SUMMARY
Histones are ubiquitinated in response to DNA double strand breaks (DSB), promoting recruitment of repair proteins to chromatin1. UBC13 (UBE2N) is an ubiquitin conjugating enzyme (E2) that heterodimerizes with UEV1a2 and synthesizes K63–linked polyubiquitin (K63Ub) chains at DSB sites in concert with the ubiquitin ligase (E3), RNF1683. K63Ub synthesis is regulated in a noncanonical manner by the deubiquitinating enzyme, OTUB1 (OTU domain-containing ubiquitin aldehyde-binding protein 1), which binds preferentially to the UBC13~Ub thiolester4. Residues N-terminal to the OTU domain, which had been implicated in ubiquitin binding5, are required for binding to UBC13~Ub and inhibition of K63Ub synthesis5. Here we describe structural and biochemical studies elucidating how OTUB1 inhibits UBC13 and other E2 enzymes. We unexpectedly find that OTUB1 binding to UBC13~Ub is allosterically regulated by free ubiquitin, which binds to a second site in OTUB1 and increases its affinity for UBC13~Ub, while at the same time disrupting interactions with UEV1a in a manner that depends upon the OTUB1 N-terminus. Crystal structures of an OTUB1-UBC13 complex and of OTUB1 bound to ubiquitin aldehyde and a chemical UBC13~Ub conjugate show that binding of free ubiquitin to OTUB1 triggers conformational changes in the OTU domain and formation of a ubiquitin-binding helix in the N-terminus, thus promoting binding of the conjugated donor ubiquitin in UBC13~Ub to OTUB1. The donor ubiquitin thus cannot interact with the E2 enzyme, which has been shown to be important for ubiquitin transfer6,7. The N-terminal helix of OTUB1 is positioned to interfere with UEV1a binding to UBC13, as well as with attack on the thiolester by an acceptor ubiquitin, thereby inhibiting K63Ub synthesis. OTUB1 binding also occludes the RING E3 binding site on UBC13, thus providing a further component of inhibition. The general features of the inhibition mechanism explain how OTUB1 inhibits other E2 enzymes4 in a non-catalytic manner.
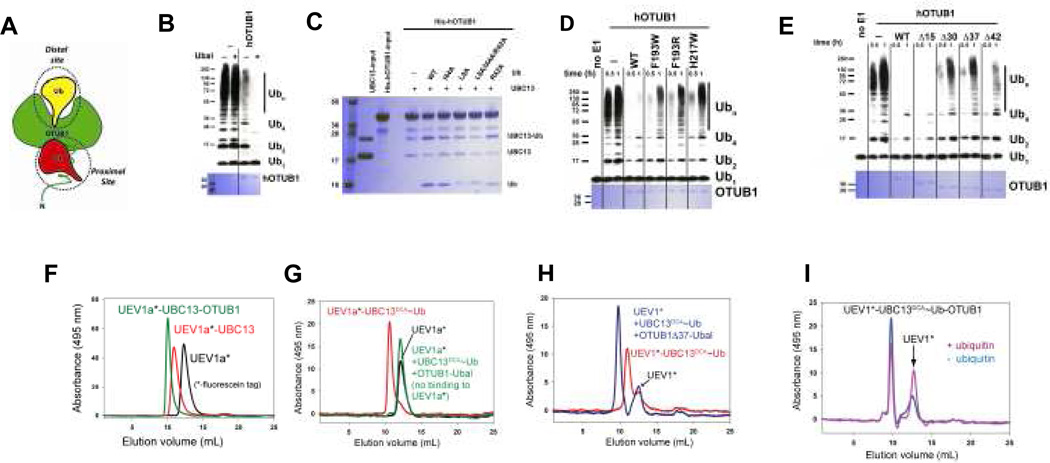
OTUB1 was previously identified as a K48 linkage-specific deubiquitinating enzyme that contains two distinct ubiquitin binding sites (Fig. 1a): a distal site and a proximal site that includes the ~45 N-terminal residues of OTUB15. These residues are important for OTUB1 inhibition of E2 activity4 and are absent in OTUB2, which does not inhibit UBC134. It was previously shown that binding of the covalent inhibitor, ubiquitin aldehyde (Ubal), to the distal ubiquitin-binding site of OTUB1 stimulates binding of ubiquitin vinyl sulfone to N-terminus5. Since the OTUB1 N-terminus was implicated in binding to the donor ubiquitin in the UBC13~Ub conjugate4, we asked whether Ubal binding to OTUB1 could enhance inhibition of UBC13 by stimulating binding of the OTUB1 N-terminus to the donor ubiquitin. The results (Fig. 1b) revealed a dramatic enhancement of the ability of OTUB1 to suppress K63Ub synthesis, suggesting that Ubal is an allosteric effector that increases the affinity of the OTUB1 N-terminus for the ubiquitin in the UBC13~Ub thiolester. This prompted us to ask whether free ubiquitin binding to the OTUB1 distal site could similarly stimulate binding of OTUB1 to UBC13~Ub conjugates. To test this, we generated a mixture of charged and uncharged UBC13C87S, which forms a more stable UBC13~Ub oxyester, purified away the free ubiquitin, and performed pull-down assays with H6-OTUB1 in the presence and absence of added free ubiquitin. Remarkably, OTUB1 shows no preference for the charged UBC13~Ub in the absence of ubiquitin, whereas addition of 100 µM free ubiquitin greatly enhances OTUB1 binding to UBC13~Ub, but not to uncharged UBC13 (Fig. 1c). By contrast, ubiquitin bearing hydrophobic patch mutations I44A, L8A or L8A/I44A/R42A (but not R42A alone) do not stimulate OTUB1 binding to UBC13~Ub like wild type ubiquitin (Fig. 1c). The relative binding of OTUB1 to UBC13~Ub increases as the concentration of free ubiquitin is increased from 2 to 50 µM (Supplementary Fig. S2). To verify that ubiquitin binding to the distal site of OTUB1 is important for inhibition of UBC13, we assayed the effect of distal site mutations, which were chosen based on structures of a covalent yOTU1-ubiquitin complex8 and of human OTUB19. Distal site substitutions F193W, F193R and H217W disrupted the ability of OTUB1 to inhibit polyubiquitination by UBC13/UEV1a (Fig. 1d) without affecting binding of OTUB1 to UBC13 (Supplementary Fig. S3). Taken together, our results indicate that the ability of OTUB1 to bind preferentially to the UBC13~Ub conjugate and inhibit ubiquitin transfer is allosterically regulated by free ubiquitin binding to the distal site of OTUB1, which triggers capture of the conjugated ubiquitin in the OTUB1 proximal site.
