Abstract
Background
Genome-wide association studies (GWAS) identified variants at 19p13.1 and ZNF365 (10q21.2) as risk factors for breast cancer among BRCA1 and BRCA2 mutation carriers, respectively. We explored associations with ovarian cancer and with breast cancer by tumor histopathology for these variants in mutation carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA).
Methods
Genotyping data for 12,599 BRCA1 and 7,132 BRCA2 mutation carriers from 40 studies were combined.
Results
We confirmed associations between rs8170 at 19p13.1 and breast cancer risk for BRCA1 mutation carriers (hazard ratio (HR)=1.17; 95%CI 1.07–1.27; p=7.42×10−4) and between rs16917302 at ZNF365 (HR=0.84; 95%CI 0.73–0.97; p=0.017) but not rs311499 at 20q13.3 (HR=1.11; 95%CI 0.94–1.31; p=0.22) and breast cancer risk for BRCA2 mutation carriers. Analyses based on tumor histopathology showed that 19p13 variants were predominantly associated with estrogen receptor (ER)-negative breast cancer for both BRCA1 and BRCA2 mutation carriers, whereas rs16917302 at ZNF365 was mainly associated with ER-positive breast cancer for both BRCA1 and BRCA2 mutation carriers. We also found for the first time that rs67397200 at 19p13.1 was associated with an increased risk of ovarian cancer for BRCA1 (HR=1.16; 95%CI 1.05–1.29; p=3.8×10−4) and BRCA2 mutation carriers (HR=1.30; 95%CI 1.10–1.52; p=1.8×10−3).
Conclusions
19p13.1 and ZNF365 are susceptibility loci for ovarian cancer and ER subtypes of breast cancer among BRCA1 and BRCA2 mutation carriers.
Impact
These findings can lead to an improved understanding of tumor development and may prove useful for breast and ovarian cancer risk prediction for BRCA1 and BRCA2 mutation carriers.
Keywords: BRCA1, BRCA2, breast cancer risk, ovarian cancer risk, 19p13.1, ZNF365
Introduction
Genome-wide association studies (GWAS) have been used to identify several loci containing common variants that are associated (p<1.0×10−7) with breast cancer risk in the general population. Variants from twelve of these loci have also been investigated as modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers (1–3). While only variants in CASP8, TOX3, 2q35, and 6q25.1 have been associated with breast cancer risk in BRCA1 mutation carriers, variants in FGFR2, TNRC9/TOX3, MAP3K1, LSP1, 2q35, SLC4A7/NEK10, 5p12 and 1p11.2 loci have been associated with breast cancer in BRCA2 mutation carriers (1–3). This is consistent with the known associations between these SNPs and estrogen receptor (ER) status of breast cancers in the general population (4).
Most recently, a GWAS of BRCA1 mutation carriers conducted through CIMBA identified five SNPs on 19p13 that were associated with breast cancer risk for BRCA1 mutation carriers (5). Two of these showed independent associations (rs8170 hazard ratio (HR)=1.26; 95%CI 1.17–1.35; Ptrend = 2.3×10−9 and rs2363956 HR=0.84; 95%CI 0.80–0.89; Ptrend=5.5×10−9). Imputation analysis of the 19p13 region, using 1000 Genomes Project data, identified several correlated SNPs with more significant associations than rs8170 and rs2363956. The 19p13.1 locus was also found to be associated with ER negative breast cancer (rs8170 OR=1.21, p=0.003) and triple negative breast cancer (tumors lacking expression of ER, progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2)) (rs8170 OR=1.28, p=1.2×10−6) in the general population (5). In addition, the 19p13.1 locus has been associated with ovarian cancer in the general population (rs8170 OR=1.12; p=3.6×10−6) (6), but was not found to be associated with ovarian cancer in BRCA1 mutation carriers (rs8170 HR=1.07; p=0.33) (5). A separate GWAS in BRCA2 mutation carriers identified two breast cancer susceptibility alleles (rs16917302 at ZNF365 (10q21.2), HR=0.75; 95%CI 0.66–0.86; p=3.8×10−5 and rs311499 at 20q13.33, HR=0.72; 95%CI 0.61–0.85; p=6.6×10−5) (7). A weakly correlated SNP at the ZNF365 locus (rs10995190) has also been associated with breast cancer overall (OR=0.83; p=5.1 × 10−15) and ER positive (p=4.1×10−6) but not ER negative breast cancer in the general population (8).
Here, we genotyped more than 12,000 BRCA1 and 7,000 BRCA2 mutation carriers from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA), for the previously genotyped variant at 19p13.1, rs8170, and one of the imputed SNPs that was found to have a stronger association with breast cancer risk for BRCA1 mutation carriers (rs67397200). We also genotyped SNPs at ZNF365 (rs16917302), and 20q13.3 (rs311499) in an effort to verify these loci as risk factors for ovarian cancer and to further validate these loci as risk factors for breast cancer in BRCA1 and BRCA2 mutation carriers.
Materials and Methods
Subjects
All mutation carriers participated in clinical or research studies at the host institutions under ethically approved protocols and provided written informed consent. Subjects were BRCA1 and BRCA2 mutation carriers recruited by 40 study centers in 22 countries and assembled through the CIMBA initiative (Supplementary Table 1). The majority were recruited through cancer genetics clinics and enrolled into national or regional studies. Others were identified in research studies of high-risk families, by population-based sampling of cases and some by community recruitment. Eligibility to participate in CIMBA is restricted to female carriers of pathogenic BRCA1 or BRCA2 mutations, defined by generally recognized criteria (Breast Cancer Information Core), who were 18 years old or older at recruitment. Information collected included the year of birth; mutation description (including nucleotide position and base change); age at last follow-up; ages at breast and ovarian cancer diagnoses; and age or date at bilateral prophylactic mastectomy. Information was also available on the country of residence. Related individuals were identified through a unique family identifier. Women with pathogenic mutations in both BRCA1 and BRCA2 were excluded from the current analysis. The primary analysis was restricted to women self-reported as “white European”. Overlap of carriers between studies was evaluated by comparing the year of birth, exact mutation description, the reported ages, and previous SNP genotype data available within the CIMBA database. Duplicated mutation carriers were included only once in the analysis.
Genotyping
Rs311499 at 20q13.3, rs16917302 at ZNF365 and both rs8170 and rs67397200 at 19p13.1 were genotyped using the iPLEX Mass Array platform at four genotyping centers as part of a larger study of 24 candidate SNPs. All centers included at least 2% duplicate samples and a random mixture of affected and unaffected carriers on each plate. Samples that failed for five or more of the SNPs genotyped were excluded from the analysis. Studies with a SNP call rate of <95% were excluded from the analysis of the SNP. The concordance between duplicates had to be at least 98%. To assess the accuracy of genotyping across genotyping centers, all centers genotyped 95 DNA samples from a standard test plate (Coriell Institute) for all SNPs. Genotyping centers with more than one concordance failure on the test plate for a SNP were excluded for analyses of that SNP. Deviation from Hardy-Weinberg equilibrium (HWE) was assessed for unrelated subjects separately for each SNP and study. The observed genotype frequencies were not significantly different from the expected under HWE for any of the SNPs and studies. After the above exclusions, a total of 19,731 unique mutation carriers (12,599 BRCA1 and 7,132 BRCA2) from 40 studies had an observed genotype for at least one SNP (Supplementary Table 1).
Tumor pathology data collection
Tumor pathology data were collected from patient pathology reports, medical records, pathology review data, tumor registry records and results from tissue microarrays. ER status was identified as negative or positive, with immunohistochemistry scoring data and methodology provided when available. Most studies applied a cut-off of >10% tumor cells stained positive for ER positive status. For a small number of cases, where other scoring methods based on the proportion and intensity of staining were applied (Allred score, Remmele score and H-score), widely-accepted cut-offs were used. Consistency checks were performed to validate receptor data against supplementary scoring information if provided.
