary flow HPLC with a diode array UV detector set at 254 nm (Agilent Technologies, Palo Alto, CA). A 4.6 mm × 25 cm Luna 5 μm C18 column (Phenomenex, Torrance, CA) was used with a gradient from 5 to 40% CH3OH in H2O over the course of 35 min at a flow rate of 10 μL/min.
LC-ESI-MS/MS analysis
This was performed with an Agilent 1100 capillary flow HPLC (Agilent Technologies) with a 250 mm × 0.5 mm 5μm particle size Polar RP column (Phenomenex) and a Vantage (Thermoelectron, San Jose, CA) triple quadrupole mass spectrometer. The solvent elution program was a 10 μL/min gradient from 5 to 40% CH3OH in 35 min at 30 °C. The ESI source was set in the positive ion mode as follows: voltage, 3.7 kV; current, 3 μA; and heated ion transfer tube, 275 °C. The collision energy was 12 eV, and the Ar collision gas pressure was 1.0 mTorr.
Adducts were quantified by MS/MS with selected reaction monitoring (SRM) at m/z 296 → m/z 180 ([M + H]+ → [BH]+) for N2-ethyl-dGuo, and at the corresponding transition m/z 301 → m/z 185 for [15N5]N2-ethyl-dGuo.
A calibration curve was constructed before each analysis using a standard solution of N2-ethyl-dGuo and [15N5]N2-ethyl-dGuo. A constant amount of [15N5]N2-ethyl-dGuo (5 fmol) was mixed with differing amounts of N2-ethyl-dGuo (0.1, 0.5, 4, 10 and 100 fmol) and analyzed by LC-ESI-MS/MS-SRM.
Statistical Analysis
To determine whether the levels of N2-ethylidene-dGuo (measured as N2-ethyl-dGuo) in oral cell DNA reached a peak after each dose of alcohol, the ratio of the maximum level reached at each dose to the baseline level of the DNA adduct for that dose was calculated on the log scale and a 95% confidence interval for the ratio was formed. Then the estimate and confidence limits were converted back to the original scale. The same method was used to determine whether the levels of N2-ethyl-dGuo returned to baseline after reaching a peak, except that the ratio was formed using the baseline DNA adduct level for the next alcohol dose, or the final N2-ethyl-dGuo level in the case of the third alcohol dose. To determine whether there was a dose-response, a repeated measures model was used, with the log of dose predicting the peak/baseline ratio, modeling the covariance matrix with dose as the repeated factor within subjects.
Results
The 10 study participants were Caucasian and 21 – 31 years old (mean ± SD, 25 ± 3). All subjects participated in the 4 weekly meetings (one introductory and 3 dosing meetings) and provided the mouthwash samples requested at all time-points for a total of 230 samples. Table 1 summarizes demographics, amounts of ethanol administered, and systemic alcohol levels. During the three dosing sessions women reached an average of 0.03 ±0.02 %, 0.05 ± 0.01 % and 0.06 ± 0.01 %, and men an average of 0.03 ±0.01 %, 0.05 ± 0.01 % and 0.07 ± 0.01 % blood alcohol.
Table 1.
Study participants’ demographic characteristics, grams of ethanol per dose and blood alcohol concentration (BAC %) attained after administration of each dose.