Figure 1. Allosteric regulation of OTUB1 by ubiquitin.
A. Schematic diagram of OTUB1 illustrating proximal and distal ubiquitin binding sites.
B. Effect of ubiquitin aldehyde (Ubal) on the ability of hOTUB1 to inhibit K63 polyubiquitin synthesis by UBC13/UEV1a. Assays include 0.1 µM E1, 0.4 µM UBC13/UEV1a, 0.5 µM hOTUB1, 5 µM ubiquitin. The 3 hr time point is shown in the presence (right) and absence (left) of hOTUB1, without (−) and with (+) 0.5 µM Ubal. Top shows detection by anti-Ub Western blot; Coomassie staining below shows level of hOTUB1.
C. Pull down assay showing binding of H6-tagged hOTUB1 to a mixture of UBC13 and UBC13~Ub oxyester in the presence and absence 100 µM free ubiquitin (WT or mutant).
D. Effect of hOTUB1 distal site mutations on inhibition of K63Ub synthesis. Assay performed as in (B) but with 1 µM OTUB1, showing 0.5 and 1 hr time points.
E. Effect of hOTUB1 N-terminal deletions of 15, 30, 37, and 42 residues on inhibition of K63Ub synthesis by UBC13/UEV1a. Assay performed as in (D).
F. Gel filtration showing complex formation between fluorescein-labelled UEV1a (UEV1a*), UBC13 and hOTUB1. Signal due to UEV1a only was monitored at 495 nm.
G. Experiment performed as in (F) showing complex formation of UEV1a*, UBC13DCA~UbG75C and hOTUB1 in the absence (red) and presence (green) of Ubal.
H. Experiment performed as in (F) but with hOTUB1Δ37. The position at which free UEV1* migrates is indicated.
I. Experiment performed as in (F) with fluorescein-labeled UEV mixed with UBC13DCA~UbG75C and OTUB1 samples prepared in the presence and absence of 200 µM ubiquitin. The position at which free UEV1* migrates is indicated.
Since ubiquitin aldehyde most likely enhances interactions between the OTUB1 N-terminus and the donor ubiquitin in UBC13~Ub, we examined the effect of N-terminal deletions in OTUB1 to delimit the minimal fragment needed for binding and inhibition. Deletion of residues 1–15 has no effect on inhibition of K63Ub synthesis by UBC13/UEV1a (Fig. 1e) while deletion of 30, 37 or 42 residues significantly disrupts inhibition. The OTUB1Δ15 deletion similarly behaves like full-length OTUB1 in pull-downs with the UBC13~Ub conjugate while larger deletions exhibit defects (Supplementary Fig. S4), indicating that N-terminal residues 16–45 are sufficient for activity.
Since a UEV (ubiquitin E2 variant) must bind to UBC13 and position the acceptor ubiquitin in order for K63Ub synthesis to occur10, we asked whether OTUB1 could bind to UBC13 in the presence of UEV1a. In gel filtration assays using fluorescently labeled UEV1a, OTUB1 and uncharged UBC13 migrate as a ternary complex with UEV1a (Fig. 1f). To assay binding to charged UBC13, we generated a non-hydrolyzable conjugate in which Ub with a C-terminal G75C is covalently linked to the active site cysteine of UBC13 with dichloroacetone (DCA) 11. UEV1a binds to UBC13DCA~Ub but OTUB1-Ubal interferes with UEV1a binding to the UBC13~Ub conjugate (Fig. 1g). By contrast, the N-terminal deletion, OTUB1Δ37, can still form a complex with UBC13DCA~Ub and labeled UEV1 in the presence of Ubal (Fig. 1h), suggesting that the N-terminus of OTUB1 competes with UEV binding when OTUB1 is bound to Ubal. We verified that free ubiquitin has a similar effect on UEV binding by comparing migration of a sample containing labeled UEV1, UBC13~Ub and OTUB1 prepared in the presence and absence of free ubiquitin and found that the ratio of free UEV1 to UEV1-UBC13DCA~Ub-OTUB1 increases when ubiquitin is present (Fig. 1i). Similarly, pull-downs with H6-OTUB1 do not show an enhancement in co-precipitation of UEV1a along with UBC13~Ub in the presence of added free ubiquitin (Supplementary Fig. S5). These results suggest that the N-terminus of OTUB1 interferes with UEV binding and thus with K63Ub synthesis, and that the ability of the N-terminus to interfere with UEV depends upon a conformational change that is triggered by binding of free ubiquitin to OTUB1.
