Abstract
Background
Plasma levels of high density lipoprotein cholesterol (HDL-C) are known to be heritable, but only a fraction of the heritability is explained. We used a high density genotyping array containing SNPs from HDL-C candidate genes selected on known biology of HDL-C metabolism, mouse genetic studies, and human genetic association studies. SNP selection was based on tagging-SNPs but also included low-frequency nonsynonymous SNPs.
Methods and Results
Association analysis in a cohort containing extremes of HDL-C (case-control, n=1733) provided a discovery phase, with replication in three additional populations for a total meta-analysis in 7,857 individuals. We replicated the majority of loci identified through genome wide association studies and present on the array (including ABCA1, APOA1/C3/A4/A5, APOB, APOE/C1/C2, CETP, CTCF-PRMT8, FADS1/2/3, GALNT2, LCAT, LILRA3, LIPC, LIPG, LPL, LRP4, SCARB1, TRIB1, ZNF664), and provide evidence suggestive of association in several previously unreported candidate gene loci (including ABCG1, GPR109A/B/81, NFKB1, PON1/2/3/4). There was evidence for multiple, independent association signals in five loci, including association with low frequency nonsynonymous variants.
Conclusions
Genetic loci associated with HDL-C are likely to harbor multiple, independent causative variants, frequently with opposite effects on the HDL-C phenotype. Cohorts composed of extreme individuals may be efficiently used in a case-control discovery of quantitative traits.
Keywords: lipids, genetic association, HDL cholesterol, cardiovascular diseases
Background
Elevated high density lipoprotein cholesterol (HDL-C) concentration is associated with decreased risk of atherosclerotic cardiovascular disease, independent of low density lipoprotein cholesterol (LDL-C) concentration1. The high density lipoprotein (HDL) particle is composed of phospholipid, cholesterol, triglyceride and a variety of proteins2 and is believed to exert its anti-atherosclerotic effects via a process called reverse cholesterol transport, where the HDL particle promotes the efflux of cholesterol from extra-hepatic tissues and cells (particularly macrophages) and delivers it to the liver for excretion in the bile3. The HDL particle is generated, matured, and catabolized4 through a complex series of processes presenting many discrete biological steps at which genetic perturbations could profoundly affect HDL concentration. While the heritability of HDL-C concentration is estimated at approximately 50%5, the precise genetic players and the extent to which each of these factors affects HDL-C concentration in the general population is largely unknown.
Genome-wide association studies (GWAS) have identified several genetic loci contributing to variation in plasma lipids, including HDL-C6. Many of the identified loci contain genes previously implicated in lipid metabolism, either through identified Mendelian disorders of lipid metabolism or by biochemical and cell biology investigations of lipid metabolism. GWAS have also identified novel loci that may elucidate previously unsuspected pathways regulating lipid metabolism. However, the commercially available GWAS platforms suffer from uneven genomic SNP coverage and SNP coverage in some of the HDL-C candidate genes is particularly poor. In addition they are designed to assay a sample of common SNPs (minor allele frequency, MAF > 0.05) across the entire genome, rather than provide comprehensive dense genotyping, including low-frequency variants. Furthermore, all of the GWASs to date have been performed in subjects unselected on the basis of lipid traits. Population based resequencing studies of HDL-C candidate genes have suggested that both common and rare associated variants are concentrated in subjects at the extremes of the HDL-C distribution7, 8, suggesting that studies utilizing subjects from the extremes of the HDL-C distribution may be more powerful than similarly sized population cohorts, particularly for identifying low-frequency variants associated with the HDL-C phenotype.
HDL-C candidate gene studies have been previously performed looking at a variable number of HDL-C candidate genes9. Many of these studies have suffered from failure of subsequent studies to replicate their results. Lack of replication is likely due to multiple causes, including studying a limited number of variants in the candidate genes and small sample sizes, with many of the studies analyzing different variants for the same candidate genes9, 10.