| subject | gender | age | Weight (kg) | dose 1
|
dose 2
|
dose 3
|
|||
|---|---|---|---|---|---|---|---|---|---|
| g of ethanol | BACa | g of ethanol | BAC | g of ethanol | BAC | ||||
| 1 | F | 30 | 59 | 16.1 | 0.02 | 23.2 | 0.04 | 30.4 | 0.04 |
| 2 | F | 23 | 65 | 17.7 | 0.02 | 25.6 | 0.05 | 33.4 | 0.06 |
| 3 | F | 23 | 64 | 17.6 | 0.03 | 25.4 | 0.05 | 33.3 | 0.05 |
| 4 | F | 26 | 92 | 25.2 | 0.04 | 36.4 | 0.05 | 47.6 | 0.07 |
| 5 | F | 28 | 74 | 20.2 | 0.05 | 29.2 | 0.06 | 38.2 | 0.07 |
| mean ± SD | 26 ± 3 | 71 ± 13 | 19.4 ± 3.6 | 0.03 ± 0.02 | 28.0 ± 5.2 | 0.05 ± 0.01 | 36.6 ± 6.8 | 0.06 ± 0.01 | |
|
| |||||||||
| subject | gender | age | Weight (kg) |
dose 1
|
dose 2
|
dose 3
|
|||
| g of ethanol | BAC | g of ethanol | BAC | g of ethanol | BAC | ||||
| 6 | M | 31 | 82 | 26.8 | 0.01 | 38.7 | 0.05 | 50.6 | 0.06 |
| 7 | M | 23 | 79 | 26.0 | 0.04 | 37.6 | 0.05 | 49.2 | 0.07 |
| 8 | M | 24 | 83 | 27.4 | 0.04 | 39.5 | 0.05 | 51.7 | 0.06 |
| 9 | M | 22 | 91 | 29.9 | 0.04 | 43.2 | 0.05 | 56.5 | 0.08 |
| 10 | M | 25 | 79 | 25.9 | 0.04 | 37.3 | 0.04 | 48.8 | 0.08 |
| mean ± SD | 25 ± 4 | 83 ± 5 | 27.2 ± 1.6 | 0.03 ± 0.01 | 39.3 ± 2.4 | 0.05 ± 0.01 | 51.4 ± 3.1 | 0.07 ± 0.01 | |
BAC. Blood alcohol concentration: mg of alcohol in 100 mL of blood expressed as %.
DNA was isolated from all samples (mean ± SD, 22 ± 22 μg). N2-Ethyl-dGuo was measured in 212 samples treated with NaBH3CN; the analysis failed in 18 samples due to poor recovery. The results are summarized in Table 2. Although the level of N2-ethyl-dGuo showed considerable intra- and inter-individual variation, it increased in all subjects after all doses, and some of the increases were large – up to 100-fold from baseline. Most of the participants reached a peak level of the DNA adduct within 4 h after the doses. Only subject 4 after the first dose and subject 10 after the second dose reached the peak level of N2-ethyl-dGuo 6 h after the dose. Levels of N2-ethyl-dGuo started to decrease after 6 h (with the same exceptions as reported above) and returned to baseline before the next dose and 168 h after the last dose. A slight increase in the level of N2-ethyl-dGuo was observed in some of the subjects, mostly at 48 or 120 h after the ethanol dose. However this increase was not statistically significant and never reached the peak level detected within 4 h after alcohol consumption.
Table 2.
Levels of N2-ethylidene-dGuo (fmol/μmol dGuo) in oral cell DNA before and after each dose of alcohol. Levels of N2-ethylidene-dGuo (as N2-ethyl-dGuo) were measured in oral cell DNA from each subject (from 1 to 10). The first row reports participants’ adduct levels measured one week before the first dose was given. This is the point at which participants began to abstain from consumption of any alcoholic beverage other than the dose given for the study. “Baseline” levels of N2-ethyl-dGuo were also measured 1 h before each dose was given. The rows that follow report the levels of N2-ethyl-dGuo at the various time points after consumption of the three increasing doses (from dose 1 to dose 3).
