Abstract
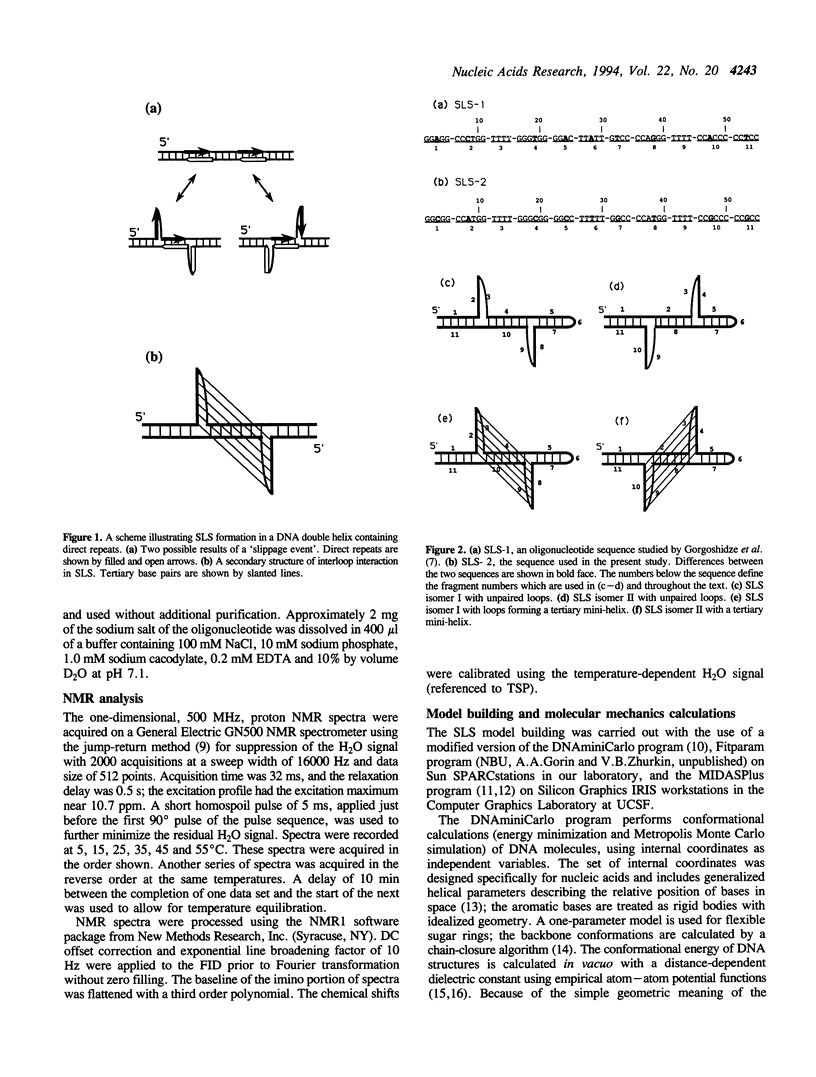
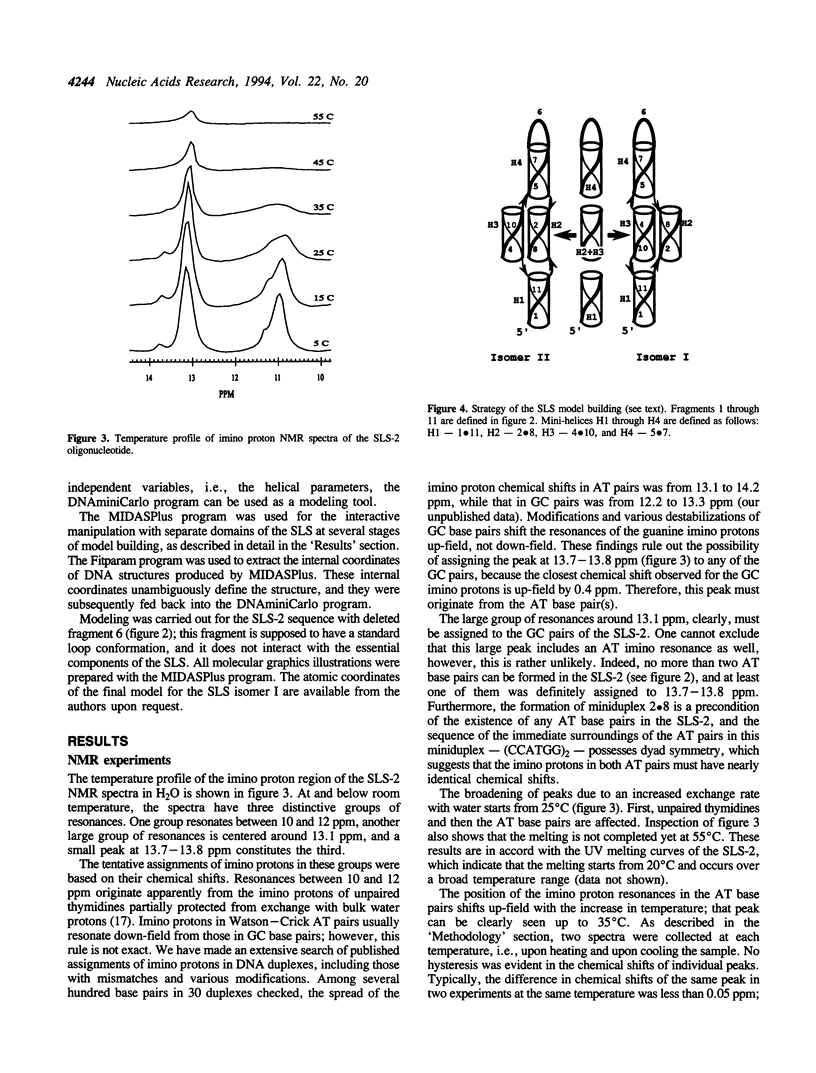
Short direct repeat sequences are often found in regulatory regions of various genes; in some cases they display hypersensitivity to S1 nuclease cleavage in supercoiled plasmids. A non-standard DNA structure (Slipped Loop Structure, or SLS) has been proposed for these regions in order to explain the S1 cleavage data; the formation of this structure may be involved in the regulation of transcription. The structure can be generally classified as a particular type of pseudoknot. To date, no detailed stereochemical model has been developed. We have applied one-dimensional 1H NMR spectroscopy to study a synthetic DNA, 55 nucleotides in length, which cannot fold as a standard hairpin but which may favor the SLS formation. AT base pairs were identified, consistent only with the formation of an additional, tertiary miniduplex in the SLS. An all-atom stereochemically sound model has been developed for the SLS with the use of conformational calculations. The model building studies have demonstrated that the tertiary miniduplex can be formed for one of the plausible SLS isomers, but not for the other.
Full text
PDF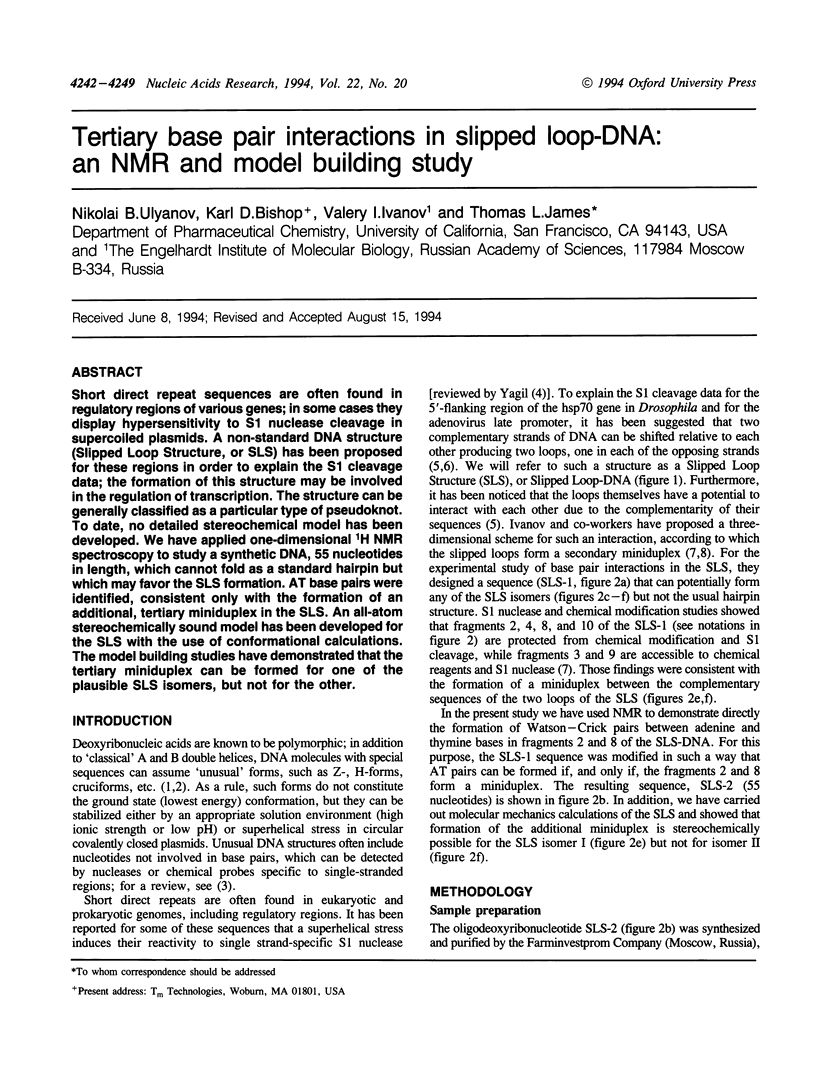



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dam E., Pleij K., Draper D. Structural and functional aspects of RNA pseudoknots. Biochemistry. 1992 Dec 1;31(47):11665–11676. doi: 10.1021/bi00162a001. [DOI] [PubMed] [Google Scholar]
- Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 1989 Jan;8(1):1–4. doi: 10.1002/j.1460-2075.1989.tb03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Schon E., Gora-Maslak G., Patterson J., Efstratiadis A. S1-hypersensitive sites in eukaryotic promoter regions. Nucleic Acids Res. 1984 Nov 12;12(21):8043–8058. doi: 10.1093/nar/12.21.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Huang C. C., Pettersen E. F., Klein T. E., Ferrin T. E., Langridge R. Conic: a fast renderer for space-filling molecules with shadows. J Mol Graph. 1991 Dec;9(4):230-6, 242. doi: 10.1016/0263-7855(91)80016-s. [DOI] [PubMed] [Google Scholar]
- Kilpatrick M. W., Torri A., Kang D. S., Engler J. A., Wells R. D. Unusual DNA structures in the adenovirus genome. J Biol Chem. 1986 Aug 25;261(24):11350–11354. [PubMed] [Google Scholar]
- Mace H. A., Pelham H. R., Travers A. A. Association of an S1 nuclease-sensitive structure with short direct repeats 5' of Drosophila heat shock genes. Nature. 1983 Aug 11;304(5926):555–557. doi: 10.1038/304555a0. [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., Lyamichev V. I., Drushlyak K. N., Dobrynin V. N., Filippov S. A., Frank-Kamenetskii M. D. DNA H form requires a homopurine-homopyrimidine mirror repeat. Nature. 1987 Dec 3;330(6147):495–497. doi: 10.1038/330495a0. [DOI] [PubMed] [Google Scholar]
- Ozoline O. N., Uteshev T. A., Masulis I. S., Kamzolova S. G. Interaction of bacterial RNA-polymerase with two different promoters of phage T7 DNA. Conformational analysis. Biochim Biophys Acta. 1993 Mar 20;1172(3):251–261. doi: 10.1016/0167-4781(93)90211-u. [DOI] [PubMed] [Google Scholar]