To address these concerns, we participated in a collaboration between the University of Pennsylvania Institute of Translational Medicine and Therapeutics (ITMAT), the Broad Institute, and the National Heart Lung and Blood Institute (NHLBI) supported Candidate-gene Association Resource (CARe) Consortium to develop the ITMAT-Broad-CARe (IBC) cardiovascular gene array11 using Illumina Infinium technology12, 13. This array was designed to include 66 HDL-C candidate gene loci (Supplemental Table 1). Candidate genes were chosen based on known biology of HDL metabolism, data from mouse genetic studies, published genetic human association studies, and the first reports of lipid-related GWAS data. The IBC cardiovascular gene array also included low-frequency and rare nonsynonymous variants with potential phenotypic consequences.
We utilized an extreme HDL-C case/control study design as a discovery cohort to assess association of 2415 SNPs in 66 HDL-C candidate genes and replicated our findings in three cohorts composed of subjects unselected on the basis of lipid traits. The dense SNP coverage of candidate genes on the IBC array and the inclusion of low-frequency non-synonymous variants enabled us to identify multiple nonsynonymous SNPs in candidate genes with suggestive evidence of association with HDL-C and which may help explain the association signal. Additionally, we were able to identify several genes with multiple, independent SNPs significantly associated with HDL-C.
Methods
Subjects
Discovery cohort
The University of Pennsylvania (UPenn) HDL Case-Control study (Penn-CC) is composed of subjects with extreme levels of HDL-C recruited through the Hospital of the UPenn clinical laboratory. The UPenn Institutional Review Board (IRB) approved the study protocol. PennCATH is composed of consecutive subjects undergoing coronary angiography at UPenn Health System hospitals and has been previously described 14. In this study, unrelated subjects of European ancestry with HDL > 90th percentile for age and gender were analyzed as cases (n=695) and subjects with HDL < 30th percentile for age and gender were analyzed as controls (n=1038).
Three Replication Cohorts
The UPenn Replication Cohort (Penn-RC), the MONItoring of trends and determinants in CArdiovascular disease/Cooperative Health Research in the Region of Augsburg (MONICA/KORA) Augsburg study, and the Genetic Regulation of Arterial Pressure of Humans in the Community (GRAPHIC) study were used for replication analysis. Genotypes and HDL-C were available on up to 2,752 Penn-RC subjects, 1,544 MONICA/KORA Augsburg participants, and 2020 GRAPHIC subjects. Each study's respective IRB approved the study protocols and informed consent was obtained from all participants.
Statistical analyses
Genotypes were obtained using the HumanCVD beadchip (Illumina, CA). For quality control, we eliminated SNPs with genotype call rate < 95%, with minor allele frequency (MAF) < 0.01 in Penn-RC controls, or if there was a significant departure from Hardy-Weinberg equilibrium (P < 1 × 10-6 in combined cases and controls). We also eliminated individuals with < 97.5% of the array SNPs successfully genotyped.
The discovery cohort was analyzed as a case/control design using multiple logistic regression to test the association of each SNP. The replication cohorts were analyzed using multiple linear regression to test the association of each SNP with HDL-C after adjusting for age and gender under an additive model of inheritance. All analyses were conducted using PLINK software 15.
All candidate gene SNPs were tested in the replication cohorts. Each study was analyzed separately and to summarize the data a meta-analysis was implemented using the METAL software16, which uses the P-value and the direction of the effect to calculate a z-statistic. Individual z-statistics were combined in an overall z-statistic which was calculated as a weighted sum of each of the individual study z-statistics (where weights were proportional to the square-root of the number of individuals examined in each sample and were selected such that the squared weights sum to 1). The corresponding P-value was then calculated. A P-value significance threshold of < 2.03 × 10-5 in the meta-analysis was determined based on the conservative Bonferroni correction (0.05/2459 tests) and a P-value of < 0.05 was specified as a suggestive association with HDL-C.