| subject | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| mean | SD | ||||||||||||
|
|
|||||||||||||
| 1 week before the dose | 2581 | 2769 | 1427 | 4047 | 2856 | 1256 | 1650 | 1661 | 1532 | 8034 | 2781 | 2041 | |
|
| |||||||||||||
| dose 1 | baseline | 2795 | 2364 | 1503 | 684 | 4892 | 2757 | 1476 | 1052 | 4195 | 3382 | 2510 | 1374 |
| 2 hours | 6149 | 1793 | 3501 | 1266 | 13420 | 2433 | 6850 | 10100 | 6378 | 8570 | 6046 | 3922 | |
| 4 hours | 7768 | 10900 | 8122 | 1458 | 5673 | 6467 | 2420 | 15919 | 6136 | 10630 | 7549 | 4233 | |
| 6 hours | 1737 | 1690 | 1824 | 2844 | 2541 | 2199 | 1911 | 1610 | 5355 | 5807 | 2752 | 1545 | |
| 24 hours | 4168 | 1672 | 2018 | 1236 | 2541 | 1362 | 1606 | 871 | 2789 | 4436 | 2270 | 1218 | |
| 48 hours | 5137 | 1087 | 4776 | 971 | 2362 | 1423 | 2006 | 5556 | 1744 | 8203 | 3326 | 2438 | |
| 120 hours | 4464 | 2664 | 2154 | 780 | 3389 | 2157 | 3792 | 815 | 1271 | 3394 | 2488 | 1277 | |
|
| |||||||||||||
| dose 2 | baseline | -a | 1666 | 1803 | 1855 | - | 2328 | 1657 | 2684 | 1108 | 1043 | 1768 | 554 |
| 2 hours | - | 3787 | 2623 | 4591 | - | 32230 | 16090 | 35330 | 8094 | 8841 | 13950 | 12960 | |
| 4 hours | - | 9532 | 1533 | 7570 | - | 8323 | 2840 | 86630 | 22040 | 1479 | 17490 | 28720 | |
| 6 hours | - | 3693 | 935 | 4195 | - | 2782 | 1960 | 3551 | 2969 | 10500 | 3823 | 2889 | |
| 24 hours | - | 1631 | 1710 | 1973 | - | 1867 | 1780 | 2271 | 15650 | 2896 | 3722 | 4837 | |
| 48 hours | - | 1598 | 419 | 2535 | - | 3264 | 2072 | 1461 | 2176 | 1582 | 1888 | 841 | |
| 120 hours | - | 1717 | 3738 | 628 | - | 1022 | 1902 | 1987 | 2959 | 762 | 1839 | 1082 | |
|
| |||||||||||||
| dose 3 | baseline | 1466 | 1108 | 2019 | 1417 | 1338 | 242 | 2341 | 443 | 1546 | 1039 | 1296 | 638 |
| 2 hours | 13610 | 4935 | 32440 | 5071 | 31890 | 9003 | 10610 | 43430 | 33090 | - | 20450 | 14640 | |
| 4 hours | 12260 | 17300 | 18360 | 4875 | 14230 | 7273 | 6338 | 11990 | 28860 | 39890 | 16140 | 10900 | |
| 6 hours | - | 452 | 15100 | 3668 | 831 | 859 | 1047 | 3723 | 20470 | 4397 | 5616 | 7181 | |
| 24 hours | 551 | 461 | 11270 | 1634 | 240 | 316 | 754 | 872 | 2028 | 1469 | 1960 | 3326 | |
| 48 hours | 1815 | 1420 | 3084 | 2201 | 853 | 1679 | 538 | 1118 | 1021 | - | 1525 | 779 | |
| 120 hours | 10020 | 369 | 861 | 1706 | 494 | 361 | 1093 | 186 | 1124 | 1156 | 1737 | 2948 | |
| 168 hours | 791 | 318 | 608 | 4087 | 361 | 264 | 600 | 326 | 426 | - | 865 | 1221 | |
Values missing due to poor recovery of N2-ethyl-dGuo.