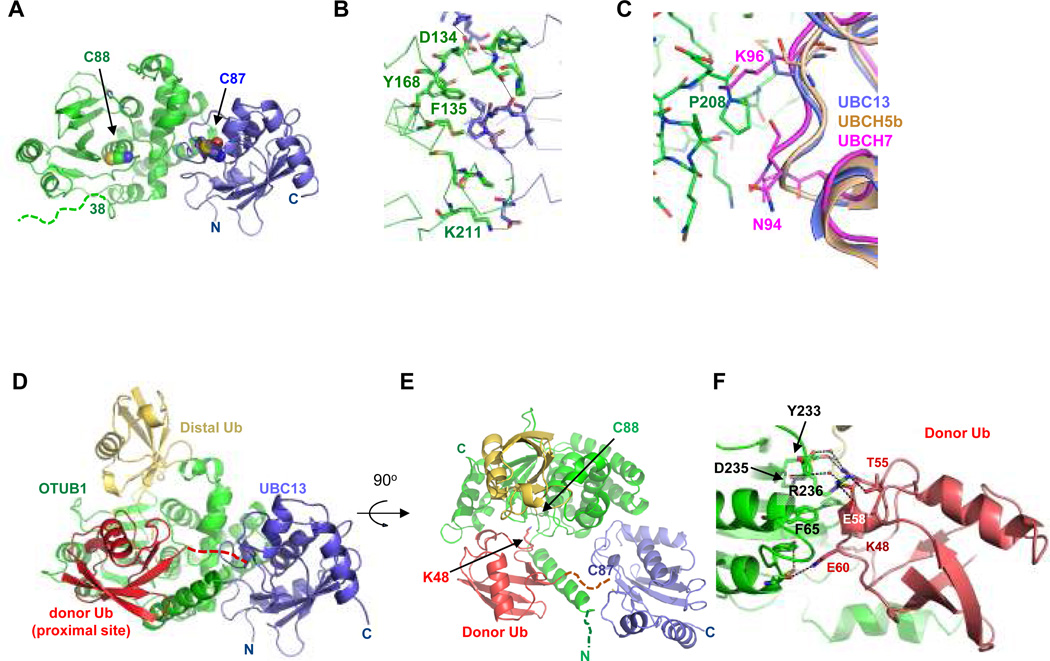
In order to determine the structural basis for OTUB1 inhibition of E2 enzymes, and how ubiquitin allosterically regulates OTUB1 activity, we determined the structure of Caenorhabditis elegans OTUB1 (ceOTUB1) bound to human UBC13 at 1.8 Å resolution (Fig. 2a), and a 2.35 Å resolution quaternary complex structure containing ceOTUB1, Ubal and a UBC13DCA~Ub conjugate generated with UbG76C. The resulting non-native linkage is four bond lengths longer than the native thioester (Supplementary Fig. S6). Human UBC13 is 89% identical to ceUBC13, while human OTUB1 (hOTUB1) shares 34% sequence identity and 56% similarity with ceOtub1 (Supplementary Fig. S7) and inhibits K63Ub chain formation by human UBC13/UEV1a (Supplementary Fig. S8a). CeOTUB1 is a weaker inhibitor of UBC13, as reflected in its higher Kd of 58.5 µM as compared to 7.04 µM for hOTUB1 (Supplemental Fig. S8b). Crystals of the ceOTUB1-Ubal-UBC13DCA~Ub complex contain four complexes in the P212121 asymmetric unit. The ubiquitin conjugated to UBC13 could be unambiguously positioned in two of the four complexes (Supplementary Fig. S9); our discussion focuses on the complex with the most well-ordered ubiquitin (complex 1). Since the N-terminus of OTUB1 that plays a key role in inhibition is poorly conserved between human and ceOTUB1, we also determined the 3.1 Å resolution structure of a quaternary complex with a hybrid OTUB1 containing the N-terminal 45 residues of human OTUB1 and the OTU domain of ceOTUB1 (Supplementary Fig. S7b). The hybrid h/ceOTUB1 inhibits K63Ub synthesis by UBC13/UEV1a (Supplementary Fig. S10). Details on all structure determinations are in Supplementary Methods and statistics are in Supplementary Table S1.
Figure 2. Structure OTUB1-UBC13 and OTUB1-Ubal-UBC13DCA~Ub.
A. Complex of ceOTUB1 (green) bound to human UBC13 (blue). Respective active site cysteines are shown as space-filling representations. Dashed line indicates disordered residues.
B. Contacts at ceOTUB1 (green) -UBC13 (blue) interface.
C. Superposition of UBCH5b (UBE2D2, PDB ID 2ESK) and UBCH7 (UBE2L3, PDB ID 1FBV) with UBC13 in the complex with ceOTUB1. UBCH7 contains an insertion (at N94) and a lysine (K96) that would interfere with binding.
D. Structure of hybrid h/ceOTUB1 (green) bound to Ubal (distal Ub, yellow), UBC13 (blue) and ubiquitin (proximal Ub, red) that is covalently linked to the active site cysteine (C87) of UBC13 by a DCA linkage. Dashed line indicated disordered C-terminal residues 73–76 of the donor ubiquitin and DCA linkage.
E. A 90° rotation compared to (D) showing positions of ceOTUB1 and UBC13 active site cysteine and modeled location of K48 of the proximal ubiquitin.
F. Contacts between the donor ubiquitin (red) and the OTU domain (green) in the ceOTUB1-Ubal-UBC13DCA~Ub complex.
In the structure of apo ceOTUB1 bound to UBC13 (Fig. 2a), the OTU domain of ceOTUB1 binds to UBC13 in an orientation that places their respective active site cysteines 28 Å apart on the same face of the complex, burying 1281 Å2 of total surface area. Of the 12 ceOTUB1 side chains at the interface with UBC13 (Fig. 2b), seven are identical in hOTUB1 and four are similar (Supplementary Fig. S7a) and can mediate comparable interactions with UBC13. Consistent with this, the double substitution Y170A/F138A in hOTUB1 (Y168A/F135A in ceOTUB1) is defective in binding to UBC13 (Supplementary Fig. S11). Similar interactions could form between OTUB1 and UBE2D2 (UBCH5b) (Fig. 2c), but clashes due to an insertion and a non-conserved lysine would arise with UBE2L3 (UBCH7), consistent with the observation that OTUB1 inhibits UBCH5b but not UBCH7 4.
An overview of the h/ceOTUB1-Ubal-UBC13DCA~Ub complex is shown in Figures 2d and 2e. Ubal binds to the OTUB1 distal site while the donor ubiquitin in the UBC13~Ub conjugate binds in the OTUB1 proximal site, which comprises residues in both the OUT domain and the N-terminus. In the absence of bound ubiquitin, the ceOTUB1 N-terminus (residues 1–37, corresponding to hOTUB1 residues 1–39) is disordered (Fig. 2a). However, in the OTUB1-Ubal-UBC13DCA~Ub complexes, part of the N-terminus of OTUB1 becomes ordered, forming an ubiquitin-binding helix that contacts the donor ubiquitin in the distal site (Fig. 2e). Additional contacts with the donor ubiquitin are mediated by the OTU domain, which, as described below, undergoes a set of conformational changes triggered by Ubal binding to the distal site.
The donor ubiquitin binds to the proximal site of OTUB1 (Fig. 2d) in an orientation that places K48 of the ubiquitin near the OTUB1 active site (Fig. 2e). A K48 isopeptide linkage can be modeled between the proximal and distal ubiquitins, consistent with OTUB1 isopeptidase specificity for K48-linked diubiquitin5. Residues 73–76 of the donor ubiquitin and the DCA linkage are not visible in the electron density map, indicating that they do not adopt a unique conformation in the crystal. The distance between the C-terminal ubiquitin residue and the active site cysteine is ~12.5 Å, which is sufficient to accommodate the four missing residues and a native thiolester linkage. The ubiquitin interface with the OTU domain buries 850 Å2 of surface area. Ubiquitin side chains that lie between residues 54–60 contact the OTU domain, forming both direct and water-mediated hydrogen bonds and van der Waals interactions (Fig. 2f). Three of the contacting ceOTUB1 side chains are R236, Y233 and D235, which are only in a position to contact ubiquitin in the quaternary complex.