Conditional analyses were performed by incorporating genotypes of the most significantly associated SNP from each associated locus as covariates simultaneously in the association analyses to identify any residual association signal from additional independent SNPs associated with HDL-C. The conditional analysis was performed as a forward step-wise regression adding the genotypes of the highest remaining associated SNPs in each locus as covariates in an iterative process until there was no longer any evidence of association with HDL. This was performed using a gene-based Bonferonni correction for the total number of candidate genes tested (P < 7.6 × 10-4, 0.05/66 genes). Each study was analyzed separately and the data was summarized using the METAL software16
Additional information is available in Supplemental Methods.
Results
Extreme Case-Control Discovery
A total of 2415 SNPs were analyzed for 66 candidate genes in a case-control cohort composed of individuals from the extremes of the HDL-C phenotypic distribution (Table 1) after removing very low frequency (MAF < 0.01 in controls) and poorly performing SNPs. 31 SNPs from three candidate genes (CETP, LIPG, LPL) reached the threshold for significance in the discovery dataset, based on the conservative Bonferroni correction (P < 2.1 × 10-5, 0.05/2415 SNPs). An additional 293 SNPs from 39 further candidate genes had suggestive evidence of association (uncorrected P < 0.05, Supplemental Table 2).
Table 1.
Baseline Characteristics
| Cohort | Penn-CC Cohort cases n=695 | Penn-CC Cohort controls n=1038 | Penn-RC n=2752 | MONICA/KORA n=1544 | GRAPHIC n= 2020 |
|---|---|---|---|---|---|
| Ascertainment | Physician referral, HDL>90th percentile | Physician referral, HDL<30th percentile | Community-based, prospective cohort | Community-based, prospective cohort | Family--based, population cohort |
| Mean Age (years) | 57.82 ± 14.75 | 60.00 ± 12.66 | 54.72 ± 10.10 | 52.49 ± 10.49 | 39.32 ± 14.50 |
| Female Gender (%) | 48.9 | 38.8 | 33.8 | 44.4 | 49.5 |
| Mean HDL (mg/dl) | 90.12 ± 20.57 | 35.06 ± 6.24 | 49.00 ± 13.51 | 55.65 ± 16.73 | 55.80 ± 14.18 |
| Range HDL (mg/dl) | 60-186 | 20-50 | 9-164 | 2-142 | 19.5-124.8 |
Values with ‘±’ are means ± standard deviation; Penn, University of Pennsylvania; CC, Case-Control; RC, Replication Cohort
Replication Study
As the association of these SNPs was identified through a case/control cohort ascertained on the extremes of a quantitative trait, an attempt was made to replicate these results in three cohorts unselected on the basis of lipid traits to verify that these SNPs are associated with the quantitative trait of HDL-C: a United States cohort of individuals of European descent (Penn-RC) and two European cohorts (MONICA/KORA and GRAPHIC). We analyzed the 324 SNPs with suggestive or significant evidence of association in the case/control discovery cohort (uncorrected P < 0.05). Results from the replication cohorts were combined together using the METAL software (individual cohort and the combined replication analysis results are available in Supplemental Table 2). All 31 of the SNPs reaching the threshold for significance in the discovery cohort were replicated at a significance threshold of P < 0.05 in the combined replication analysis and 40 SNPs from 4 genes (CETP, LPL, APOE, LIPC) were replicated with P < 2.1 × 10-5 in the combined replication analysis.
Meta-Analysis
A meta-analysis was performed for the HDL candidate genes across all four cohorts, discovery and three replication cohorts, in a total of 7,857 subjects. 72 SNPs from six candidate genes (APOE, CETP, FADS1/2/3, LIPC, LIPG, LPL) reached our Bonferroni corrected level of significance (Supplemental Table 3). All of these candidate loci have been previously reported to be significantly associated with HDL-C. Of the 31 SNPs significantly associated in the discovery cohort, only one failed to reach the significance threshold in the meta-analysis (a CETP SNP, rs11076174, meta P = 1.0 × 10-4). Similarly, of the 72 SNPs significantly associated with HDL-C in the meta-analysis, only a single SNP did not exhibit suggestive association in the discovery cohort (a LIPC SNP, rs261338, meta-analysis P = 1.27 × 10-5, Penn-CC P = 0.16). 374 SNPs in 42 candidate genes had suggestive evidence of association after meta-analysis (P < 0.05, Supplemental Table 3). The most significant SNP for each of the candidate genes with at least one SNP exhibiting a P < 0.01 is presented in Table 2. Notably, the most significantly associated SNP for several of the loci (APOA5, APOB, PON2, LRP2) were nonsynonymous variants I44M (rs12287066), A4481T (rs1801695), S311C (rs7493), and G669D (rs34291900); respectively. In addition, many of the candidate genes had nonsynonymous SNPs associated with HDL-C, although these SNPs may not have been the most significantly associated SNP. Supplemental Table 4 lists all of the nonsynonymous SNPs in the candidate genes suggestively associated with HDL-C (P < 0.05).