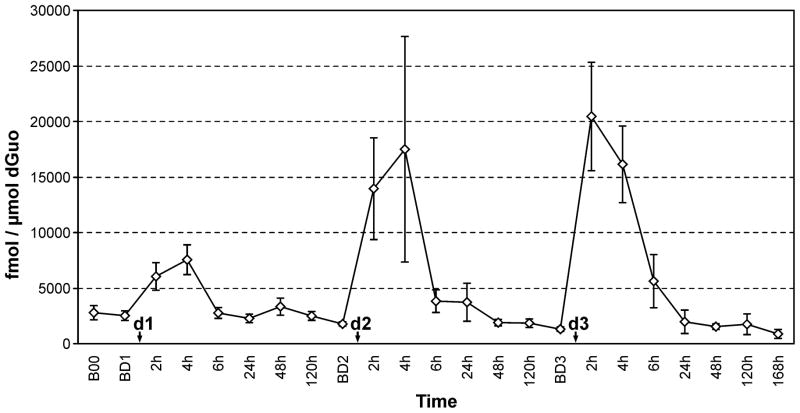
Statistical analysis is summarized in Table 3. The ratio (on the log scale) of the maximum N2-ethyl-dGuo level measured within 6 h of each dose to the baseline level of the adduct for that dose is presented. After the first dose of alcohol, the peak/baseline ratio of N2-ethyl-dGuo was 3.8, with the peak occurring 3.6 h post-dose. The ratio was 8.6 after the 2nd (higher) dose, with the peak occurring at 3.5 h post-dose. The ratio was 17.5 after the 3rd (highest) dose of alcohol, with the peak occurring 2.4 h post-dose. The peak/baseline ratios were significant at all doses, indicating that even a single administration of alcohol resulted in a statistically significant increased level of N2-ethyl-dGuo in oral cell DNA, which returned to the baseline level before the next alcohol dose. The dose-response trend was significant (p = 0.001). We also calculated the ratio of the maximum level of N2-ethyl-dGuo measured within 6 h of each dose to the baseline DNA adduct level for the next dose (or the final N2-ethyl-dGuo level in the case of the third dose). The results demonstrated that N2-ethyl-dGuo levels in oral cell DNA returned to baseline after each dose administered. We also observed a significant decrease from the initial baseline levels of N2-ethyl-dGuo to those measured 168 h after the final dose of alcohol (p < 0.001). The results of the study are presented graphically in Figure 1.
Table 3.
Variation of peak/baseline ratio and peak/next baseline ratio after increasing doses of alcohol.
| Dose 1 | Dose 2 | Dose 3 | p for trend | |
|---|---|---|---|---|
| Peak/Baselinea Mean (95% CI) | 3.8 (2.4, 5.9) | 8.6 (3.8, 19.3) | 17.5 (8.5, 35.9) | p = 0.001 |
| Peak timing (h) b Mean (SD) | 3.6 (1.3) | 3.5 (1.4) | 2.4 (0.8) | |
| Peak/Next Baseline c Mean (95% CI) | 4.5 (2.8, 7.4) | 14.1 (3.5, 56.7) | 31.9 (10.9, 93.6) | p < 0.001 |
The ratio between the average maximum N2-ethyl-dGuo level reached after each dose of alcohol and the average baseline level of for that dose was calculated and a 95% confidence interval for the ratio was formed.
The average time at which the maximum N2-ethyl-dGuo was reached after each dose was calculated.
To determine whether the levels of N2-ethyl-dGuo returned to baseline after reaching a peak, the ratio between the average maximum N2-ethyl-dGuo level reached after each alcohol dose and the average baseline DNA adduct level measured for the next alcohol dose was calculated. The final N2-ethyl-dGuo level measured at 168h was used as next baseline level for the third alcohol dose.
Figure 1. Summary of the mean levels of N2-ethyl-dGuo at various intervals before and after the three doses.
The graph summarizes the mean levels of N2-ethyl-dGuo (fmol/μmol dGuo) measured in the oral cell DNA of the 10 subjects at the time-points analyzed in the study. The first time-point reported on the left (B00) refers to the orientation meeting, 1 week before consumption of the first dose. This time-point represents the point at which participants began to abstain from consuming any alcoholic beverage other than the dose administered for the study. The next time-point (BD1) refers to the baseline level detected 1 week later, 1 h before consumption of the first dose (d1, lowest). Subsequently, the graph shows the levels of N2-ethyl-dGuo measured at the various time-points considered after each dose (2h – 120h). The DNA adduct levels were measured at the same time-points before and after exposure to the next 2 doses (d2, intermediate, and d3, highest). The baseline time points measured 1h before the dose (BD1, BD2 and BD3) are seven days apart. Values are means and standard errors.
Discussion
The results of this study clearly demonstrate that alcohol consumption increases levels of the major acetaldehyde-DNA adduct N2-ethylidene-dGuo, measured as N2-ethyl-dGuo, in oral cells. The effect was significant within 4 – 6 h even after consumption of the lowest dose of alcohol, comparable to roughly one drink. To our knowledge this is the first study to investigate the effects of alcohol consumption on the time course of DNA adduct formation in healthy volunteers. All previous published studies reporting effects of alcohol consumption on N2-ethyl-dGuo levels were performed with heavy drinkers or alcoholics (8,9) and no time-dependence of DNA adduct formation was reported.