Statistical analysis
The aim of the primary analysis was to evaluate the association between each genotype and breast cancer risk. We conducted the analysis by modelling the retrospective likelihood of the observed genotypes conditional on the disease phenotypes as previously described (9). The phenotype of each individual was defined by age at diagnosis of breast cancer or age at last follow-up. Individuals were censored at the earliest of age of first breast cancer diagnosis, ovarian cancer diagnosis, bilateral prophylactic mastectomy or age at last observation. Mutation carriers censored at ovarian cancer diagnosis were considered unaffected in the analysis of breast cancer. The effect of each SNP was modelled either as a per-allele HR (multiplicative model) or as separate HRs for heterozygotes and homozygotes. We used a Cox proportional-hazards model and tested the assumption of proportional hazards by adding a “genotype×age” interaction term in order to fit models in which the HR changed with age. We examined heterogeneity across studies by comparing models that allowed for study-specific log-hazard ratios against models in which the same log-hazard ratio was assumed to apply to all studies. All analyses were stratified by country of residence and applied cohort specific breast cancer incidence rates for BRCA1 and BRCA2 (10). A robust variance-estimation approach was used to adjust for the non-independence among related carriers.
To evaluate the evidence of replication for each of the SNPs, analyses were restricted to mutation carriers who had not been used in any of the previous BRCA1 and BRCA2 studies. The number of new samples used in each of the SNP analyses are shown in Supplementary Table 2. In addition, analysis were performed using all available BRCA1 and BRCA2 carriers. The combined effects of the SNPs on breast cancer risk, were evaluated by fitting retrospective likelihood models while allowing for linkage disequilibrium between the loci. To test for potential effects of survival bias, prevalent cases, defined as mutation carriers diagnosed more than five years prior to the age at recruitment, were excluded. Associations with specific functional class of mutation were also assessed. Class 1 mutations are predicted to undergo nonsense mediated RNA decay resulting in reduced levels of mutant transcript while Class 2 mutations are predicted to generate stable mutant proteins (11). The associations with breast cancer subtypes defined by the estrogen receptor (ER) status of the tumors in BRCA1 and BRCA2 mutation carriers were assessed by an extension of the retrospective likelihood approach that models the simultaneous effect of each SNP on more than one tumor subtype (12). Associations with ovarian cancer risk were evaluated within a competing risk analysis framework (13) by estimating HRs simultaneously for breast and ovarian cancers. Since each mutation carrier was at risk of breast and ovarian cancer we assumed that the probabilities of developing each disease were independent conditional on the underlying genotype. In this analysis, individuals were followed to the age of the first breast or ovarian cancer diagnosis and were considered to have developed the corresponding disease. Individuals were censored for breast cancer at the age of bilateral prophylactic mastectomy and for ovarian cancer at the age of bilateral oophorectomy and were assumed to be unaffected for the corresponding disease. The remaining individuals were censored at the age at last observation and were assumed to be unaffected for both diseases.
Results
After quality control exclusions, genotype data from 12,599 BRCA1 and 7,132 BRCA2 mutation carriers including 5,408 BRCA1 and 3,780 BRCA2 mutation carriers not studied in the original GWAS were available for analysis. Of the BRCA1 mutation carriers, 6,390 were affected with breast cancer and 6,209 were considered unaffected in the breast cancer analysis (censored at bilateral prophylactic mastectomy, ovarian cancer, or age at last follow up). Similarly, among the BRCA2 mutation carriers, 3,810 were affected with breast cancer and 3,322 were unaffected. The characteristics of these mutation carriers are shown in Table 1 and the origins of the samples are summarized in Supplementary Table 1.
Table 1.
Summary characteristics for the 19,731 eligible BRCA1 and BRCA2 mutation carriers* used in the analysis
| Characteristic | BRCA1 | BRCA2 | ||
|---|---|---|---|---|
| Unaffected | Breast Cancer |
Unaffected | Breast Cancer |
|
| Number | 6209 | 6390 | 3322 | 3810 |
| Person-Years follow-up | 264903 | 263068 | 147053 | 168201 |
| Median Age at Censure (IQR1) | 42 (34–50) | 40 (34–47) | 43 (34–53) | 43 (37–50) |
| Age at Censure, N (%) | ||||
| <30 | 1189 (19.2) | 691 (10.8) | 611 (18.4) | 306 (8.0) |
| 30–39 | 1661 (26.8) | 2445 (38.3) | 834 (25.1) | 1141 (30.0) |
| 40–49 | 1765 (28.4) | 2191 (34.3) | 865 (26.0) | 1394 (36.6) |
| 50–59 | 1058 (17.0) | 812 (12.7) | 566 (17.0) | 687 (18.0) |
| 60–69 | 380 (6.1) | 198 (3.1) | 302 (9.1) | 226 (5.9) |
| 70+ | 156 (2.5) | 53 (0.8) | 144 (4.3) | 56 (1.5) |
| Year of birth, N (%) | ||||
| <1920 | 28 (0.5) | 30 (0.5) | 23 (0.7) | 44 (1.2) |
| 1920–29 | 131 (2.1) | 196 (3.1) | 99 (3.0) | 167 (4.4) |
| 1930–39 | 369 (5.9) | 516 (8.1) | 232 (7.0) | 430 (11.3) |
| 1940–49 | 832 (13.4) | 1341 (21.0) | 458 (13.8) | 896 (23.5) |
| 1950–59 | 1409 (22.7) | 1989 (31.1) | 691 (20.8) | 1160 (60.5) |
| 1960–69 | 1703 (27.4) | 1666 (26.1) | 902 (27.2) | 868 (22.8) |
| 1970+ | 1737 (28.0) | 652 (10.2) | 917 (27.6) | 245 (6.4) |
| Mutation Class, N (%) | ||||
| Class 12 | 4063 (65.4) | 3878 (60.7) | 3114 (93.7) | 3520 (92.4) |
| Class 22 | 1780 (28.7) | 1973 (30.9) | 72 (2.2) | 100 (2.6) |
| Other | 366 (5.9) | 539 (8.4) | 136 (4.1) | 190 (5.0) |
IQR: Interquartile range
See methods for definitions
Carriers of self reported European ancestry only.
The associations between breast cancer risk in BRCA1 and BRCA2 mutation carriers and the minor alleles of rs8170 and rs67397200 (19p13.1), rs16917302 (ZNF365), and rs311499 (20q13.33) are summarized in Table 2. The minor allele of rs8170 at 19p13.1 was strongly associated with risk of breast cancer in BRCA1 mutation carriers (HR=1.20; 95%CI 1.13–1.28; p=8.7×10−9) but not BRCA2 mutation carriers. This result for 12,599 BRCA1 mutation carriers was consistent with the original finding in the BRCA1 GWAS using 8,363 BRCA1 mutation carriers (HR=1.26; 95%CI 1.17–1.35; p=2.3×10−9). A separate analysis restricted to carriers not used in the BRCA1 GWAS also confirmed the association (HR=1.17; 95%CI 1.07–1.27; p=7.42×10−4) (Supplementary Table 2). Similarly, rs67397200 at 19p13.1, which was imputed in the BRCA1 GWAS, was strongly associated with breast cancer risk in BRCA1 carriers (HR=1.17; 95%CI 1.11–1.23; p=2.4×10−8) (Table 2). There was no evidence of heterogeneity in the HRs across studies for BRCA1 mutation carriers (Figure 1). However, there was evidence that the per-allele HRs in BRCA1 mutation carriers for rs8170 (p=0.015) and rs67397200 (p=0.007) at 19p13.1 decreased with increasing age of diagnosis of breast cancer. Since rs8170 and rs67397200 are located in the same region of 19p13.1 (r2=0.58), we conducted an analysis for the joint effects of these SNPs on breast cancer risk in BRCA1 mutation carriers (n=10,173). When accounting for haplotype structure, rs67397200 remained significant (P for inclusion=2.75×10−3) and was retained in the model, whereas rs8170 was excluded (P for inclusion=0.18). Rs8170 and rs67397200 were not associated with breast cancer risk for BRCA2 mutation carriers (Table 2).
Table 2.