Table 2.
Most significant SNP for each Candidate Locus in Meta-analysis
| Penn-CC | Penn-RC | MONICA/KORA | GRAPHIC | Meta-analysis | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | SNP | A1 | A2 | MAF | T | P | N | T | P | N | T | P | N | Z | P | N | Z | P | N |
| CETP | rs17231506 | A | G | 0.30 | 7.57 | 3.82E-14 | 1600 | 6.25 | 4.68E-10 | 2691 | 6.84 | 1.12E-11 | 1536 | 7.83 | 5.00E-15 | 2020 | 14.04 | 8.85E-45 | 7847 |
| LPL | rs264 | A | G | 0.15 | 3.57 | 3.60E-04 | 1599 | 3.54 | 4.13E-04 | 2691 | 2.73 | 6.39E-03 | 1543 | 3.63 | 2.87E-04 | 2019 | 6.73 | 1.75E-11 | 7852 |
| LIPG | rs2156552 | A | T | 0.14 | -4.07 | 4.80E-05 | 1600 | -2.30 | 2.16E-02 | 2693 | -0.95 | 3.40E-01 | 1543 | -3.37 | 7.48E-04 | 2020 | -5.31 | 1.09E-07 | 7856 |
| APOE/C1/C2 | rs2075650 | G | A | 0.13 | -2.34 | 1.93E-02 | 1600 | -4.15 | 3.46E-05 | 2693 | -2.31 | 2.12E-02 | 1544 | -1.36 | 1.73E-01 | 2020 | -5.19 | 2.07E-07 | 7857 |
| FADS1/2/3 | rs174570 | A | G | 0.11 | -2.95 | 3.22E-03 | 1600 | -2.92 | 3.52E-03 | 2693 | -2.65 | 8.16E-03 | 1543 | -1.41 | 1.58E-01 | 2020 | -4.93 | 8.37E-07 | 7856 |
| LIPC | rs588136 | G | A | 0.22 | 2.36 | 1.83E-02 | 1585 | 2.10 | 3.63E-02 | 2670 | 3.38 | 7.58E-04 | 1531 | 2.19 | 2.89E-02 | 2020 | 4.89 | 1.01E-06 | 7806 |
| APOA1/A4/A5/C3 | rs12287066* | A | C | 0.08 | -2.81 | 5.03E-03 | 1600 | -2.05 | 4.03E-02 | 2691 | -1.61 | 1.07E-01 | 1540 | -1.65 | 9.89E-02 | 2019 | -4.02 | 5.87E-05 | 7850 |
| ABCG1 | rs914189 | C | G | 0.21 | 2.34 | 1.93E-02 | 1600 | 2.78 | 5.50E-03 | 2693 | 1.20 | 2.29E-01 | 1544 | 1.51 | 1.32E-01 | 2020 | 3.98 | 6.97E-05 | 7857 |
| APOB | rs1801695* | A | G | 0.03 | 2.87 | 4.05E-03 | 1600 | 2.72 | 6.59E-03 | 2693 | 0.50 | 6.14E-01 | 1544 | 1.65 | 9.80E-02 | 2020 | 3.95 | 7.80E-05 | 7857 |
| ABCA1 | rs3905001 | G | C | 0.28 | 1.53 | 1.27E-01 | 1598 | 1.74 | 8.17E-02 | 2693 | 1.85 | 6.52E-02 | 1540 | 2.43 | 1.51E-02 | 2020 | 3.76 | 1.71E-04 | 7851 |
| GPR109A/B/81 | rs3922628 | T | A | 0.22 | 2.61 | 8.97E-03 | 1597 | 1.13 | 2.58E-01 | 2684 | 0.56 | 5.73E-01 | 1434 | 3.12 | 1.81E-03 | 2017 | 3.69 | 2.23E-04 | 7732 |
| SCARB1 | rs838878 | A | G | 0.34 | 1.82 | 6.93E-02 | 1600 | 1.70 | 8.85E-02 | 2693 | 1.94 | 5.23E-02 | 1543 | 1.59 | 1.12E-01 | 2020 | 3.48 | 4.94E-04 | 7856 |