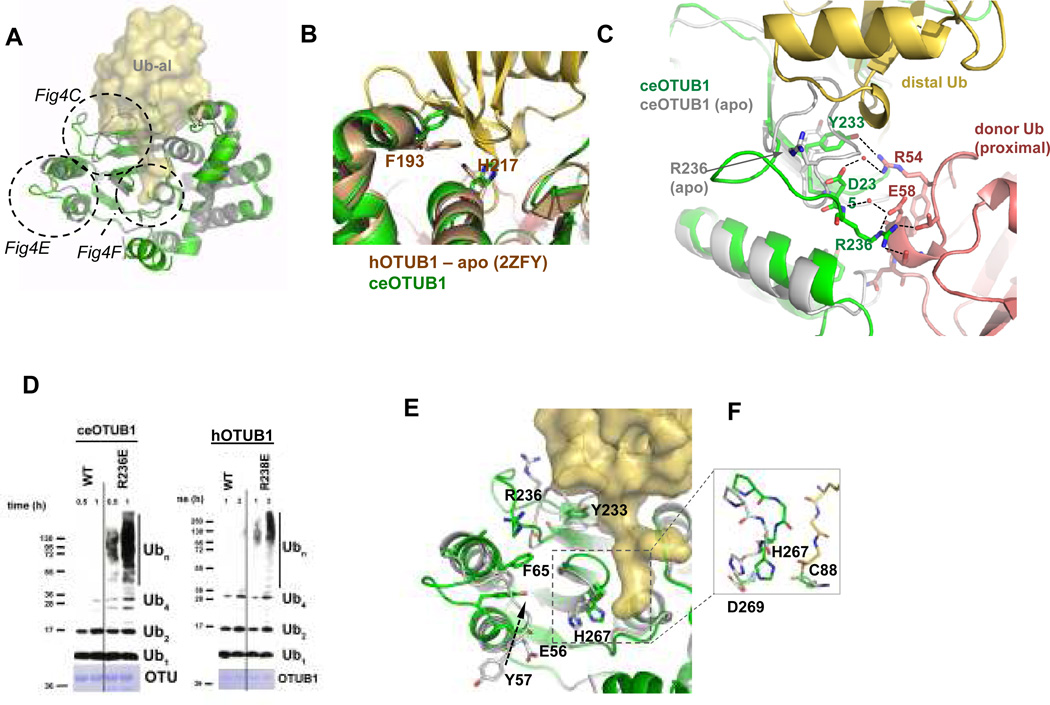
The observed contacts between the donor ubiquitin and the OTU domain depend upon distal site binding of Ubal, which forms a covalent bond with the active site cysteine (Supplementary Fig. S12) and triggers conformational changes in three regions of the globular OTU domain (Fig. 3a). Ubal binds to the distal ubiquitin binding site of OTUB1 (Fig. 3b) in a manner similar to yeast8 and viral12–14 OTU enzymes, and accounts for the effects of mutations in the OTUB1 distal site (Fig. 1d). A loop (residues 235–245) that partially occludes the distal site in the absence of ubiquitin undergoes a large rearrangement that relieves steric clashes with the distal ubiquitin and positions R236 of ceOTUB1 to make multiple contacts with the donor ubiquitin bound in the proximal site of OTUB1 (Figs. 3c). In the structure of apo hOTUB19, this residue is disordered (backbone and side chain atoms) and lies in a loop that presumably changes conformation upon distal ubiquitin binding. Mutating the conserved arginine to glutamic acid in both human (R238E) and C. elegans (R236E) OTUB1 disrupts inhibition (Fig. 3d), consistent with its role in binding the donor ubiquitin. Interestingly, the corresponding residue is a glutamic acid in OTUB2, which lacks an N-terminal arm and does not inhibit UBC134. Y233, which occludes the distal site in apo ceOTUB1 and undergoes a conformational change to hydrogen bond with the distal Ub (Fig. 3c), is conserved in hOTUB1 (Supplementary Fig. S7a). Another set of conformational changes in the loop connecting helices 1 and 2 of OTUB1 flips the solvent exposed Y57 side chain into the interior of the OTU domain, where it stacks between F65 and E56 (Fig. 3e). The altered loop conformation relieves steric clashes with the donor ubiquitin that would otherwise occur. Binding of the distal ubiquitin is accompanied by additional local rearrangements that narrow the binding cleft around the ubiquitin C-terminal tail (Fig. 3e) and moves the ceOTUB1 active site histidine, H267, into a position between D269 and C88 to activate the cysteine for catalysis (Fig. 3f).
Figure 3. Conformational changes in the OTU domain triggered by Ubal binding.
A. Superposition of ceOTUB1 (green) bound to Ubal (yellow surface) with the structure of apo ceOTUB1(gray). Dotted circles indicate regions of conformational change, which are illustrated in the figure panels noted.
B. Location of hOTUB1 distal site mutations that affect inhibition. The structure of hOTUB1 (2ZFY; brown) is superimposed on ceOTUB1 (green) – Ubal (yellow). Ubiquitin residues L8 and I44, where substitutions with alanine disrupt allosteric effect of ubiquitin binding, are shown. View is 180° rotation about vertical compared with (A).
C. Structural differences in the OTU domain in the presence (green) and absence (gray) of distal Ub that affect contacts with the donor Ub. Side chain conformations for ceOTUB1 in presence and absence of Ubal are shown for R236 and Y233. Dotted lines indicate hydrogen bonds and salt bridges. View shown is from “top” of complex as shown on right of panel (A), rotated 90° counterclockwise.
D. Effect of mutating OTUB1 conserved arginine, ceOTUB1-R236E and hOTUB1-R238E, on inhibition of UBC13/UEV1a. Assay performed as in panel 1A, with 1 µM hOTUB1 and 15 µM ceOTUB1.