Although DNA adduct formation is universally accepted as a crucial event in carcinogenesis, remarkably few previous studies have evaluated the extent and kinetics of DNA adduct formation after deliberate human exposure to carcinogens. Rothman et al. investigated the formation of “PAH-DNA adducts” in peripheral white blood cells of 4 subjects who consumed charcoal-broiled beef and found an increase in adduct levels in 2 of them, using samples collected 2, 5, 8, 12 and 24 days after the beginning of the controlled feeding (18). Dingley at al. investigated the effects of a dietary-relevant oral dose of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in a group of patients with colon cancer and showed an increase in PhIP-DNA adduct levels in peripheral blood DNA after exposure (19). The adduct levels reached a peak 2–4 h after exposure and decreased significantly over 24 h. Investigation of the kinetics of formation of DNA adducts after exposure to DNA binding compounds in humans has been mostly limited to studies testing clinical response to platinum-based therapies in cancer patients (20,21).
The rationale for this study originated in our recent observation of a dose-response effect of ethanol consumption on levels of N2-ethylidene-dGuo in leukocyte DNA (11). In that study some of the subjects were hospitalized, and a decreasing trend in adduct levels was observed according to the length of the subjects’ stay in the hospital. This observation suggested the need to understand the kinetics of N2-ethylidene-dGuo formation and removal. Furthermore, although the cells obtained from mouthwashes are comprised mostly of desquamated epithelial cells, we hypothesized that oral cell DNA would be a more relevant target for the investigation of alcohol-related DNA adduct formation and its potential role in head and neck carcinogenesis than peripheral blood cells. All previous studies quantified N2-ethyl-dGuo in leukocyte DNA. Therefore, this study investigated the role of alcohol consumption on DNA adduct formation in oral cells of healthy subjects who consumed increasing doses of ethanol in a controlled setting. We targeted blood alcohol levels below intoxication (considered to arise at a blood alcohol level of 0.08 %) (15). The three doses selected for the study can roughly be described as 1 drink, 2 drinks and 3 drinks per subject. The possible influence of acetaldehyde already present in the alcoholic beverage was reduced by our choice of 100 proof vodka for the study. Among hard liquors, vodka generally has relatively low acetaldehyde content. It is manufactured to have no distinctive aroma or taste and residual congeners are present at levels much lower than those found in other spirits (1,2). Our data are consistent with the results reported by Linderborg et al. showing that even a single sip of a strong alcoholic beverage (40% ethanol) can lead to elevated concentrations of acetaldehyde in the mouth (22).
Since food can interfere with alcohol absorption and can also be a source of acetaldehyde, our subjects were asked to eat only a light breakfast at least one h before administration of the alcohol dose and they were not allowed to eat until 4 h after the dose. Additionally, to reduce influences of other sources of alcohol on the results, study participants were asked to refrain from using mouthwash containing alcohol for the duration of the study.
Clear peak levels of N2-ethylidene-dGuo were observed in all subjects within 6 h from each alcohol dose. Adduct levels returned to baseline after each dose. These results support a direct causal effect of alcohol consumption on N2-ethylidene-dGuo formation. These results are consistent with studies showing that salivary acetaldehyde reaches a peak between 0 and 30 min after alcohol consumption, then decreases slowly over a 3 h period (23).
We observed large inter-individual variations in N2-ethylidene-dGuo levels. This finding is consistent with the high inter-individual variation in salivary acetaldehyde content observed after alcohol exposure in another study (14). We also observed great variability in levels of the DNA adduct 24 h after the alcohol dose, and a slight increase in its levels 2 or 5 days after the dose in some subjects. Endogenously produced acetaldehyde is known to have an effect on levels of this adduct in leukocyte DNA (24) and this could also affect levels of the adduct in oral cells. Additionally, acetaldehyde is present in a number of foods including yogurt and cheese (25). Since food sources provide exposure to acetaldehyde in amounts lower than the amounts found in the saliva after ethanol exposure, the participants’ diet was not restricted. Thus contribution from exposure to acetaldehyde in foods to the levels of N2-ethylidene-dGuo observed in the samples taken more than 6 h after the alcohol dose cannot be ruled out.