| PON1/2/3/4 | rs7493* | G | C | 0.24 | 2.03 | 4.22E-02 | 1600 | 0.66 | 5.10E-01 | 2693 | 2.36 | 1.86E-02 | 1543 | 2.20 | 2.81E-02 | 2019 | 3.46 | 5.42E-04 | 7855 |
| NFKB1 | rs3774964 | G | A | 0.36 | 2.62 | 8.90E-03 | 1600 | 2.03 | 4.30E-02 | 2693 | -0.06 | 9.55E-01 | 1539 | 2.05 | 4.00E-02 | 2020 | 3.38 | 7.19E-04 | 7852 |
| PCSK6 | rs1471656 | A | G | 0.45 | 2.02 | 4.31E-02 | 1600 | 1.35 | 1.77E-01 | 2693 | 2.39 | 1.72E-02 | 1538 | 1.19 | 2.35E-01 | 2020 | 3.36 | 7.75E-04 | 7851 |
| PCSK5 | rs1340510 | G | A | 0.28 | 1.41 | 1.59E-01 | 1599 | 2.42 | 1.54E-02 | 2692 | 2.94 | 3.39E-03 | 1542 | -0.49 | 6.24E-01 | 2019 | 3.10 | 1.91E-03 | 7852 |
| GALNT2 | rs2144300 | G | A | 0.44 | -2.07 | 3.90E-02 | 1600 | -2.32 | 2.02E-02 | 2692 | -0.43 | 6.64E-01 | 1542 | -1.18 | 2.37E-01 | 2020 | -3.08 | 2.05E-03 | 7854 |
| LCAT | rs2292318 | A | G | 0.14 | 0.00 | 9.96E-01 | 1600 | 1.22 | 2.24E-01 | 2693 | 2.48 | 1.33E-02 | 1509 | 2.37 | 1.76E-02 | 2020 | 3.01 | 2.62E-03 | 7822 |
| PPARD | rs2016520 | G | A | 0.23 | 1.83 | 6.70E-02 | 1600 | 2.05 | 4.06E-02 | 2693 | 0.73 | 4.67E-01 | 1544 | 0.97 | 3.31E-01 | 2020 | 2.84 | 4.51E-03 | 7857 |
| PLTP | rs378114 | A | G | 0.29 | 1.91 | 5.60E-02 | 1598 | 1.17 | 2.43E-01 | 2693 | 0.65 | 5.18E-01 | 1543 | 1.97 | 4.83E-02 | 2020 | 2.83 | 4.60E-03 | 7854 |
| SOAT1 | rs4421551 | C | A | 0.14 | 2.55 | 1.08E-02 | 1600 | 2.12 | 3.40E-02 | 2690 | 0.90 | 3.68E-01 | 1530 | 0.06 | 9.56E-01 | 2020 | 2.82 | 4.81E-03 | 7840 |
| LRP2 | rs34291900* | A | G | 0.03 | -0.66 | 5.11E-01 | 1600 | -3.16 | 1.59E-03 | 2693 | -1.49 | 1.38E-01 | 1544 | 0.10 | 9.17E-01 | 2020 | -2.75 | 5.95E-03 | 7857 |
A, allele; MAF, Minor allele frequency from Penn-RC; T, T-statistic; P, P-value; Z, Z-statistic; N, Sample size
denotes nonsynonymous SNP
Many of the most significant SNPs associated with HDL-C in first generation GWAS were included on the IBC array. A few additional SNPs from late first-generation GWAS were included on the second version of the IBC array, which was only genotyped in one of the subcohorts of Penn-RC, PennCAC. Previous GWAS SNPs were examined in all cohorts and in the meta-analysis (Supplemental Table 5). We replicated 47 of 65 testable SNPs, and 17 of 28 testable loci at P < 0.05, including the recently reported loci near the LILRA3, LRP4, SCARB1, TRIB1, and ZNF648 genes6.