Evaluation of associations between SNPs and breast cancer risk among BRCA1 and BRCA2 mutation carriers of European ancestry
|
SNP/ |
Unaffected | Affecteda | ||||
|---|---|---|---|---|---|---|
| Mutation | Genotype | N (%) | N (%) | HR | 95% CI | p-value |
| rs8170 – 19p13.1 | ||||||
| BRCA 1 | GG | 3870 (67.5) | 3755 (63.3) | 1.00 | ||
| AG | 1689 (29.4) | 1950 (32.9) | 1.22 | 1.14 – 1.31 | ||
| AA | 178 (3.1) | 227 (3.8) | 1.35 | 1.13 – 1.62 | ||
| per allele | 1.20 | 1.13 – 1.28 | 8.7×10−9 | |||
| BRCA 2 | GG | 2047 (66.3) | 2501 (68.2) | 1.00 | ||
| AG | 931 (30.2) | 1026 (28.0) | 0.93 | 0.84 – 1.03 | ||
| AA | 108 (3.5) | 138 (3.8) | 1.16 | 0.89 – 1.52 | ||
| per allele | 0.98 | 0.90 – 1.07 | 0.67 | |||
| rs67397200 – 19p13.1 | ||||||
| BRCA 1 | CC | 2536 (51.0) | 2455 (46.0) | 1.00 | ||
| GC | 2022 (40.6) | 2397 (44.9) | 1.24 | 1.16 – 1.34 | ||
| GG | 415 (8.4) | 487 (9.1) | 1.25 | 1.10 – 1.43 | ||
| per allele | 1.17 | 1.11 – 1.23 | 2.4×10−8 | |||
| BRCA 2 | CC | 1553 (49.8) | 1871 (50.7) | 1.00 | ||
| GC | 1302 (41.7) | 1494 (40.5) | 0.95 | 0.87 – 1.04 | ||
| GG | 265 (8.5) | 323 (8.8) | 1.07 | 0.91 – 1.27 | ||
| per allele | 1.00 | 0.93 – 1.07 | 0.97 | |||
| rs311499 – 20q13.3 | ||||||
| BRCA 1 | GG | 5346 (86.2) | 5484 (85.9) | 1.00 | ||
| AG | 816 (13.2) | 873 (13.7) | 1.03 | 0.94 – 1.13 | ||
| AA | 41 (0.7) | 28 (0.4) | 0.67 | 0.42 – 1.08 | ||
| per allele | 1.00 | 0.91 – 1.09 | 0.94 | |||
| BRCA 1 | GG | 2873 (86.6) | 3312 (87.0) | 1.00 | ||
| AG | 429 (13.0) | 475 (12.5) | 0.94 | 0.82 – 1.07 | ||
| AA | 16 (0.5) | 21 (0.6) | 0.97 | 0.60 – 1.57 | ||
| per allele | 0.95 | 0.84 – 1.07 | 0.36 | |||
| rs16917302 - 10q21.2 | ||||||
| BRCA 1 | AA | 4913 (79.3) | 5084 (79.7) | 1.00 | ||
| CA | 1216 (19.6) | 1222 (19.2) | 0.96 | 0.88 – 1.01 | ||
| CC | 71 (1.1) | 73 (1.1) | 0.94 | 0.69 – 1.27 | ||
| per allele | 0.96 | 0.89 – 1.03 | 0.27 | |||
| BRCA 2 | AA | 2583 (77.9) | 3101 (81.5) | 1.00 | ||
| CA | 691 (20.8) | 674 (17.7) | 0.82 | 0.74 – 0.92 | ||
| CC | 41 (1.2) | 32 (0.8) | 0.78 | 0.49 – 1.23 | ||
| per allele | 0.83 | 0.75 – 0.93 | 7.0×10−4 | |||
Breast cancer
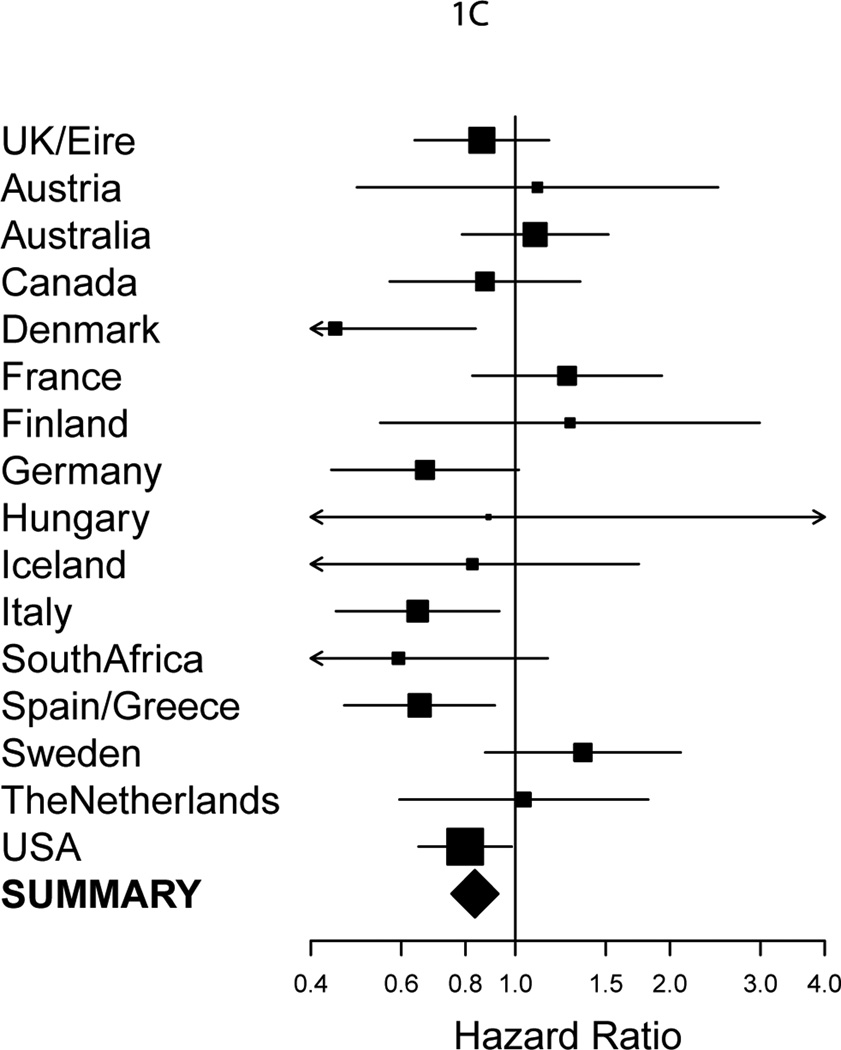
Figure 1. Forest plots of the associations by country of residence of BRCA1 and BRCA2 mutation carriers with breast cancer risk overall.
(A–C) Squares indicate the country specific per-allele HR estimates for SNPs (A) rs8170 for BRCA1 mutation carriers, (B) rs67397200 for BRCA1 mutation carriers and (C) rs16917302 for BRCA2 mutation carriers. The area of the square is proportional to the inverse of the variance of the estimate. Horizontal lines indicate 95% Confidence Intervals (CIs).
Among SNPs identified from the original BRCA2 GWAS, an analysis of genotype data from 7,132 BRCA2 mutation carriers confirmed that rs16917302 at the ZNF365 locus was associated with a decreased risk of breast cancer (HR=0.83; 95%CI 0.75–0.93; p=7.0×10−4). The association also replicated in the additional carriers, not previously included in the BRCA2 GWAS (HR=0.84; 95%CI 0.73–0.97; p=0.017) (Supplementary Table 2). In contrast, rs311499 from 20q13.3, which was associated with breast cancer risk in the BRCA2 GWAS (HR=0.72; 95%CI 0.61–0.85; p=6.6×10−5) (7), was not associated with risk of breast cancer in BRCA2 carriers in the overall analysis (HR=0.95; 95%CI 0.84–1.07; p=0.36 (Table 2) nor the replication study (HR=1.11; 95%CI 0.94–1.31; p=0.22) (Supplementary Table 2). There was no evidence for heterogeneity in the HRs across studies for BRCA2 mutation carriers (Figure 1). HRs for rs16917302 and rs311499 did not vary by age at diagnosis.