E. View of OTUB domain structural rearrangements colored as in (C). View as in panel (A); proximal ubiquitin not shown.
F. Detailed view of catalytic triad in the presence and absence of Ubal (carbon colored as in (C).
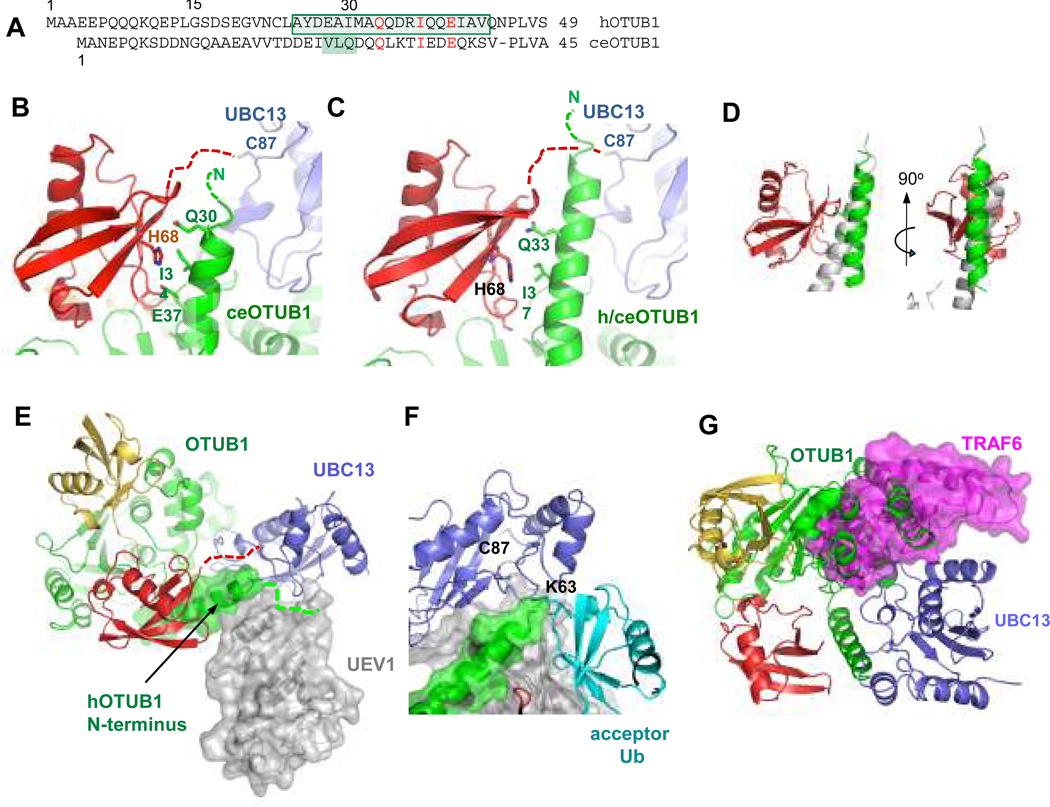
The OTUB1 N-terminal ubiquitin-binding helix seen in the structure spans residues 28 – 39 of ceOTUB1 (complex 1) and 25 – 44 of hOTUB1 (Figs. 4a–4c), burying 542 Å2 and 626 Å2, respectively on the donor ubiquitin (electron density shown in Supplementary Fig. S13). The helix interacts with the donor ubiquitin in a manner reminiscent of the RAP80 UIM15 (Fig. 4d). Despite limited sequence identity between the ceOTUB1 and hOTUB1 N-terminus (Fig. 4a), the three side chains that contact the donor ubiquitin in the 2.35 Å resolution structure of ceOTUB1 (Fig. 4b) are conserved in hOTUB1 (Fig. 4a) and are oriented towards ubiquitin in the same manner in the 3.1 Å resolution h/ceOTUB1 structure (Fig. 4c). In the ceOTUB1 complex (Fig. 4b), residues E37 and I34 contact donor ubiquitin residue H68 while Q33 interacts with backbone atoms. In the structure containing the human N-terminus, the helix extends beyond the donor ubiquitin and approaches the UBC13 active site cysteine (Fig. 4c). It is possible that additional residues may be ordered when the complex is in solution, as nine residues from the minimal OTUB1Δ15 fragment that exhibits full activity (Fig. 1d) are missing from the h/ceOTUB1 complex structure. It is not clear whether the shorter helix observed in the ceOTUB1 complex reflects a structural difference in solution, or whether crystal contacts interfere with helix formation. The close approach of the OTUB1 N-terminus to the donor ubiquitin C-terminus in both complexes (Figs. 4b and 4c) leaves open the possibility that additional contacts may form with the donor ubiquitin tail linked to UBC13 via a native thioester.
Figure 4. OTUB1 N-terminal arm and the mechanism of E2 inhibition.
A. Sequence alignment of N-terminal arms of hOTUB1 (top) and ceOTUB1 (bottom). Boxed residues form a helix in the quaternary complex structures containing Ubal and UBC13DCA~Ub; additional shaded residues in ceOTUB1 are ordered in complex 1 but are not helical.
B. Donor Ub (red) interactions with the ceOTUB1 N-terminal helix (green); Ubc13 shown in blue. Dashed lines indicate disordered residues.
C. Interactions with the hOTUB1 N-terminal helix of the h/ceOTUB1 hybrid, depicted as in (B).
D. Superposition comparing RAP80 (3A1Q) binding to ubiquitin with hOTUB1 N-terminal helix. Two views are shown.
E. Superposition of h/ceOTUB1-Ubal-UBC13DCA~Ub with UBC13/UEV1 (1J7D) showing predicted position of UEV1 (gray). The solvent-accessible surface of the human N-terminal arm residues of OTUB1 is depicted.
F. Superposition with quaternary complex showing relative position of the TRAF6 E3 ligase (3HCT).
The structures show how OTUB1 interferes with UEV binding and positioning of the acceptor ubiquitin, and also occludes the RING E3 binding site. Figure 4e shows a superposition with the structure of a UBC13/UEV1 complex16 showing that N-terminal helix of hOTUB1 clashes with the expected location of UEV1. Modeling of the predicted position of the acceptor ubiquitin based on the structure of yeast Ubc13~Ub/Mms217 shows the N-terminus of OTUB1 in a position to interfere with attack by the acceptor ubiquitin lysine on the thiolester (Fig. 4f). Since OTUB1 also inhibits UBCH5b4, which does not function with a UEV, we speculate that the OTUB1 N-terminus may also interfere with acceptor ubiquitin binding for other E2s. The repositioning of the donor ubiquitin away from the E2 likely also contributes to inhibition, in light of evidence that the donor ubiquitin in the E2~Ub thiolester interacts specifically with the E218,19 and that this is essential for ubiquitin transfer 6,7. In addition, superposition with the structure of UBC13 bound to TRAF620 shows that the OTUB1 binding site overlaps with the E3 RING-binding site (Fig. 4g), indicating that competition between OTUB1 and RNF168 would further suppress UBC13 activity in vivo. Competition with E3 binding is likely to be particularly important for OTUB1 inhibition of UBCH5b, which, unlike UBC13, is strictly dependent upon an E3 ligase for activity.