Conditional Analyses
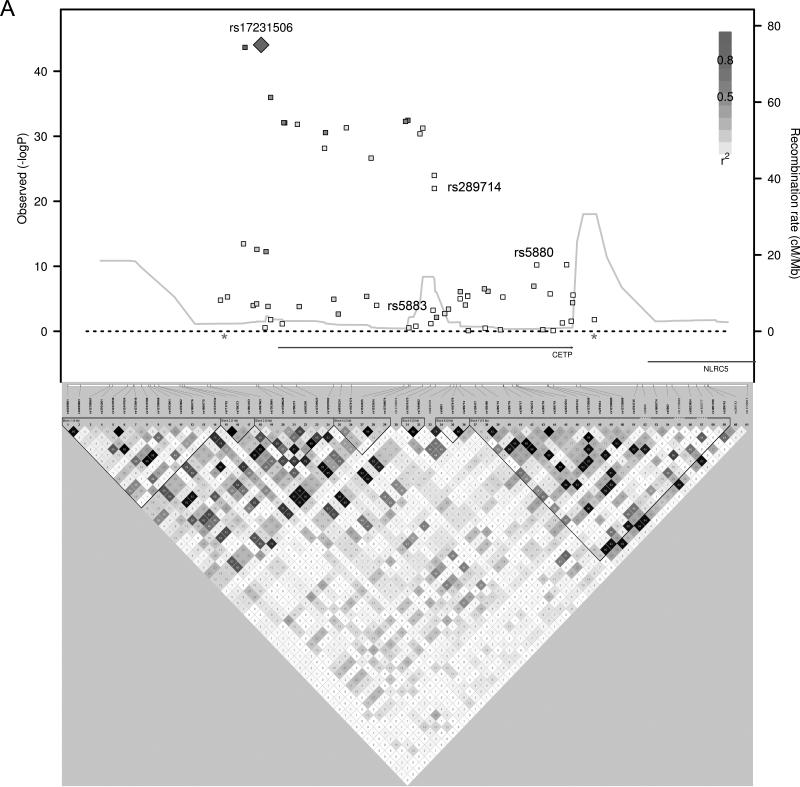
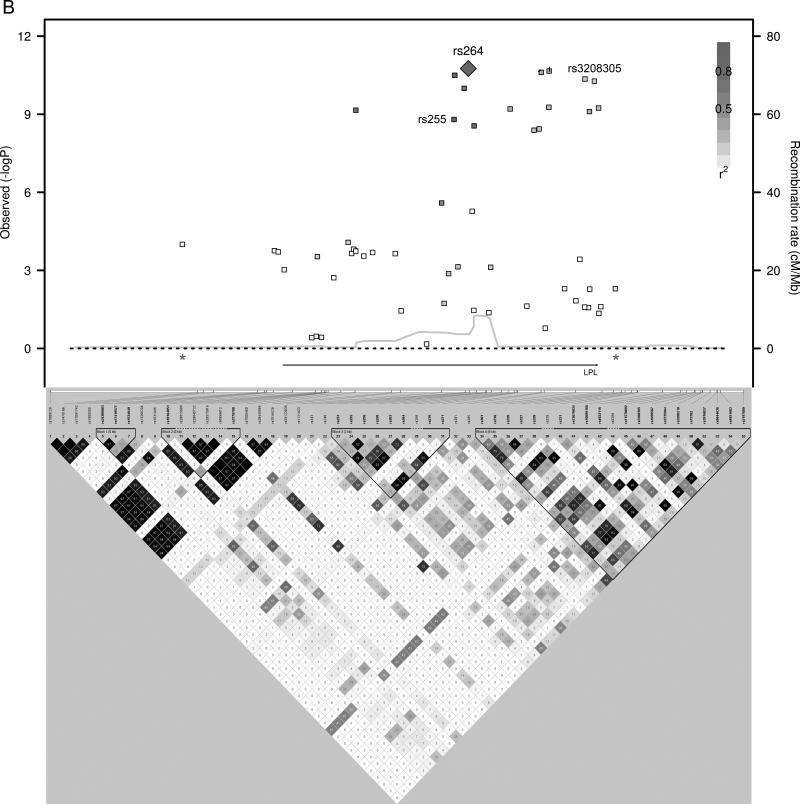
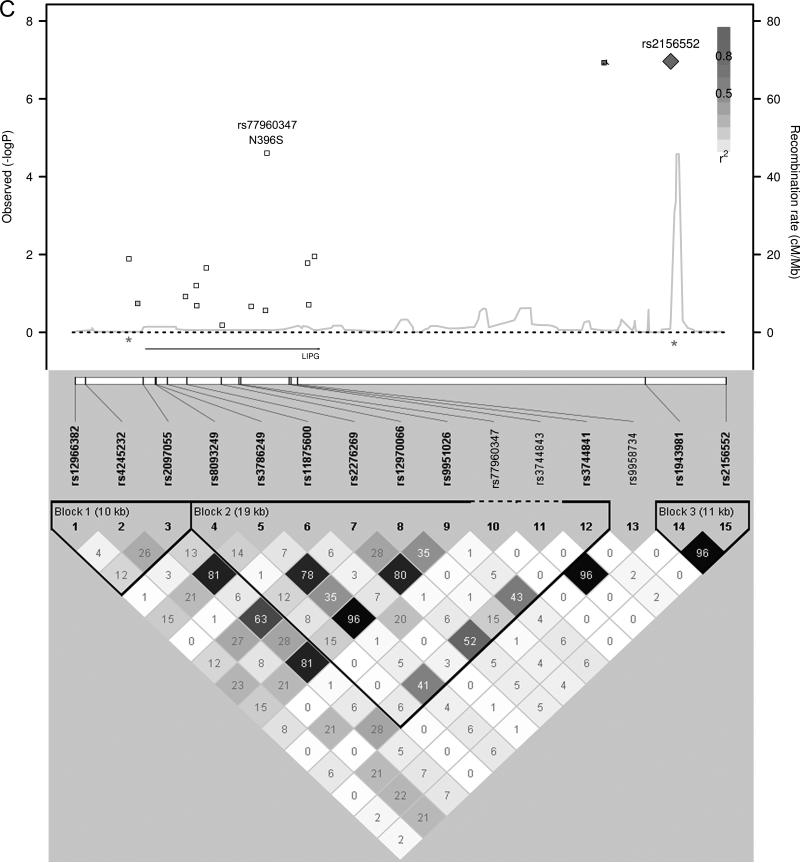
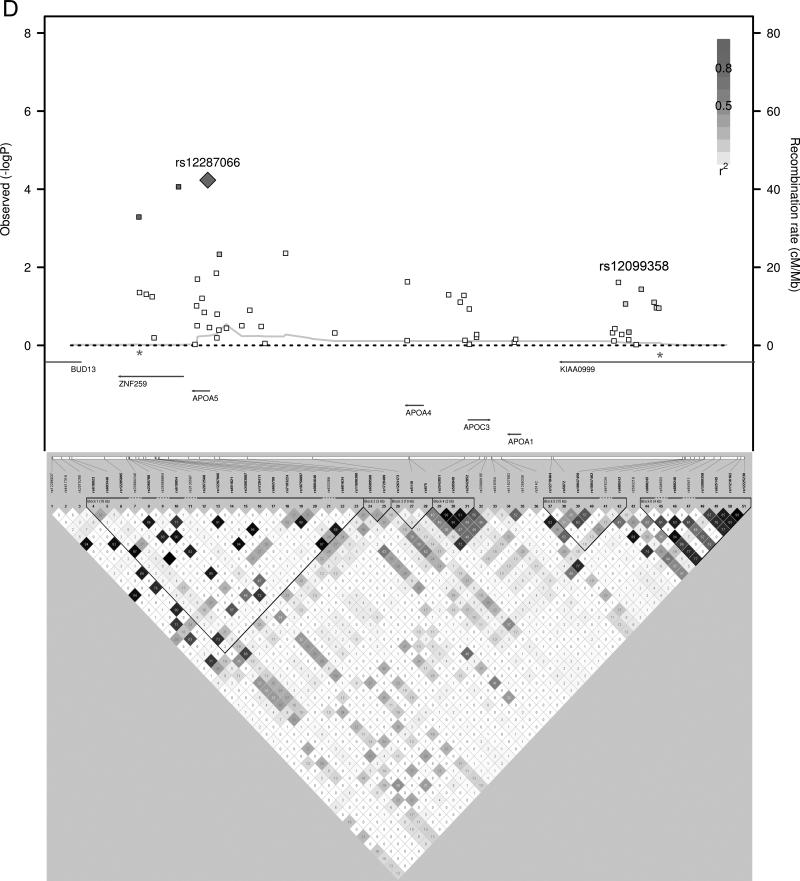
We used a forward step-wise regression analysis to identify additional independent association signals at the most significantly associated loci. Using a gene-based Bonferonni correction for the total number of candidate genes tested (P < 7.6 × 10-4, 0.05/66 genes), 14 candidate loci contained SNPs associated with HDL-C. Genotypes of the most significantly associated SNPs from each of these 14 loci were simultaneously included as covariates in the association analyses. Additional independently associated SNPs were identified and their genotypes were included as covariates in the association analyses in an iterative process to