To determine whether the inclusion of long-term survivors influenced the results, we repeated our analyses of the four SNPs, excluding BRCA1 and BRCA2 mutation carriers diagnosed with breast cancer more than five years before recruitment (prevalent cases). The strength of the associations for rs16917302 at ZNF365 (per-allele HR=0.85) for BRCA2 mutation carriers, and for rs8170 (per-allele HR=1.19) and rs67397200 at 19p13.1 (per-allele HR=1.16) for BRCA1 mutation carriers were essentially unchanged (Supplementary Table 3). There was no influence of mutation type for BRCA1 mutation carriers on breast cancer risk in the associations between mutations conferring susceptibility to nonsense mediated RNA decay (NMD) (Class 1) and missense or truncating mutations not triggering NMD (Class 2) for any of the SNPs (Supplementary Table 4).
Breast tumors in BRCA1 mutation carriers are predominantly ER-negative (14) and rs8170 from 19p13.1 is strongly associated with ER-negative but not ER-positive breast cancer in the general population (5). Because of these previous findings, we evaluated whether rs8170 and rs67397200 at 19p13.1, as well as rs311499 at 20q13.3 and rs16917302 at ZNF365, were differentially associated with ER-positive and/or ER-negative tumor status in BRCA1 and BRCA2 mutation carriers. Although the stratified results suggested a slightly stronger association for the 19p13.1 rs67397200 SNP with ER-negative disease than with ER-positive disease in BRCA1 mutation carriers (per allele ER-negative HR=1.22; 95%CI 1.14–1.30; p=4.4×10−9; per allele ER-positive HR=1.14; 95%CI 1.01–1.30; p=0.040), the difference was not significant (p=0.41) (Table 3). Rs67397200, however, was associated with ER-negative disease (per allele HR=1.29; 95%CI 1.11–1.49; p=8.7×10−4) but not ER-positive disease (per allele HR=0.92; 95%CI 0.85–1.01; p=0.074) in BRCA2 mutation carriers (p-heterogeneity= 1.5×10−4) (Table 3). The lack of association with rs311499 at 20q13.3 did not vary by ER-status in BRCA1 or BRCA2 mutation carriers. For BRCA2 mutation carriers, the minor allele of rs16917302 at ZNF365 was inversely associated with both ER-positive (per allele HR=0.86; 95%CI 0.75–0.97; p=0.016) and ER-negative tumors (per allele HR=0.79; 95%CI 0.62–1.00; p=0.048) (p-heterogeneity=0.56) (Table 3). However, in BRCA1 mutation carriers rs16917302 was associated with ER-positive (per allele ER-positive HR=0.77; 95%CI 0.62–0.95; p=0.016), but not ER-negative status (p-heterogeneity=0.028) (Table 3).
Table 3.
Associations between SNPs and breast cancer risk by estrogen receptor (ER) status of breast cancer cases among women with BRCA1 and BRCA2 mutations
|
SNP/ |
Unaff |
Affecteda (N) |
ER+ |
ER− |
Case het p-valueb |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | N | ER− | ER+ | HR | 95% CI | p-trend | HR | 95% CI | p-trend | p-trend |
| rs8170 – 19p13.1 | ||||||||||
| BRCA1 | 4483 | 1820 | 541 | 1.12 | 0.96 – 1.29 | 0.15 | 1.23 | 1.14 – 1.33 | 2.0×10−7 | 0.26 |
| BRCA2 | 2738 | 401 | 1343 | 0.94 | 0.85 – 1.05 | 0.26 | 1.18 | 0.99 – 1.40 | 0.058 | 0.026 |
| rs67397200 – 19p13.1 | ||||||||||
| BRCA1 | 4486 | 1821 | 542 | 1.14 | 1.01 – 1.30 | 0.040 | 1.22 | 1.14 – 1.30 | 4.4×10−9 | 0.41 |
| BRCA2 | 2733 | 401 | 1349 | 0.92 | 0.85 – 1.01 | 0.074 | 1.29 | 1.11 – 1.49 | 8.7×10−4 | 1.5×10−4 |
| rs311499 – 20q13.3 | ||||||||||
| BRCA1 | 4898 | 1890 | 559 | 1.07 | 0.87 – 1.31 | 0.51 | 0.95 | 0.85 – 1.06 | 0.35 | 0.31 |
| BRCA2 | 2930 | 406 | 1372 | 0.95 | 0.82 – 1.09 | 0.48 | 0.83 | 0.63 – 1.10 | 0.19 | 0.40 |
| rs16917302 - 10q21.2 | ||||||||||
| BRCA1 | 4897 | 1888 | 558 | 0.77 | 0.62 – 0.95 | 0.016 | 1.01 | 0.92 – 1.11 | 0.85 | 0.028 |
| BRCA2 | 2927 | 406 | 1372 | 0.86 | 0.75 – 0.97 | 0.016 | 0.79 | 0.62 – 1.00 | 0.048 | 0.56 |
Breast cancer
p-value for heterogeneity in the associations with ER−positive and ER−negative breast cancer
ER+: ER positive, ER−: ER negative, Unaff: Unaffected
BRCA1 and BRCA2 mutations are associated with elevated risk of ovarian cancer. In this CIMBA study 1,465 BRCA1 mutation carriers and 453 BRCA2 mutation carriers who developed ovarian cancer were also genotyped for the four SNPs under study. To assess the influence of these SNPs on ovarian cancer risk in BRCA1 and BRCA2 mutation carriers we used a competing risk analysis that evaluated the associations with breast and ovarian cancer risk simultaneously. While previous studies did not detect an association between rs8170 at 19p13.1 and ovarian cancer in BRCA1 or BRCA2 mutation carriers (5), in this competing risk analysis with larger numbers of BRCA1 and BRCA2 mutation carriers, rs8170 was significantly associated with ovarian cancer risk in both BRCA1 (HR=1.15; 95%CI 1.03–1.29; p=0.015) and BRCA2 (HR=1.34; 95%CI 1.12–1.62; p=1.9×10−3) mutation carriers (Table 4). Similarly rs67397200 at 19p13.1 was associated with ovarian cancer risk in both BRCA1 (HR=1.16; 95%CI 1.05–1.29; p=3.8×10−4) and BRCA2 (HR=1.30; 95%CI 1.10–1.52; p=1.8×10−3) mutation carriers (Table 4). Rs311499 at 20q13.3 and rs16917302 at ZNF365 were not associated with ovarian cancer risk for either BRCA1 or BRCA2 mutation carriers (Table 4).
Table 4.
