Abstract
A primary goal of genetic association studies is to elucidate genes and novel biological mechanisms involved in disease. Recently, genome-wide association studies have identified many common genetic variants that are significantly associated with complex diseases such as cancer. In contrast to Mendelian disorders, a sizable fraction of the variants lies outside known protein-coding regions; therefore, understanding their biological consequences presents a major challenge in human genetics. Here we describe an integrated framework to allow non-protein coding loci to be annotated with respect to regulatory functions. This will facilitate identification of target genes as well as prioritize variants for functional testing.
Keywords: GWAS, cancer risk, SNP, functionality
Genome wide association studies (GWAS) have successfully identified common genetic risk factors that drive human diseases. Several GWAS of adult cancers have revealed many variants (usually single nucleotide polymorphisms—SNPs) associated with sporadic cancers. In contrast to Mendelian disorders where most disease-causing mutations result in loss-of-function or truncated proteins, most complex disease-associated variants are located in non-protein coding loci.
Knowledge of the non-protein coding genome is rudimentary compared to the protein coding genome. Unlike protein coding regions, where the genetic code provides a handbook on how triplets of nucleotides predict the coding of amino acids, non-protein coding regions have no such reference manual. The ENCODE data strongly demonstrate that the non-protein part of the genome is much more than a “space filler’ for the protein coding part.1 This opinion piece presents a description of a systematic approach on how to characterize non-protein coding risk regions, which will facilitate the identification of target genes and provide a rational basis for prioritizing putative causal variants.
Most risk-associated SNPs discovered in GWAS are unlikely to be the causal variants that are actually initiating disease. Two factors can explain this phenomenon: (1) Only a subset of variation is actually being tested directly. Whereas many of the current GWAS genotyping arrays are designed to capture and test a large fraction of common variation genome-wide, they do so only indirectly by way of SNP tagging;2 and (2) The polymorphisms associated with risk may lie in regions of strong linkage disequilibrium (LD), and remain highly correlated (based on pair-wise r2) with multiple other polymorphisms spanning relatively large genomic distances, some spanning hundreds of kilobases. LD and r2 are pair-wise measures of association between SNPs and provide a quantitative metric to estimate how often alleles on the same chromosome are co-inherited. Because of the relatively young age of the human species, LD can be quite extensive in the human population and thus, r2 may be high between many SNP pairs. In this situation it is difficult (or impossible) to determine which ones are biologically important solely based on their statistical association with disease risk.
What are the steps to determine which risk-associated SNPs are biologically relevant?
Once a region is significantly associated with a particular phenotype (e.g., prostate cancer), a necessary step is to sequence the region in a set of chromosomes to enumerate all the variation in the region. Creating such a catalogue allows the determination of all polymorphisms that are correlated with the index signal. Data generated from the 1,000 Genome Project (http://www.1000genomes.org), which is estimated to provide a comprehensive catalogue of variation down to 1% in frequency, will soon make this a much easier task.3 As stated above, because of the extensive LD intrinsic to the human genome, many variants will be highly correlated and thus genetically indistinguishable from each other. It should be kept in mind that other classes of variation (e.g., insertion/deletions, copy number variation) will also likely contribute to complex traits.
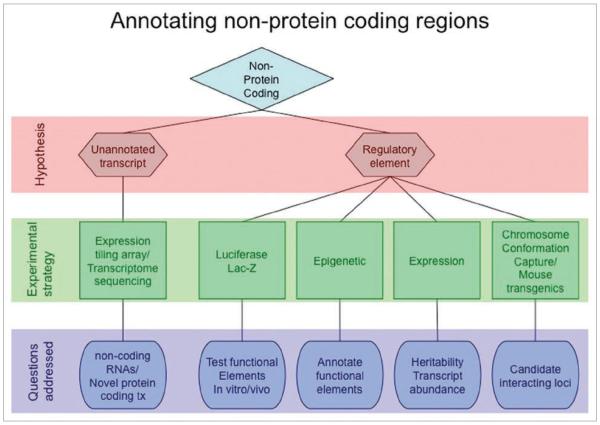
Here we outline an approach to localize biologically functional variation at risk loci revealed through GWAS, particularly when they occur in DNA not associated with annotated genes. Our approach is based on two main hypotheses. First, there may be as yet un-annotated transcripts (non protein-coding or protein-coding) at the regions, and second, the regions may contain regulatory elements, such as enhancers. Identifying biologically relevant regions will inform our understanding of the connection between these regions and their target genes and will aid in the identification of causal alleles. See Figure 1 for a schematic of the process.
Figure 1.
Strategy to annotate non-protein coding regions.