The ability of OTUB1 to serve as both an isopeptidase and an inhibitor of E2 enzyme activity arises from its ability to bind to selected E2s, while taking advantage of the allosteric communication between the proximal and distal ubiquitin binding sites of OTUB1 and the distinctive features of its N-terminus. Given the high degree of coupling between the multiple binding interactions within the OTUB1-Ub-UBC13~Ub complex, the degree of inhibition in vivo will clearly depend upon the relative concentrations of OTUB1, E2~Ub thiolester, E3 and free ubiquitin in the cell. An interesting question is whether the dependence of OTUB1 repression on ubiquitin binding to the distal site is exploited to modulate OTUB1 activity in response to fluctuations in the concentration of free ubiquitin or of free chains, whose C-terminal subunits could similarly bind to the distal site of OTUB1. Our findings establish new directions for investigating how the allosteric regulation of OTUB1 may be exploited to regulate ubiquitination in the DNA damage response.
Methods Summary
Cloning, expression, protein purification and crystallization are described in Methods and in accompanying references5. The DCA linkage between the active site cysteine of UBC13 and a C-terminal cysteine in UbG75C or UbG76C was generated by a modification of the published method 11. The hybrid h/ceOTUB1 protein contains residues 1–45 of hOTUB1 and residues 43–276 of ceOTUB1. Structures were determined by molecular replacement as described in full Methods. Free ubiquitin chain synthesis was assayed by gel electrophoresis and products were detected by Western blot with anti-ubiquitin antibody or by Coomassie staining. Pull-down assays were performed with purified recombinant protein. Assays of complex formation between OTUB1, UBC13, UBC13DCA~Ub and UEV1a were performed by gel filtration with fluorescein-labeled UEV1a or UEV1, monitoring fluorescein absorbance at 495 nm. Binding of OTUB1 to UBC13 was measured by fluorescence anisotropy using fluorescently labeled UBC13, and equilibrium dissociation constants were calculated using SigmaPlot (SPSS).
Methods
Cloning and mutagenesis
Cloning of human and C. elegans OTUB1 (hOTUB1 and ceOTUB1, respectively) was performed as described previously5. The human UBC13 open reading frame was amplified from a human cDNA library and cloned into a pET vector containing N-terminal His6-SUMO-2 tag (pETSUMO-2) The human UEV1a ORF was synthesized (IDT) and subcloned into the pETSUMO-2 vector as above. The human UEV1 (missing the first 30 residues of UEV1a) expression plasmid was purchased from Addgene (www.addgene.org)
Mutants of hOTUB1 were generated by site-directed mutagenesis using the Quick Change® mutagenesis kit (Stratagene Inc.) following the manufacturer’s protocol. The hybrid h/ceOTUB1 was generated by swapping the first 41 residues of ceOTUB1 with the first 45 residues of hOTUb1 using Infusion® ligase-free cloning (Clonetech Inc., USA) hOTUB1 with an N-terminal 41-residue truncation (OTUB1ΔN41) was generated as previously described5, all other OTUB1 deletions were generated using Infusion® ligase-free cloning (Clonetech Inc., USA)
Protein expression and purification
All proteins were expressed in E. coli Rosetta-2 (DE3) cells grown in Luria-Bertani (LB) medium. Cultures (6×1L) were inoculated using 1% (v/v) overnight saturated cultures and were grown at 37°C to an OD600 of 0.8. Proteins were induced at 16°C overnight by addition of 1 mM isopropyl-β-D-thio-galactoside (IPTG). Cells were harvested by centrifugation (8000×g, 15 min) and either lysed immediately or stored at −80°C for later use.
hOTUB1, ceOTUB1, human E1 enzyme and ubiquitin were purified as previously described 5,21. Deletions and mutants of human and C. elegans OTUB1 and of ubiquitin were purified according to the same protocol as the WT proteins. UBC13 and UEV1a were purified by resuspending cell pellets in lysis buffer (20 mM Hepes pH 7.3, 300 mM NaCl, 10 mM imidazole, 2 mM β-mercaptoethanol) after adding 0.1 mM phenyl-methyl sulfonyl fluoride (PMSF). Cells were disrupted using a Microfluidizer (Microfluidics Inc., Waltham, MA) and the lysate was centrifuged to remove cell debris. The lysate was subjected to immobilized metal affinity chromatography (IMAC) using 5ml His-Trap columns (GE Biosciences) developed with a linear imidazole gradient of 25–400 mM in 20 column volumes. Fractions containing purified protein were pooled, SENP-2 protease was added in a ratio of 1:100 to cleave off the His-SUMO-2 tag, and pooled fractions were dialyzed overnight at 4°C against lysis buffer. Cleaved protein was then subjected to a second round of IMAC and the cleaved protein was collected from the flow-through. Proteins were then purified by gel filtration on a Superdex 75 column (GE Healthcare), dialyzed into 20 mM HEPES, pH 7.3, 150mM NaCl, and 1mM DTT, concentrated, and stored at −80°C. Proteins for crystallization, enzyme assays and binding studies were > 98% pure as visualized on a Coomassie-stained gel. His6-tagged hOTUB1 used in pull-down assays was ~90% pure.
Protein modifications
UBC13, UEV1a and UEV1 were labeled with fluorescein-5-maleimide (Invitrogen) as described in the manufacturer’s protocol. Ubiquitin aldehyde was prepared as described22.
Preparation of UBC13~Ub conjugates
UBC13C87S~Ub oxyester was prepared as previously described 17. The UBC13DCA~Ub covalent conjugate was prepared according to a modification of the protocol by Yin et al.11. Purified ubiquitin containing the substitution G76C (UbG76C) or G75C (UbG75C) and UBC13 were dialyzed separately overnight into 20 mM sodium borate buffer, pH 8.0. and 2 mM TCEP (tris(2-carboxyethyl)phosphine), mixed in the proportion of 1 mM UbG76C or UbG75C to 330 µM UBC13, and incubated on ice for 15 minutes. A stock of 20 mM 1,3-dichloroacetone (DCA) was prepared in dimethylformamide (DMF) and added to the conjugation reaction to a final concentration of 0.8 mM DCA. The reaction was stopped after 1 hr by addition of 10 mM β-mercaptoethanol. The coupling efficiency was approximately 50%. For the UbG76C reaction, mix was diluted 10-fold with 10 mM Tris, pH 8, loaded onto a mono Q column (GE Healthcare) pre equilibrated with 10 mM Tris, pH 8. Free UbG76C eluted in the flow-through and UBC13DCA~Ub eluted together with unconjugated UBC13 with 180 mM NaCl in 20 mM Tris, pH 8. For the UbG75C reaction, UBC13DCA~Ub(G75C) was purified by gel filtration on a Superdex 75 column pre-equilibrated with 20 mM HEPES pH 7.3, 100 mM NaCl and 2 mM DTT. The separation efficiency was about 10 % of the total amount of UBC13DCA~Ub(G75C) in the reaction mix.