Associations with SNPs and breast and ovarian cancer risk using a competing risk analysis model among BRCA1 and BRCA2 mutation carriers of European ancestry
|
SNP/ |
Unaffected | Breast Cancer |
Ovarian Cancer |
Breast Cancer | Ovarian Cancer | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | Genotype | N (%) | N (%) | N (%) | HR | 95% CI | p-value | HR | 95% CI | p-value |
| rs8170 – 19p13.1 | ||||||||||
| BRCA1 | GG | 2972 (67.9) | 3730 (63.3) | 923 (66.0) | 1.00 | 1.00 | ||||
| AG | 1269 (29.0) | 1936 (32.9) | 434 (31.0) | 1.26 | 1.17 – 1.36 | 1.23 | 1.08 – 1.42 | |||
| AA | 139 (3.2) | 224 (3.8) | 42 (3.0) | 1.34 | 1.10 – 1.63 | 1.04 | 0.72 – 1.50 | |||
| per allele | 1.22 | 1.14 – 1.30 | 2.1×10−9 | 1.15 | 1.03 – 1.29 | 0.015 | ||||
| BRCA2 | GG | 1788 (67.0) | 2494 (68.2) | 266 (62.2) | 1.00 | 1.00 | ||||
| AG | 796 (29.9) | 1024 (28.0) | 137 (32.0) | 0.95 | 0.85 – 1.05 | 1.17 | 0.93 – 1.47 | |||
| AA | 83 (3.1) | 138 (3.8) | 25 (5.8) | 1.37 | 1.05 – 1.80 | 2.72 | 1.65 – 4.48 | |||
| per allele | 1.02 | 0.94 – 1.12 | 0.62 | 1.34 | 1.12 – 1.62 | 1.9×10−3 | ||||
| rs67397200 – 19p13.1 | ||||||||||
| BRCA1 | CC | 1903 (51.5) | 2436 (46.0) | 652 (49.7) | 1.00 | 1.00 | ||||
| GC | 1498 (40.5) | 2381 (44.9) | 540 (41.2) | 1.28 | 1.18 – 1.38 | 1.16 | 1.01 – 1.33 | |||
| GG | 298 (8.1) | 484 (9.1) | 120 (9.2) | 1.33 | 1.16 – 1.53 | 1.36 | 1.07 – 1.73 | |||
| per allele | 1.20 | 1.13 – 1.27 | 4.5×10−10 | 1.16 | 1.05 – 1.29 | 3.8×10−4 | ||||
| BRCA2 | CC | 1363 (50.5) | 1866 (50.7) | 194 (45.2) | 1.00 | 1.00 | ||||
| GC | 1123 (41.6) | 1489 (40.5) | 184 (42.9) | 0.96 | 0.87 – 1.06 | 1.15 | 0.92 – 1.44 | |||
| GG | 214 (7.9) | 323 (8.8) | 51 (11.9) | 1.18 | 0.99 – 1.41 | 1.95 | 1.37 – 2.77 | |||
| per allele | 1.03 | 0.96 – 1.11 | 0.39 | 1.30 | 1.10 – 1.52 | 1.8×10−3 | ||||
| rs311499 – 20q13.3 | ||||||||||
| BRCA1 | GG | 4115 (86.0) | 5442 (85.9) | 1273 (86.9) | 1.00 | 1.00 | ||||
| AG | 637 (13.3) | 869 (13.7) | 183 (12.5) | 1.01 | 0.92 – 1.12 | 0.88 | 0.74 – 1.05 | |||
| AA | 32 (0.7) | 28 (0.4) | 9 (0.6) | 0.70 | 0.42 – 1.17 | 1.16 | 0.47 – 2.87 | |||
| per allele | 0.99 | 0.90 – 1.08 | 0.77 | 0.91 | 0.77 – 1.07 | 0.25 | ||||
| BRCA2 | GG | 2492 (86.7) | 3303 (87.0) | 390 (86.1) | 1.00 | 1.00 | ||||
| AG | 372 (12.9) | 474 (12.5) | 58 (12.8) | 0.93 | 0.82 – 1.07 | 0.92 | 0.68 – 1.26 | |||
| AA | 11 (0.4) | 21 (0.6) | 5 (1.1) | 1.09 | 0.68 – 1.74 | 2.23 | 0.80 – 6.22 | |||
| per allele | 0.95 | 0.84 – 1.08 | 0.44 | 1.02 | 0.76 – 1.37 | 0.88 | ||||
| rs16917302 - 10q21.2 | ||||||||||
| BRCA1 | AA | 3784 (79.2) | 5044 (79.7) | 1169 (79.8) | 1.00 | 1.00 | ||||
| CA | 937 (19.6) | 1216 (19.2) | 285 (19.5) | 0.96 | 0.88 – 1.04 | 0.97 | 0.84 – 1.13 | |||
| CC | 60 (1.3) | 73 (1.2) | 11 (0.8) | 0.86 | 0.62 – 1.19 | 0.47 | 0.25 – 0.92 | |||
| per allele | 0.95 | 0.88 – 1.03 | 0.21 | 0.92 | 0.80 – 1.05 | 0.20 | ||||
| BRCA2 | AA | 2237 (77.9) | 3094 (81.5) | 353 (78.1) | 1.00 | 1.00 | ||||
| CA | 601 (20.9) | 671 (17.7) | 93 (20.6) | 0.81 | 0.72 – 0.92 | 0.93 | 0.72 – 1.21 | |||
| CC | 35 (1.2) | 32 (0.8) | 6 (1.3) | 0.80 | 0.50 – 1.30 | 1.21 | 0.46 – 3.18 | |||
| per allele | 0.83 | 0.74 – 0.92 | 5.8×10−4 | 0.96 | 0.76 – 1.22 | 0.76 | ||||
Discussion
GWAS of BRCA1 and BRCA2 mutation carriers previously identified variants at 19p13.1, ZNF365 and 20q13.3 as candidate breast cancer risk modifiers (5, 7). In this study, we further evaluated associations between variants at these loci and both breast and ovarian cancer in BRCA1 and BRCA2 mutation carriers. For the first time, we found that both rs8170 and the previously imputed rs67397200 at 19p13.1 were strongly associated with ovarian cancer in both BRCA1 and BRCA2 mutation carriers. In addition, we found that rs8170 and rs67397200 at 19p13.1 were associated with breast cancer risk for BRCA1 and rs16917302 at ZNF365 was associated with breast cancer in BRCA2 mutation carriers in this replication study using an independent set of mutation carriers and in the combined analyses of data from the original study and the replication study. In contrast, rs311499 at 20q13.3 showed no association with breast cancer in the replication study. We also report for the first time that the BRCA1 GWAS SNP rs67397200 is associated with ER-negative breast cancer in BRCA2 mutation carriers and that the BRCA2 GWAS SNP rs16917302 is associated with ER-positive disease in BRCA1 mutation carriers.
The GWAS for breast cancer in BRCA1 mutation carriers originally identified significant associations between variants at the 19p13.1 locus and risk of breast cancer. Five SNPs including rs8170 from a 39 kb region were associated with risk of disease. In an analysis of joint effects of these SNPs on breast cancer risk, the best model included rs8170 or rs4808611 and rs8100241 or rs2363956 (P for inclusion=7.7×10−5 and P=6.7×10−5 for rs8170 and rs8100241, respectively) (5), suggesting that the associations were driven by a single causative variant partially correlated with all five SNPs. Imputation of additional SNPs in the region from the 1000 Genome Project identified eight perfectly correlated SNPs within a 13-kb region that were more significantly associated with breast cancer risk. Of these, we chose rs67397200, which has an r2=0.58 with rs8170 and r2=0.37 with rs8100241/rs2363956, for further genotyping in an effort to determine whether this SNP (or one of the seven other highly correlated SNPs) exhibited stronger associations with breast cancer. In an analysis of rs8170 in 11,669 and rs67397200 in 10,312 BRCA1 mutation carriers, we observed similarly strong associations with breast cancer for BRCA1 mutation carriers. In a joint analysis of rs8170 and rs67397200, allowing for haplotype structure, only rs67397200 remained significant. We were unable to genotype some of the original GWAS SNPs (rs2363956/rs8100241) in the present study and, as a consequence, could not evaluate the joint associations with rs67397200. It is therefore still unclear whether rs67397200 accounts solely for the association signal. The 35kb region containing rs8170 and rs67397200 includes the ABHD8 (abhydrolase domain containing 8), ANKLE1 (ankyrin repeat and LEM domain containing 1) and C19orf62 genes. C19orf62, encodes MERIT40 (Mediator of Rap80 Interactions and Targeting 40 kD), a BRCA1 interacting protein that forms a complex with BRCA1-BARD1, Abraxas1, RAP80, BRCC36 and BRCC45 and is required for recruitment and retention of the BRCA1-BARD1 ubiquitin ligase at sites of DNA damage (15). Because alterations in MERIT40 expression or function may modify BRCA1 activity, variants in the C19orf62 locus are attractive candidate breast cancer risk modifiers. However, since rs67397200 and the seven other imputed SNPs, that showed the most significant associations with breast cancer risk in BRCA1 mutation carriers, are located at the 3’ end of ANKLE1 near ABHD8 it is also possible that one of these genes rather than C19orf62 is influenced by the underlying causative variants in this region. Further comprehensive genotyping of other common variants and/or rare SNPs from this locus and detailed functional studies will be required to resolve this issue.