Transcript discovery can be achieved either by using tiling arrays4 of the region or by transcriptome sequencing using “next-generation” sequencing platforms, such as Illumina/Solexa or AB Solid5 to capture both protein-coding (but un-annotated) and regulatory RNA, such as lincRNA.7 If new transcripts are discovered, association of the transcript abundance with risk allele status can be tested. The heritability of transcript levels can be quite high making this a logical and powerful approach to connect risk variant status with target gene (reviewed in ref. 6).
Discovery of regulatory elements is also challenging but as outlined below, we have recently developed a systematic approach to annotate non-protein coding regions. The main objective of this approach is to prioritize regions within an LD block using markers of chromatin ‘activity’ to guide selection of smaller sub-regions rendering them amenable to biochemical analyses and to help focus efforts on a subset of variants within a larger LD block. Thus, we advocate the use of chromatin structural and occupancy information to inform gene regulatory activity. Throughout the rest of the primer, we will focus on enhancers, although the rationale applies equally well to other regulatory elements (e.g., promoters, silencers, locus-control regions, insulators).
Enhancers activate gene expression independent of their orientation and are commonly scattered across large non-coding intervals as well as in introns. It is also important to note that enhancers (as opposed to promoters) are often cell-type specific.9 Recently, chromatin marks have proven to be a powerful method for annotating regulatory elements.
Although evolutionary conservation is commonly used to predict enhancers, the sensitivity and specificity of this strategy alone is low due to the fact that many functional/regulatory elements can be unconstrained across mammalian evolution.2 We suggest that chromatin annotation by using DNase1 hypersensitivity10 and/or histone modifications, which reflect accessibility and histone modifying enzymatic activities, are more informative in identifying functional enhancers. Among histone modifications, mono-methylation of histone H3 at lysine 4 (H3K4me1) identifies specifically, and apparently selectively, enhancers across the board.9,11 On the other hand, trimethylation of histone H3 at the same lysine, (H3K4me3) predominately marks promoters. Thus, the ratio of H3K4 mono-/tri-methylation at specific loci can be diagnostic for enhancers. The acetylation of histone H3 at lysines 9 and 14 (H3K9,14Ac) along with the occupancy of the most common histone acetylator, p300 are additionally a highly accurate means for identifying enhancers and their associated activities.12,13 Finally, RNA polymerase II (RNAPII) occupancy normally demarcates genomic areas where transcription starts and elongation of transcripts occurs, or where RNAPII is poised for this activity.7 However, if occupancy is detected at sites where no transcript is apparent, it may point to the site as being an engaged enhancer mediating transcription at a distant gene. This was shown to be the case in other systems,8 as well as at the well-studied distant enhancer of the PSA gene (reviewed in ref. 9). Performing these analyses (RNA expression and chromatin demarcation) in several tissue specific culture models (possibly adding to them data from ENCODE2) or similar global studies will reveal a joint epigenomic and regulatory profile at the targeted regions and their surrounding chromatin. The resulting rich genomic and epigenomics landscape can then be systematically analyzed using unsupervised methods10 or identified using prior knowledge.7 Specifically, unique modes can be associated with enhancer activity, allowing the dissection of large target regions into well-localized (~1 kb) units with putative function and transforming the functional map problem into a target validation effort for a limited number of loci.
Once the putative enhancers are epigenetically defined, subsequent biochemical/cell biological approaches can be used to functionally validate them as such. The ~1 kb DNA segments encompassing particular epigenetic regions can be cloned into luciferase and β-galactosidase reporter vectors to be tested directly for enhancer activity11 in cell culture and mouse models,12 respectively. Furthermore, differential transcription factor occupancy at specific alleles (defined by SNPs) may be assessed by direct ChIP and electro-mobility shift assays. Once enhancers are validated, the target genes of the enhancers can be sought. The genetic targets of the enhancers may be discovered by utilizing mouse knock-in or knock-out models, followed by genome-wide expression analysis, or by chromatin conformation capture assays, which trap the enhancer (bait) in close proximity of its target genes in cis and/or trans. Newer methodologies, (e.g., zinc finger nucleases and recombinant adeno-associated viruses) that allow an investigator to accurately engineer genomic changes in somatic cells may also prove useful. Once target genes are identified, the transcript abundance of the candidate gene can be tested for association with risk allele status as outlined above. Ideally, mRNA levels will be associated with risk allele status, although this may not always be the case.19,20
Once functional variation in enhancers, their mechanisms of action and target genes have been revealed (as formulated above), it will be important to confirm their association with cancer risk. To increase the likelihood of identification of the causal allele, we propose integration of the re-sequencing and epigenetic data and conducting fine mapping studies in multiple populations. Most of the GWAS conducted to date have been performed in populations of White European ancestry, with the initial signal often showing considerable heterogeneity in its association when examined in other racial/ethnic populations, which may be the result of variability in LD between the tag SNP and the biologically relevant SNP.21,22 If variation in LD patterns is responsible for signal heterogeneity in the tag SNP between populations, then the expectation is that the biologically relevant SNP will be more strongly associated with risk across multiple populations. In this way variants may be moved from functionality to causality, thereby facilitating and complementing fine mapping efforts. Finally, putative causal variation may be tested in mouse models of the particular disease in question.