- Poltev V. I., Shulyupina N. V. Simulation of interactions between nucleic acid bases by refined atom-atom potential functions. J Biomol Struct Dyn. 1986 Feb;3(4):739–765. doi: 10.1080/07391102.1986.10508459. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Wyatt J. R., Tinoco I., Jr Conformation of an RNA pseudoknot. J Mol Biol. 1990 Jul 20;214(2):437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Dinh S., Neumann J. M., Huynh-Dinh T., Allard P., Lallemand J. Y., Igolen J. DNA fragment conformations IV - Helix-coil transition and conformation of d-CCATGG in aqueous solution by 1H-NMR spectroscopy. Nucleic Acids Res. 1982 Sep 11;10(17):5319–5332. doi: 10.1093/nar/10.17.5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Qiu Q. Q., Deuel T. F. An S1 nuclease sensitive region in the PDGFA-chain gene promoter contains a positive transcriptional regulatory element. Biochem Biophys Res Commun. 1994 Jan 14;198(1):103–110. doi: 10.1006/bbrc.1994.1015. [DOI] [PubMed] [Google Scholar]
- Yagil G. Paranemic structures of DNA and their role in DNA unwinding. Crit Rev Biochem Mol Biol. 1991;26(5-6):475–559. doi: 10.3109/10409239109086791. [DOI] [PubMed] [Google Scholar]
- Yu Y. T., Manley J. L. Structure and function of the S1 nuclease-sensitive site in the adenovirus late promoter. Cell. 1986 Jun 6;45(5):743–751. doi: 10.1016/0092-8674(86)90788-9. [DOI] [PubMed] [Google Scholar]