Our GWAS for breast cancer in BRCA2 mutation carriers previously identified strong associations between rs16917302 in the ZNF365 (dbGENE id: 22891) locus and breast cancer (7). We have now replicated this association for BRCA2 mutation carriers. Rs16917302 is located within intron 4 of ZNF365 and is unique to isoform C, the longest of the four isoforms created by alternative splicing sites (16). In independent studies, rs10995195 in ZNF365, which is 27kb upstream from and only weakly correlated (r2=0.1) with rs16917302, has been associated with breast cancer risk (8) and with mammographic density (17) in the general population. In addition, a cluster of SNPs located 154kb from rs16917302 in isoform D of ZNF365 has been associated with Crohn’s disease (18–20), and the region has also been implicated in family-based linkage analyses with uric acid nephrolithiasis (21) and hypotrichosis (22). It is unclear whether there is genetic or biologic linkage between these seemingly disparate phenotypes. Further fine-mapping of the ZNF365 region and functional analyses will be needed to identify the causative variants for each phenotype and to understand the downstream biological effects.
Likewise, rs311499 at 20q13.3 was associated with breast cancer risk in BRCA2 mutation carriers in the BRCA2 GWAS (per allele HR=0.72; 95%CI 0.61–0.85; p=6.6×10−5). However, this association was not confirmed in the replication study described above (HR=1.11; 95%CI 0.94–1.31; p=0.22) or in the combined analysis of the discovery and replication stages (HR=0.95; 95%CI 0.84–1.07; p=0.36). This result was not unexpected because the association between rs311499 and breast cancer did not reach significance (p<0.05) in Stage 2 of the BRCA2 GWAS (HR=0.86; 95%CI 0.67–1.06; p=0.13) (7).
To further characterize the influence of the 19p13.1 and ZNF365 loci on breast cancer risk, we assessed the strength of association with ER-negative and ER-positive breast cancer in BRCA1 and BRCA2 mutation carriers. As reported above, rs67397200 at 19p13.1 was associated with both ER-negative and ER-positive breast cancer in BRCA1 mutation carriers, whereas rs8170 at 19p13.1 was only associated with ER-negative disease. Interestingly, rs67397200 and rs8170 were also associated with ER-negative breast cancer but not ER-positive breast cancer in BRCA2 mutation carriers. This is consistent with our previous finding that rs8170 at 19p13.1 is more strongly associated with ER-negative than ER-positive breast cancer in the general population (5). Given that the majority of BRCA1 breast tumors exhibit a basal breast cancer phenotype (14), it remains to be determined whether ER-positive basal cases account for the mild association with ER-positive disease in BRCA1 mutation carriers.
In contrast, we found that rs16917302 in the ZNF365 locus was associated with both ERpositive and ER-negative disease in BRCA2 mutation carriers. This was consistent with associations for both ER-positive and ER-negative breast cancer in a recent GWAS of breast cancer cases with a family history of the disease (8). In contrast, among BRCA1 mutation carriers the association with breast cancer risk was restricted to ER-positive cases. This suggests that refinement of phenotype, perhaps in specific subpopulations, may result in detection of previously hidden associations.
Ovarian cancer is an important component of the cancer phenotype in both BRCA1 and BRCA2 mutation carriers. Because breast and ovarian cancer can occur in the same mutation carriers, it has been suggested that susceptibility SNPs common to breast and ovarian cancer may exist in these populations. However, to date none of the SNPs associated with breast cancer risk in BRCA1 or BRCA2 mutation carriers have been associated with ovarian cancer risk. Similarly, SNPs in the BCN2 locus that are associated with ovarian cancer risk in BRCA1 and BRCA2 mutation carriers are not associated with breast cancer risk (13). Furthermore, in the general population, only SNPs in the 8q24 locus are known to influence both breast and ovarian cancer, and these appear to be independent disease-specific effects (23). Thus, the recent finding that SNPs at 19p13.1 were associated with breast cancer in BRCA1 mutation carriers (5) and also with ovarian cancer in the general population (6) raised the possibility that a locus with common influences on breast and ovarian cancer does exist. However, the BRCA1 GWAS failed to detect any association for 19p13.1 SNPs with ovarian cancer among BRCA1 mutation carriers (843 ovarian cases) (HR=1.07; 95%CI 0.93–1.24; p=0.33) (5). Here we re-evaluated associations between 19p13.1 SNPs and ovarian cancer using larger numbers of BRCA1 (n=1312) and BRCA2 (n=429) carriers diagnosed with ovarian cancer. Rs67397200 at 19p13.1 was associated with ovarian cancer risk in both BRCA1 (HR=1.16; 95%CI 1.05–1.29; p=3.8×10−4) and BRCA2 (HR=1.30; 95%CI 1.10–1.52; p=1.8×10−3) mutation carriers. The magnitude of the effect on ovarian cancer risk in BRCA1 carriers (HR=1.16) was similar to that observed for breast cancer. This is the first locus found to influence both breast and ovarian cancer risk in either BRCA1 or BRCA2 mutation carriers.
Including the SNPs from the present study, six loci are now known to modify the risk of breast cancer for BRCA1 mutation carriers (CASP8, TOX3, 2q35, 6q25.1, 19p13, and ZNF365 (ER-positive disease only)) (1–3, 5, 10, 24) and ten loci are known to modify the risk of breast cancer for BRCA2 mutation carriers (FGFR2, TOX3, MAP3K1, LSP1, 2q35, SLC4A7, 5p12, ZNF365, 1p11.2 and 19p13.1 (ER-negative only)) (5, 7, 10, 24). Taken together, these SNPs result in large variation in the absolute risk of breast cancer for BRCA1 and BRCA2 mutation carriers and may further improve our ability to provide individualized risks of breast cancer for BRCA1 and BRCA2 mutation carriers.
Supplementary Material
Acknowledgements
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. In particular, we thank (BFBOCC) Laima Tikhomirova, (CBCS) Bent Ejlertsen, (CNIO) R.M. Alonso, G Pita and R.M. Milne, the Molecular Genetics Laboratory from Hospital de Cruces, (CONSIT) Marco A. Pierotti, Gaia Roversi, Elisa Cattaneo Carla B. Ripamonti and Marilena Morganti (Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy), Bernardo Bonanni (Istituto Europeo di Oncologia, Milan, Italy); (EMBRACE) Cambridge: Susan Peock, Debra Frost, Steve D. Ellis, Elena Fineberg, Radka Platte. North of Scotland Regional Genetics Service, Aberdeen: Zosia Miedzybrodzka, Helen Gregory. Northern Ireland Regional Genetics Service, Belfast: Patrick Morrison, Lisa Jeffers. West Midlands Regional Clinical Genetics Service, Birmingham: Trevor Cole, Kai-ren Ong, Jonathan Hoffman. South West Regional Genetics Service, Bristol: Alan Donaldson, Margaret James. East Anglian Regional Genetics Service, Cambridge: Joan Paterson, Sarah Downing, Amy Taylor. Medical Genetics Services for Wales, Cardiff: Alexandra Murray, Mark T. Rogers, Emma McCann. St James’s Hospital, Dublin & National Centre for Medical Genetics, Dublin: M. John Kennedy, David Barton. South East of Scotland Regional Genetics Service, Edinburgh: Mary Porteous, Sarah Drummond. Peninsula Clinical Genetics Service, Exeter: Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman, Kathryn Hill. West of Scotland Regional Genetics Service, Glasgow: Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt, Sarah Gibson, Eshika Haque, Ed Tobias, Alexis Duncan. South East Thames Regional Genetics Service, Guy’s Hospital London: Louise Izatt, Chris Jacobs, Caroline Langman, Anna Whaite. North West Thames Regional Genetics Service, Harrow: Huw Dorkins. Leicestershire Clinical Genetics Service, Leicester: Julian Barwell. Yorkshire Regional Genetics Service, Leeds: Julian Adlard, Gemma Serra-Feliu. Cheshire & Merseyside Clinical Genetics Service, Liverpool: Ian Ellis, Catherine Houghton. Manchester Regional Genetics Service, Manchester: D Gareth Evans, Fiona Lalloo, Jane Taylor. North East Thames Regional Genetics Service, NE Thames, London: Lucy Side, Alison Male, Cheryl Berlin. Nottingham Centre for Medical Genetics, Nottingham: Jacqueline Eason, Rebecca Collier. Northern Clinical Genetics Service, Newcastle: Fiona Douglas, Oonagh Claber, Irene Jobson. Oxford Regional Genetics Service, Oxford: Lisa Walker, Diane McLeod, Dorothy Halliday, Sarah Durell, Barbara Stayner. The Institute of Cancer Research and Royal Marsden NHS Foundation Trust: Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Lucia D’Mello, Elizabeth Page, Audrey Ardern-Jones, Kelly Kohut, Jennifer Wiggins, Elena Castro, Anita Mitra, Lisa Robertson. North Trent Clinical Genetics Service, Sheffield: Jackie Cook, Oliver Quarrell, Cathryn Bardsley. South West Thames Regional Genetics Service, London: Shirley Hodgson, Sheila Goff, Glen Brice, Lizzie Winchester, Charlotte Eddy, Vishakha Tripathi, Virginia Attard. Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton: Diana Eccles, Anneke Lucassen, Gillian Crawford, Donna McBride, Sarah Smalley (EMBRACE); (GOG) the investigators of the Australia New Zealand Gynaecological Oncology Group (ANZGOG); (GEMO) Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Centre Hospitalier Universitaire de Lyon / Centre Léon Bérard, & Equipe «Génétique du cancer du sein», Centre de Recherche en Cancérologie de Lyon: Olga Sinilnikova, Sylvie Mazoyer, Laure Barjhoux, Carole Verny-Pierre, Sophie Giraud, Mélanie Léone; and Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Bruno Buecher, Claude Houdayer, Virginie Moncoutier, Muriel Belotti, Carole Tirapo, Antoine de Pauw. Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Audrey Remenieras, Véronique Byrde, Olivier Caron, Gilbert Lenoir. Centre Jean Perrin, Clermont–Ferrand: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet. Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, François Eisinger. Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier. CHU de Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier, Pascal Pujol. Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin, Claude Adenis. Hôpital René Huguenin/Institut Curie, St Cloud: Etienne Rouleau, Rosette Lidereau, Liliane Demange, Catherine Nogues. Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker. Institut Bergonié, Bordeaux: Emmanuelle Barouk-Simonet, Françoise Bonnet, Virginie Bubien, Nicolas Sevenet, Michel Longy. Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel. CHU de Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung, Magalie Peysselon. CHU de Dijon: Fanny Coron, Laurence Faivre. CHU de St-Etienne: Fabienne Prieur, Marine Lebrun, Caroline Kientz. Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice: Marc Frénay. CHU de Limoges: Laurence Vénat-Bouvet. CHU de Nantes: Capucine Delnatte. CHU Bretonneau, Tours: Isabelle Mortemousque. Creighton University, Omaha, USA: Henry T.Lynch, Carrie L.Snyder ; (HCSC) Red Tematica de Investigacion Corporativa en Cancer (ISCIII); (HEBCS) Kristiina Aittomäki, Kirsimari Aaltonen and Carl Blomqvist and Tuomas Heikkinen and research nurse Irja Erkkilä; (HEBON) Netherlands Cancer Institute, Amsterdam, NL: F.B.L. Hogervorst, S. Verhoef, M. Verheus, L.J. van ‘t Veer, F.E. van Leeuwen, M.A. Rookus; Erasmus Medical Center, Rotterdam, NL: M. Collée, A.M.W. van den Ouweland, A. Jager, M.J. Hooning, M.M.A. Tilanus-Linthorst, C. Seynaeve; Leiden University Medical Center, NL, Leiden: C.J. van Asperen, J.T. Wijnen, M.P. Vreeswijk, R.A. Tollenaar, P. Devilee; Radboud University Nijmegen Medical Center, Nijmegen, NL: M.J. Ligtenberg, N. Hoogerbrugge; University Medical Center Utrecht, Utrecht, NL: M.G. Ausems, R.B. van der Luijt; Amsterdam Medical Center, NL: C.M. Aalfs, T.A. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz, H.E.J. Meijers-Heijboer; University Hospital Maastricht, Maastricht, NL: E.B. Gomez-Garcia, C.E. van Roozendaal, Marinus J. Blok, B. Caanen; University Medical Center Groningen University, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits; The Netherlands Foundation for the detection of hereditary tumours, Leiden, NL: H.F. Vasen; (INHERIT) Stéphane Dubois, Martine Dumont, Martine Tranchant (Cancer Genomics Laboratory, CRCHUQ) and Sylvie Desjardins and Marc-André Rodrigue (Plateforme de séquençage et de génotypage des génome du CRCHUL/CHUQ); (kConFab)
Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study; (KUMC) JoEllen Weaver; (OCGN) Teresa Selander, Nayana Weerasooriya and members of the Ontario Cancer Genetics Network; (OSU-CCG) Leigha Senter and Kevin Sweet and the Ohio State University Human Genetics Sample Bank; (SEABASS) Cancer Research Initiatives Foundation (Malaysia), University Malaya (Malaysia), National University Hospital (Singapore), University Kebangsaan Malaysia (Malaysia), Hospital Kuala Lumpur (Malaysia) and Putrajaya Hospital (Malaysia); (SWE-BRCA) Per Karlsson, Margareta Nordling, Annika Bergman and Zakaria Einbeigi, Gothenburg, Sahlgrenska University Hospital, Marie Stenmark-Askmalm and Sigrun Liedgren, Linköping University Hospital, Åke Borg, Niklas Loman, Håkan Olsson, Maria Soller, Helena Jernström, Katja Harbst and Karin Henriksson, Lund University Hospital, Annika Lindblom, Brita Arver, Anna von Wachenfeldt, Annelie Liljegren, Gisela Barbany-Bustinza and Johanna Rantala, Stockholm, Karolinska University Hospital, Beatrice Melin, Henrik Grönberg, Eva-Lena Stattin and Monica Emanuelsson, Umeå University Hospital, Hans Ehrencrona, Richard Rosenquist and Niklas Dahl, Uppsala University Hospital; (UKGRFOCR) Roswell Park Alliance Foundation for their continued support of the Gilda Radner Ovarian Family Cancer Registry, Carole Pye, Patricia Harrington, Eva Wozniak, Kirsten Moysich and Lara Sucheston
Grant Support
This research was supported by NIH grant CA128978, an NCI Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a U.S. Department of Defence Ovarian Cancer Idea award (W81XWH-10-1-0341) and grants from the Breast Cancer Research Foundation and the Komen Foundation for the Cure. This work was also supported by Cancer Research UK grants C12292/A11174 and C1287/A10118. The research leading to these results has received funding from the European Community's Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175). Support was also provided by the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program and by the Canadian Breast Cancer Research Alliance-grant #019511. A.C.A. is a CR-UK Senior Cancer Research Fellow. D.F.E. is CR-UK Principal Research Fellow. GCT is a NHMRC Senior Principal Research Fellow. BFBOCC was supported by the Research Council of Lithuania grant LIG-19/2010 to RJ. BMBSA was supported by grants from the Cancer Association of South Africa (CANSA) to Elizabeth J. van Rensburg. BCFR was supported by the National Cancer Institute, National Institutes of Health under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Columbia University (U01 CA69398), Fox Chase Cancer Center (U01 CA69631), Huntsman Cancer Institute (U01 CA69446), Cancer Prevention Institute of California (formerly the Northern California Cancer Center) (U01 CA69417), University of Melbourne (U01 CA69638), and Research Triangle Institute Informatics Support Center (RFP No. N02PC45022-46). CBCS was supported by The Neye Foundation. CNIO was partially supported by Fundación Mutua Madrileña, Asociación Española Contra el Cáncer, the Spanish Ministry of Science and Innovation (FIS PI08 1120) and the Basque Foundation for Health Innovation and Research (BIOEF): BIO07/CA/006. CONSIT TEAM was supported by grants from Ministero della Salute (Extraordinary National Cancer Program 2006 “Alleanza contro il Cancro” to LV and PR, and “Progetto Tumori Femminili” to PR), Ministero dell’Universita’ e Ricerca (RBLAO3-BETH to PR), Fondazione Italiana per la Ricerca sul Cancro (Special Project “Hereditary tumors” to PR), Associazione Italiana per la Ricerca sul Cancro (4017 to PP) and by funds from Italian citizens who allocated the 5×1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5×1000”). The DKFZ study was supported by funds from the DKFZ. EMBRACE was supported by Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo were supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust were supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles, Elizabeth Bancroft and Lucia D’Mello were supported by Cancer Research UK Grant C5047/A8385. GC-HBOC was supported by a grant of the German Cancer Aid (grant 109076) and by the Centre of Molecular Medicine Cologne (CMMC). The GEMO study was supported by the Ligue National Contre