The target genes under control of functional/causal SNP-containing enhancers may have important roles in the cancer phenotype, such as proliferation, migration and apoptosis. Their cDNAs may be cloned into human expression vectors to study their protein products’ functions. Endpoints of the cancer phenotype, such as cell division, migration and apoptosis rates and protease secretion may be measured in cultured cancer cells and mouse xenografts after the overexpression of vectors encoding the genes of interest, or selected siRNA knockdown of the endogenous genes.
In recent studies from our laboratories we have started down the road outlined above to identify two functional SNPs at chromosome 8q24 respectively associated with prostate and colorectal cancer. In the one study,13 we identified several transcriptional enhancers at 8q24. Two of them, in a prostate cancer risk region, were occupied by the androgen receptor and responded to androgen treatment; one contained a single nucleotide polymorphism (rs11986220) that resides within a FoxA1 binding site, with the prostate cancer risk allele facilitating both stronger FoxA1 binding and stronger androgen responsiveness. In another study,14 we showed that rs6983267, which is significantly associated with colorectal cancer pathogenesis, is situated in another transcriptional enhancer at 8q24. This enhancer activity is affected by the SNP, it physically interacts with the MYC proto-oncogene, and the alleles differentially bind transcription factor 7-like 2 (TCF7L2). Another group also published functional data on rs6983267 in colorectal cancer.15
Importantly and more generally, the approaches formulated above may be applied to any non-protein coding region, as they emerge from genome-wide association studies of any complex phenotype to better understand such risk-associated disease mechanisms. A further benefit is that physiologically relevant, distant-acting enhancers may be investigated; such elements are particularly challenging to identify, let alone study, since they are scattered among the vast non-protein coding portion of the genome.
Acknowledgements
Work from the authors referred to in this paper was supported by the NIH R01 CA109147 (G.A.C.) and R01 CA129435 (M.L.F.), the Prostate Cancer Foundation (G.A.C.), the Whittier Foundation (G.A.C. and C.A.H.), the American Cancer Society Institutional Research Grant IRG-58-007-48 (L.J.), the Mayer Foundation (M.L.F.) the H.L. Snyder Medical Foundation (M.L.F.), the Dana-Farber/Harvard Cancer Center Prostate Cancer SPORE (National Cancer Institute Grant No. 5P50CA90381), the Israeli Science Foundation (A.T.), the J. Harold and Edna L. LaBriola Chair in Genetic Orthopaedic Research (held by B.F.). M.L.F. is a Howard Hughes Medical Institute Physician-Scientist Early Career Awardee.
References
- 1.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haiman CA, Stram DO. Utilizing HapMap and tagging SNPs. Methods Mol Med. 2008;141:37–54. doi: 10.1007/978-1-60327-148-6_3. [DOI] [PubMed] [Google Scholar]
- 3.Curtin K, Iles MM, Camp NJ. Identifying rarer genetic variants for common complex diseases: diseased versus neutral discovery panels. Ann Hum Genet. 2009;73:54–60. doi: 10.1111/j.1469-1809.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yazaki J, Gregory BD, Ecker JR. Mapping the genome landscape using tiling array technology. Curr Opin Plant Biol. 2007;10:534–42. doi: 10.1016/j.pbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet. 2009;10:595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–9. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 9.Jia L, Shen HC, Wantroba M, Khalid O, Liang G, Wang Q, et al. Locus-wide chromatin remodeling and enhanced androgen receptor-mediated transcription in recurrent prostate tumor cells. Mol Cell Biol. 2006;26:7331–41. doi: 10.1128/MCB.00581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaschek R, Tanay A. Spatial Clustering of Multivariate Genomic and Epigenomic Information. RECOMB. 2009:170–83. [Google Scholar]
- 11.Jia L, Berman BP, Jariwala U, Yan X, Cogan JP, Walters A, et al. Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE. 2008;3:3645. doi: 10.1371/journal.pone.0003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–90. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]