Purification of ceOTUB1-Ubal-UBC13DCA~UbG76C quaternary complex
ceOTUb1 was incubated on ice with Ubal in a 1:4 molar ratio for 15 min and added to the purified apo hUBC13 and UBC13DCA~Ub mixture such that UBC13DCA~Ub was in two-fold excess over ceOTUB1, as estimated by gel electrophoresis. The mixture was incubated for another 15 min on ice and loaded onto a Superdex 200 column (GE Healthcare) pre-equilibrated with 20 mM Tris, pH 7.45, 150 mM NaCl and 2 mM DTT. The OTUB1-Ubal-UBC13DCA~Ub complex eluted as a single peak and was concentrated to 10 mg/ml and stored at −80°C.
Crystallization
All crystals were grown by the hanging drop vapor diffusion method at 20°C. A ceOTUB1/hUBC13 complex was prepared by incubating ceOTUB1 and hUBC13 at a molar ratio of 1:1 and total protein concentration of 26 mg/ml for 10 min at room temperature. Crystals were grown from a 1:1 mix of protein and well solution containing 100 mM sodium cacodylate, pH 6.5 and 1M trisodium citrate and appeared in about 2–3 days. Crystals were transferred to cryoprotectant consisting of well solution plus 20% ethylene glycol and then flash frozen in liquid nitrogen.
Crystals of the ceOTUB1-Ubal/hUBC13DCA-Ub complex were grown from a 1:1 mix of complex (10 mg/ml) with well solution containing 100 mM Bis-Tris pH 6.5, 23% PEG 3350 and 0.26–0.3M sodium chloride. Crystals appeared in about 1–2 days, were cryoprotected by well solution with added 15% ethylene glycol and then flash-frozen in liquid nitrogen.
Crystals of the h/ceOTUB1-Ubal/hUBC13DCA-Ub complex were grown from a 1:1 mix of complex (10 mg/ml) with well solution containing 100 mM MES pH 6.5, 21% PEG 10,000 and 0.1M sodium chloride. Crystals appeared in about 2–3 days, were cryoprotected by well solution with added 15% ethylene glycol and then flash frozen in liquid nitrogen
Structure Determination
Diffraction data were recorded at the GM/CA-CAT beamline 23-ID-D/B at the Advanced Photon Source under standard cryogenic conditions and processed with iMOSFLM23 for ceOTUB1/hUBC13 crystals and HKL200024 for the ceOTUB1-Ubal/hUBC13DCA~Ub crystals. For the ceOTUB1-Ubal/hUBC13DCA~Ub structure two data sets were collected from a single crystal, merged and processed with HKL200024. All data were collected at a wavelength of 1.033 Å. The structure of ceOTUB1/hUBC13 was determined by molecular replacement with Phaser25 using structures of UBC13 (1J7D) and apo hOTUB1 (2ZFY). The structure of ceOTUB1-Ubal/hUBC13-Ub was determined by molecular replacement with Molrep26 using structures of the ceOTUB1/hUBC13 complex and ubiquitin from 2GMI. The initial molecular replacement search was performed with the ceOTUB1/hUBC13 complex and located four complexes found in the asymmetric unit. The resulting positions of the ceOtub1/hUBC13 complexes were then fixed and ubiquitin was used as search model to locate the four ubiquitin aldehydes in the crystal. The position of the ceOTUB1-Ubal/hUBC13 complex was fixed and another search with ubiquitin (1–71) located two molecules of donor ubiquitin in the asymmetric unit. The structure of h/ceOTUB1-Ubal/hUBC13-Ub was determined by molecular replacement with Phaser using one complex of ceOTUB1-Ubal/hUBC13-Ub lacking the first 42 residues of ceOTUB1.
All structures were subjected to multiple rounds of manual correction and refinement using COOT27 and Phenix28. The final stages of refinement for the ceOTUB1-Ubal/hUBC13DCA~Ub complex and h/ceOTUB1-Ubal/hUBC13DCA~Ub ternary complex were done using REFMAC529. Simulated annealing omit maps were calculated with CNS30 and used to verify selected portions of the model.
The final model of ceOTUb1-hUbc13 complex includes residues 38–275 of ceOTUb1 and 3–152 of hUBC13. The final model of ceOTUB1-Ubal/hUBC13-Ub includes four complexes in the asymmetric unit: two containing all four proteins (ceOTUB1, Ubal, hUBC13 and Ub) the two lacking the donor ubiquitin conjugated to UBC13. There is no density in any of the complexes corresponding to the five C-terminal amino acids of ubiquitin or to the DCA linkage, which connects ubiquitin to the hUBC13 active site cysteine. The number of ceOTUB1 N-terminal residues visible in the map differs among the four complexes as follows : complex 1, 28–275, complex 2, 31–275, complex 3, 36–276, complex 4, 38–276. The final model of the h/ceOTUb1-Ubal/hUBC13-Ub complex includes residues 20–275 of h/ceOTUb1, 3–151 of hUBC13, 1–76 of Ubal and 1–72 of ubiquitin.
Protein-protein interaction surfaces were analyzed using the PISA server at EBI (www.pdbe.org/PISA) and manually inspected using COOT and PYMOL (www.pymol.org). Figures were generated with PYMOL.
Fluorescence polarization binding assay
Fluorescein-labeled hUBC13 (20 nM) was incubated with increasing concentrations of hOTUB1 WT or mutants in 20 mM Tris, pH 7.6, 150 mM NaCl and 10 mM β-mercaptoethanol. Polarization measurements were recorded at 25 °C with an ISS Chronos Fluorescence Lifetime Spectrometer at excitation and emission wavelengths of 492 and 520 nm, respectively. Binding data were analyzed and KD values were calculated by nonlinear regression in SigmaPlot (SPSS).