le Cancer ; Association for International Cancer Research Grant (AICR-07-0454) ; and the Association “Le cancer du sein, parlons-en!” Award. The Georgetown study was supported by the Familial Cancer Registry at Georgetown University (NIH/NCI grant P30-CA051008), the Cancer Genetics Network (HHSN261200744000C), and Swing Fore the Cure. GOG was supported through funding provided by both intramural (Clinical Genetics Branch, DCEG) and extramural (Community Oncology and Prevention Trials Program—COPTRG) NCI programs. KAP is the Cancer Council Victoria, Colebatch Clinical Research Fellow. HEBCS was supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (132473), the Finnish Cancer Society, and the Sigrid Juselius Foundation. The HEBON study was supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088, NKI2007-3756 and the ZonMW grant 91109024. HUNBOCS was supported by the Hungarian Research Grant KTIA-OTKA CK-80745. ICO was supported by Asociación Española Contra el Cáncer, Spanish Health Research Fund; Carlos III Health Institute; Catalan Health Institute and Autonomous Government of Catalonia; contract grant numbers ISCIIIRETIC RD06/0020/1051, PI10/01422, PI10/31488 and 2009SGR290. IHCC was supported by a Polish Foundation of Science award to K.J., a fellow of International PhD program, Postgraduate School of Molecular Medicine, Warsaw Medical University. ILUH was supported by the Icelandic Association “Walking for Breast Cancer Research” and by the Landspitali University Hospital Research Fund. INHERIT was supported J.S., Chairholder of the Canada Research Chair in Oncogenetics. IOVHBOCS was supported by Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) and "Ministero della Salute" (“Progetto Tumori Femminili and grant numbers RFPS 2006-5-341353, ACC2/R6.9”). kConFab was supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC [145684, 288704, 454508]. ABS is supported by a NHMRC Senior Research Fellowship. A.K.G. was funded by U01CA69631, 5U01CA113916, and the Eileen Stein Jacoby Fund while at FCCC. The author acknowledges support from The University of Kansas Cancer Center and the Kansas Bioscience Authority Eminent Scholar Program. A.K.G. is the Chancellors Distinguished Chair in Biomedical Sciences endowed Professor. The McGill study was supported by the Jewish General Hospital Weekend to End Breast Cancer. MT holds a Fonds de la Recherche en Santé du Québec clinician-scientist award. The MSKCC study was supported by the Starr Cancer Consortium, the Breast Cancer Research Foundation, the Norman and Carol Stone Cancer Research Initiative, the Kate and Robert Niehaus Clinical Cancer Research Initiative, the Lymphoma Foundation, and the Sabin Family Research Initiative. The NCI study was supported by the Intramural Research Program of the US National Cancer Institute, and by support services contracts NO2-CP-11019-50 and N02-CP-65504 with Westat, Inc, Rockville, MD. NNPIO was supported by the Russian Federation for Basic Research (grants 10-04-92601, 10-04-92110, 11-04-00227) and the Federal Agency for Science and Innovations (contract 16.512.11.2237). OCGN was supported by Cancer Care Ontario and the US National Cancer Institute, National Institutes of Health under RFA # CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators. OSU-CCG was supported by the Ohio State University Comprehensive Cancer Center. PBCS was supported by an Instituto Toscano Tumori grant to MAC. SEABASS was supported by CARIF and University Malaya. The UCSF study was supported by the Helen Diller Family Comprehensive Cancer Center at UCSF, the Avon Foundation, and the Center for Translational and Policy Research in Personalized Medicine (TRANSPERS), NIH/NCI P01 CA130818-02A1. UKFOCR was supported by a project grant from CRUK to Paul Pharoah. The UPENN study was supported Komen Foundation for the Cure to SMD, the Breast Cancer Research Foundation to KLN, and National Institutes of Health grants R01-CA083855 and R01-CA102776 to TRR. WCRI was supported by the American Cancer Society Clinical Research Professorship #SIOP-06-258-06-COUN.
Footnotes
Disclosure of Potential Conflicts of Interest
The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. No potential conflicts of interest are disclosed.
References
- 1.Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL, et al. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniou AC, Kartsonaki C, Sinilnikova OM, Soucy P, McGuffog L, Healey S, et al. Common alleles at 6q25.1 and 1p11.2 are associated with breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 2011;20:3304–3321. doi: 10.1093/hmg/ddr226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engel C, Versmold B, Wappenschmidt B, Simard J, Easton DF, Peock S, et al. Association of the variants CASP8 D302H and CASP10 V410I with breast and ovarian cancer risk in BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers Prev. 2010;19:2859–2868. doi: 10.1158/1055-9965.EPI-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2011;20:3289–3303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudet MM, Kirchhoff T, Green T, Vijai J, Korn JM, Guiducci C, et al. Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS Genet. 2010;6:e1001183. doi: 10.1371/journal.pgen.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbull C, Ahmed S, Morrison J, Pernet D, Renwick A, Maranian M, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antoniou AC, Sinilnikova OM, Simard J, Leone M, Dumont M, Neuhausen SL, et al. RAD51 135G-->C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet. 2007;81:1186–1200. doi: 10.1086/522611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou AC, Cunningham AP, Peto J, Evans DG, Lalloo F, Narod SA, et al. The BOADICEA model of genetic susceptibility to breast and ovarian cancers: updates and extensions. Br J Cancer. 2008;98:1457–1466. doi: 10.1038/sj.bjc.6604305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 12.Mulligan AM, Couch FJ, Barrowdale D, Domchek SM, Eccles D, Nevanlinna H, et al. Common breast cancer susceptibility alleles are associated with tumor subtypes in BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2. Breast Cancer Research BCR. 2011;13:R110. doi: 10.1186/bcr3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramus SJ, Kartsonaki C, Gayther SA, Pharoah PD, Sinilnikova OM, Beesley J, et al. Genetic variation at 9p22.2 and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers. J Natl Cancer Inst. 2010;103:105–116. doi: 10.1093/jnci/djq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, et al. The pathology of familial breast cancer: predictive value of immunohistochemical markers estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol. 2002;20:2310–2318. doi: 10.1200/JCO.2002.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, et al. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gianfrancesco F, Esposito T, Casu G, Maninchedda G, Roberto R, Pirastu M. Emergence of Talanin protein associated with human uric acid nephrolithiasis in the Hominidae lineage. Gene. 2004;339:131–138. doi: 10.1016/j.gene.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 17.Lindstrom S, Vachon CM, Li J, Varghese J, Thompson D, Warren R, et al. Common variants in ZNF365 are associated with both mammographic density and breast cancer risk. Nat Genet. 2011;43:185–187. doi: 10.1038/ng.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gianfrancesco F, Esposito T, Ombra MN, Forabosco P, Maninchedda G, Fattorini M, et al. Identification of a novel gene and a common variant associated with uric acid nephrolithiasis in a Sardinian genetic isolate. Am J Hum Genet. 2003;72:1479–1491. doi: 10.1086/375628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naz G, Ali G, Naqvi SK, Azeem Z, Ahmad W. Mapping of a novel autosomal recessive hypotrichosis locus on chromosome 10q11.23–22.3. Hum Genet. 2010;127:395–401. doi: 10.1007/s00439-009-0784-9. [DOI] [PubMed] [Google Scholar]
- 23.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antoniou AC, Sinilnikova OM, McGuffog L, Healey S, Nevanlinna H, Heikkinen T, et al. Common variants in LSP1, 2q35 and 8q24 and breast cancer risk for BRCA1 and BRCA2 mutation carriers. Hum Mol Genet. 2009;18:4442–4456. doi: 10.1093/hmg/ddp372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.