A potentially interesting and unexplained observation was the decrease in baseline levels of N2-ethylidene-dGuo over the course of the study. This suggests that refraining from ethanol consumption other than the experimental dose during the four weeks of the study reduced background levels of the DNA adduct.
One limitation of this study was the relatively small number of participants. Therefore, we were unable to investigate possible effects of gender, age, polymorphisms in genes involved in alcohol and acetaldehyde metabolism, or usual drinking habits on levels of the DNA adduct. Another limitation was the potential contamination of mammalian with bacterial DNA in the DNA samples isolated from the mouthwash (26). Participants were asked to brush their teeth before providing the mouthwash sample, but no additional precaution was taken to reduce possible bacterial contamination of the samples. Interference related to the presence of ethanol and acetaldehyde in commercial mouthwashes was avoided by using saline solution without antibacterial additives. Methods for the separation of bacterial DNA from human DNA are available, but these techniques would have reduced the yields of DNA preventing quantitation of N2-ethylidene-dGuo for which at least 5 μg of DNA are required. Further studies will be required to confirm our findings using DNA for which bacterial contamination has been completely excluded and on a larger number of subjects to investigate possible effects of covariates on N2-ethylidene-dGuo levels.
The biological consequences of N2-ethylidene-dGuo are unknown. Studies in vitro indicate that the closely related adduct N2-ethyl-dGuo blocks trans-lesion DNA synthesis catalyzed by a variety of DNA polymerases potentially resulting in failure of replication or frameshift deletion mutations. Studies on mammalian cells showed that the lesion can be bypassed by other specific polymerases, suggesting that mammalian cells can accurately replicate past N2-ethyl-dGuo which is thus considered non-mutagenic (27–29). However, this lesion could be mutagenic in particular contexts when polymerases are defective or reduced in their activity. N2-Ethylidene-dGuo is the major adduct formed after reaction of acetaldehyde with DNA but several other adducts resulting from this reaction are formed in lower amounts. Results from a recent study showed that exposure of human cells to levels of acetaldehyde comparable to those detected in the oral cavity after alcohol consumption resulted in formation of 1,N2-propano-dGuo (30), which can cause miscoding. Because of its higher levels, N2-ethylidene-dGuo is easier to detect and measure than other acetaldehyde DNA adducts and thus could become a marker for detection of acetaldehyde-induced damage to DNA.
In summary, we present conclusive evidence linking alcohol drinking and the kinetics of acetaldehyde-DNA adduct formation in the human oral cavity. Our results demonstrate that even low amounts of alcohol produce significant increases of acetaldehyde-DNA damage in the oral cavity within 4–6 h. Since DNA damage is a critical step in carcinogenesis, these results provide important new leads for understanding mechanisms of head and neck cancer caused by exposure to ethanol and acetaldehyde, and developing relevant biomarkers potentially applicable in cancer prevention. Furthermore, our results provide some of the first conclusive evidence linking exposure to a lifestyle-associated carcinogen and kinetics of DNA damage in humans.
Acknowledgments
This study was supported by grant ES-11297 from the National Institute of Environmental Health Sciences. Mass spectrometry was carried out in the Analytical Biochemistry core facility of the Masonic Cancer Center, supported in part by Cancer Center Support Grant CA-77598.