UEV Binding assay
UEV1a and UEV1 were fluorescein-labeled; all other proteins are unlabeled. The experiment was performed with analytical Superdex 75 column pre-equilibrated with 20 mM HEPES pH 7.3, 100 mM NaCl and 2 mM DTT. Absorbance was detected at 495 nm to monitor the presence of labeled UEV1a or UEV1. For each run, proteins were incubated for 20 min on ice before loading onto the column with 100 µL loop. The protein concentrations used in the different experiments were:
In vitro ubiquitination assay
Ubiquitination assays were performed in 25 mM Tris-HCl (pH 8.0) buffer containing 0.1 mM DTT, 1 mM ATP, 2.5 mM MgCl2, 5 mM creatine phosphate, 0.3 units ml−1 inorganic pyrophosphatase, and 0.3 units ml−1 creatine kinase. Proteins in the amounts of 0.4 µM UBC13, 0.4 µM UEV1a and 5 µM ubiquitin were mixed with hOTUB1 (1µM) or ceOTUB1(15 µM). Reactions were initiated by the addition of 0.1 µM E1 enzyme, incubated at 37 °C, and stopped at different time points by adding denaturing SDS-PAGE loading dye containing β-mercaptoehtanol (BME). For Figure 1b, 0.5 µM hOTUB1 was incubated with 0.5 µM Ubal for 15 min before addition to the reaction. Reaction products were separated on a 4–12% Bis-Tris NuPAGE (Invitrogen) gel and transferred to a Polyvinylidene fluoride (PVDF) membrane. Membranes were denatured in a 6 M guanidine HCl, 20 mM Tris-HCl, pH 7.5, 1 mM PMSF, 5 mM β-mercaptoethanol solution for 30 min at 4 °C and then washed extensively in Tris-buffered saline and Tween 20 (TBST). Membrane were blocked overnight at 4 °C with 5% BSA in TBST and incubated for 1h with ubiquitin antibody (P4D1 Santa Cruz) 1:1000 at room temperature followed by anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibody. OTUB1 was detected with Coomassie brilliant blue or SimplyBlue™ SafeStain (Invitrogen).
Pull-down assays
Ni2+-NTA beads were equilibrated in Buffer A (50 mM phosphate buffer pH 8.0, 100 mM NaCl, 5 mM β-mercaptoethanol and 10 mM Imidazole). 6XHis-hOTUB1 (30 µg) was incubated with pre-equilibrated beads in 200 µl of buffer A for 30 min. Beads were washed with 400 µL Buffer A and incubated with a mixture of hUBC13 and hUBC13(C87S)~Ub with and without the indicated concentration of free ubiquitin (2–100 µM) in 200 µL buffer A for 1 hour. Beads were washed with 400 µL Buffer A for 10 min and eluted with 25 µL of buffer A plus 250 mM imidazole. Eluates were analyzed by gel electrophoresis and staining with Coomassie Brilliant Blue or SimplyBlue™ SafeStain (Invitrogen). The pull-down in Figure S2 was performed as above except for the addition of 6XHis-hOTUB1 (7̣µg), hUBC13(C87S)~Ub (7̣µg) and ubiquitin as indicated in the figure.
Supplementary Material
Acknowledgements
We thank Elizabeth Henderson for generating the h/ceOTUB1 clone, Chris Berndsen, Anthony DiBello, Ajit Datta and Mario Bianchet for helpful discussions and an anonymous reviewer for suggesting the binding experiment with UEV1a in the presence of charged UBC13 and Ubal. GM/CA-CAT has been funded in whole or in part with funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357.
Footnotes
Author contributions
R.W. and C.W. designed the experiments and R.W. performed all biochemical experiments. Cloning, expression and protein purification were carried out by X.Z., T.W. and R.W. Complexes were prepared for crystallization and crystals were grown by X.Z. and R.W.; R.W. determined the crystal structure with guidance from C.W. R.W. and C.W. wrote the manuscript.
Data deposition
Coordinates and diffraction amplitudes are deposited in the Protein Data Bank under accession numbers 4DHI (ceOTUB1-UBC13), 4DHJ (ceOTUB1-Ubal-UBC13DCA~Ub) and 4DHZ (h/ceOTUB1-Ubal-UBC13DCA~Ub).
References
- 1.Al-Hakim A, et al. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 2010;9:1229–1240. doi: 10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 3.Stewart GS, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 4.Nakada S, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, et al. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messick TE, et al. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelmann MJ, et al. Structural basis and specificity of human otubain 1-mediated deubiquitination. Biochem J. 2009;418:379–390. doi: 10.1042/BJ20081318. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 11.Yin L, Krantz B, Russell NS, Deshpande S, Wilkinson KD. Nonhydrolyzable diubiquitin analogues are inhibitors of ubiquitin conjugation and deconjugation. Biochemistry. 2000;39:10001–10010. doi: 10.1021/bi0007019. [DOI] [PubMed] [Google Scholar]
- 12.James TW, et al. Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease. Proc Natl Acad Sci U S A. 2011;108:2222–2227. doi: 10.1073/pnas.1013388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capodagli GC, et al. Structural analysis of a viral ovarian tumor domain protease from the Crimean-Congo hemorrhagic fever virus in complex with covalently bonded ubiquitin. J Virol. 2011;85:3621–3630. doi: 10.1128/JVI.02496-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akutsu M, Ye Y, Virdee S, Chin JW, Komander D. Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains. Proc Natl Acad Sci U S A. 2011;108:2228–2233. doi: 10.1073/pnas.1015287108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato Y, et al. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009;28:2461–2468. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moraes TF, et al. Crystal structure of the human ubiquitin conjugating enzyme complex, hMms2-hUbc13. Nat Struct Biol. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- 17.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 18.Wenzel DM, Stoll KE, Klevit RE. E2s: structurally economical and functionally replete. Biochem J. 2010;433:31–42. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna S, et al. An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2.Ubc13. The structural basis for lysine 63 chain catalysis. The Journal of biological chemistry. 2003;278:13151–13158. doi: 10.1074/jbc.M212353200. [DOI] [PubMed] [Google Scholar]
- 20.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berndsen CE, Wolberger C. A spectrophotometric assay for conjugation of ubiquitin and ubiquitin-like proteins. Anal Biochem. 2011;418:102–110. doi: 10.1016/j.ab.2011.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson KD, Gan-Erdene T, Kolli N. Derivitization of the C-terminus of ubiquitin and ubiquitin-like proteins using intein chemistry: methods and uses. Methods Enzymol. 2005;399:37–51. doi: 10.1016/S0076-6879(05)99003-4. [DOI] [PubMed] [Google Scholar]
- 23.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.