Reference List
- 1.Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, et al. Carcinogenicity of alcoholic beverages. Lancet Oncology. 2007;8:292–93. doi: 10.1016/s1470-2045(07)70099-2. [DOI] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 96. Lyon, FR: IARC; 2010. Alcohol Consumption and Ethyl Carbamate. [PMC free article] [PubMed] [Google Scholar]
- 3.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nature Reviews Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 4.Zhang S, Villalta PW, Wang MY, Hecht SS. Analysis of crotonaldehyde- and acetaldehyde-derived 1,N2-propanodeoxyguanosine adducts in DNA from human tissues using liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem Res Toxicol. 2006;19:1698. doi: 10.1021/tx060154d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M, McIntee EJ, Cheng G, Shi YL, Villalta PW, Hecht SS. Identification of DNA adducts of acetaldehyde. Chem Res Toxicol. 2000;13:1149–57. doi: 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]
- 6.Wang MY, Yu NX, Chen L, Villalta PW, Hochalter JB, Hecht SS. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N-2-ethyldeoxyguanosine. Chem Res Toxicol. 2006;19:319–24. doi: 10.1021/tx0502948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang JL, Vaca CE. Development of a 32P-postlabelling method for the analysis of adducts arising through the reaction of acetaldehyde with 2′-deoxyguanosine-3′-monophosphate and DNA. Carcinogenesis. 1995;16:2177–85. doi: 10.1093/carcin/16.9.2177. [DOI] [PubMed] [Google Scholar]
- 8.Fang JL, Vaca CE. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis. 1997;18:627–32. doi: 10.1093/carcin/18.4.627. [DOI] [PubMed] [Google Scholar]
- 9.Matsuda T, Yabushita H, Kanaly RA, Shibutani S, Yokoyama A. Increased DNA damage in ALDH2-deficient alcoholics. Chem Res Toxicol. 2006;19:1374–78. doi: 10.1021/tx060113h. [DOI] [PubMed] [Google Scholar]
- 10.Wang MY, Shi YL, Cheng G, Villalta PW, Sturla SJ, Jalas JR, et al. Reactions of formaldehyde plus acetaldehyde with deoxyguanosine and DNA. Chem Res Toxicol. 2002;15:1675. doi: 10.1021/tx025614r. [DOI] [PubMed] [Google Scholar]
- 11.Balbo S, Hashibe M, Gundy S, Brennan P, Canova C, Simonato L, et al. N-2-Ethyldeoxyguanosine as a Potential Biomarker for Assessing Effects of Alcohol Consumption on DNA. Cancer Epidemiol Biomarkers Prev. 2008;17:3026–32. doi: 10.1158/1055-9965.EPI-08-0117. [DOI] [PubMed] [Google Scholar]
- 12.Jokelainen K, Heikkonen E, Roine R, Lehtonen H, Salaspuro M. Increased acetaldehyde production by mouthwashings from patients with oral cavity, laryngeal, or pharyngeal cancer. Alcohol Clin Exp Res. 1996;20:1206–10. doi: 10.1111/j.1530-0277.1996.tb01113.x. [DOI] [PubMed] [Google Scholar]
- 13.Kurkivuori J, Salaspuro V, Kaihovaara P, Kari K, Rautemaa R, Gronroos L, et al. Acetaldehyde production from ethanol by oral streptococci. Oral Oncol. 2007;43:181–86. doi: 10.1016/j.oraloncology.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Lachenmeier DW, Monakhova YB. Short-term salivary acetaldehyde increase due to direct exposure to alcoholic beverages as an additional cancer risk factor beyond ethanol metabolism. J Exp Clin Cancer Res. 2011:30. doi: 10.1186/1756-9966-30-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher HR, Simpson RI, Kapur BM. Calculation of Blood-Alcohol Concentration (Bac) by Sex, Weight, Number of Drinks and Time. Canadian Journal of Public Health-Revue Canadienne de Sante Publique. 1987;78:300–04. [PubMed] [Google Scholar]
- 16.Jones AW, Andersson L. Comparison of ethanol concentrations in venous blood and end-expired breath during a controlled drinking study. Forensic Sci Int. 2003;132:18–25. doi: 10.1016/s0379-0738(02)00417-6. [DOI] [PubMed] [Google Scholar]
- 17.Lum A, Le Marchand L. A simple mouthwash method for obtaining genomic DNA in molecular epidemiological studies. Cancer Epidemiol Biomarkers Prev. 1998;7:719–24. [PubMed] [Google Scholar]
- 18.Rothman N, Poirier MC, Baser ME, Hansen JA, Gentile C, Bowman ED, et al. Formation of Polycyclic Aromatic Hydrocarbon Dna Adducts in Peripheral White Blood-Cells During Consumption of Charcoal-Broiled Beef. Carcinogenesis. 1990;11:1241–43. doi: 10.1093/carcin/11.7.1241. [DOI] [PubMed] [Google Scholar]
- 19.Cheng G, Shi YL, Sturla SJ, Jalas JR, McIntee EJ, Villalta PW, et al. Reactions of formaldehyde plus acetaldehyde with deoxyguanosine and DNA: Formation of cyclic deoxyguanosine adducts and formaldehyde cross-links. Chem Res Toxicol. 2003;16:145–52. doi: 10.1021/tx025614r. [DOI] [PubMed] [Google Scholar]
- 20.Motzer RJ, Reed E, Perera F, Tang DL, Shamkhani H, Poirier MC, et al. Platinum-Dna Adducts Assayed in Leukocytes of Patients with Germ-Cell Tumors Measured by Atomic Absorbency Spectrometry and Enzyme-Linked-Immunosorbent-Assay. Cancer. 1994;73:2843–52. doi: 10.1002/1097-0142(19940601)73:11<2843::aid-cncr2820731130>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 21.Pieck AC, Drescher A, Wiesmann KG, Messerschmidt J, Weber G, Strumberg D, et al. Oxaliplatin-DNA adduct formation in white blood cells of cancer patients. Br J Cancer. 2008;98:1959–65. doi: 10.1038/sj.bjc.6604387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linderborg K, Joly JP, Visapaa JP, Salaspuro M. Potential mechanism for Calvados-related oesophageal cancer. Food Chem Toxicol. 2008;46:476–79. doi: 10.1016/j.fct.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Mizukami T, et al. Salivary acetaldehyde concentration according to alcoholic beverage consumed and aldehyde dehydrogenase-2 genotype. Alcohol Clin Exp Res. 2008;32:1607–14. doi: 10.1111/j.1530-0277.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Wang MY, Villalta PW, Luo XH, Feuer R, Jensen J, et al. Quantitation of an acetaldehyde adduct in human leukocyte DNA and the effect of smoking cessation. Chem Res Toxicol. 2007;20:108–13. doi: 10.1021/tx060232x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu SQ, Pilone GJ. An overview of formation and roles of acetaldehyde in winemaking with emphasis on microbiological implications. Int J Food Sci Technol. 2000;35:49–61. [Google Scholar]
- 26.Garcia-Closas M, Egan KM, Abruzzo J, Newcomb PA, Titus-Ernstoff L, Franklin T, et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10:687–96. [PubMed] [Google Scholar]
- 27.Pence MG, Blans P, Zink CN, Hollis T, Fishbein JC, Perrino FW. Lesion Bypass of N(2)-Ethylguanine by Human DNA Polymerase i. J Biol Chem. 2009;284:1732–40. doi: 10.1074/jbc.M807296200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrino FW, Blans P, Harvey S, Gelhaus SL, McGrath C, Akman SA, et al. The N-2-ethylguanine and the O-6-ethyl- and O-6-methylguanine lesions in DNA: Contrasting responses from the “bypass” DNA polymerase eta and the replicative DNA polymerase alpha. Chem Res Toxicol. 2003;16:1616–23. doi: 10.1021/tx034164f. [DOI] [PubMed] [Google Scholar]
- 29.Upton DC, Wang XY, Blans P, Perrino FW, Fishbein JC, Akman SA. Replication of N-2-ethyldeoxyguanosine DNA adducts in the human embryonic kidney cell line 293. Chem Res Toxicol. 2006;19:960–67. doi: 10.1021/tx060084a. [DOI] [PubMed] [Google Scholar]
- 30.Garcia CCM, Angeli JPF, Freitas FP, Gomes OF, de Oliveira TF, Loureiro APM, et al. [(13)C(2)]-Acetaldehyde Promotes Unequivocal Formation of 1,N(2)-Propano-2′-deoxyguanosine in Human Cells. J Am Chem Soc. 2011;133:9140–43. doi: 10.1021/ja2004686. [DOI] [PubMed] [Google Scholar]