identify all the independently associated SNPs in these 14 loci. Five of the candidate gene loci had evidence of multiple independent HDL-C association signals, sometimes with the minor alleles of the independently associated SNPs having opposite directions of effect (Table 3 and Figure 1). Combining the 22 independent SNPs identified in this manner, the proportion of variance of HDL-C explained was 6.4% in Penn-RC, 8.2% in MONICA/KORA, and 8.0% in the GRAPHIC founders, or approximately 13% - 16% of the heritability was explained.
Table 3.
Conditional Analyses
| Gene | 1st | 2nd | 3rd | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | P | D | MAF | AA Change | SNP | P | D | MAF | AA Change | SNP | P | D | MAF | AA Change | |
| CETP | rs289714 | 2.19 × 10-15 | - | 0.21 | - | rs5883 | 1.98 × 10-5 | + | 0.06 | F287F - |
rs5880 | 4.40 × 10-4 | - | 0.04 | P390A |
| LPL | rs3208305 | 1.19 × 10-4 | + | 0.32 | - | rs255 | 2.36 × 10-4 | + | 0.16 | ||||||
| LIPG | rs77960347 | 7.46 × 10-4 | + | 0.01 | N396S | ||||||||||
| APOA1/A4/A5/C3 | rs12099358 | 4.11 × 10-6 | + | 0.18 | - | ||||||||||
| ABCA1 | rs2740487 | 6.36 × 10-4 | + | 0.42 | - | ||||||||||
D, Direction; MAF, Minor allele frequency (derived from Penn-RC), Genotypes of the most significantly associated SNP from each of the top 14 loci were included as covariates in the association analysis, rs17231506 (CETP), rs264 (LPL), rs2156552 (LIPG), rs2075650 (APOE/C1/C2), rs174570 (FADS1/2/3), rs588136 (LIPC), rs12287066 (APOA1/A4/A5/C3), rs914189 (ABCG1), rs1801695 (APOB), rs3905001 (ABCA1), rs3922628 (GPR109A/B/81), rs838878 (SCARB1), rs7493 (PON1/2/3/4), rs3774964 (NFKB1).
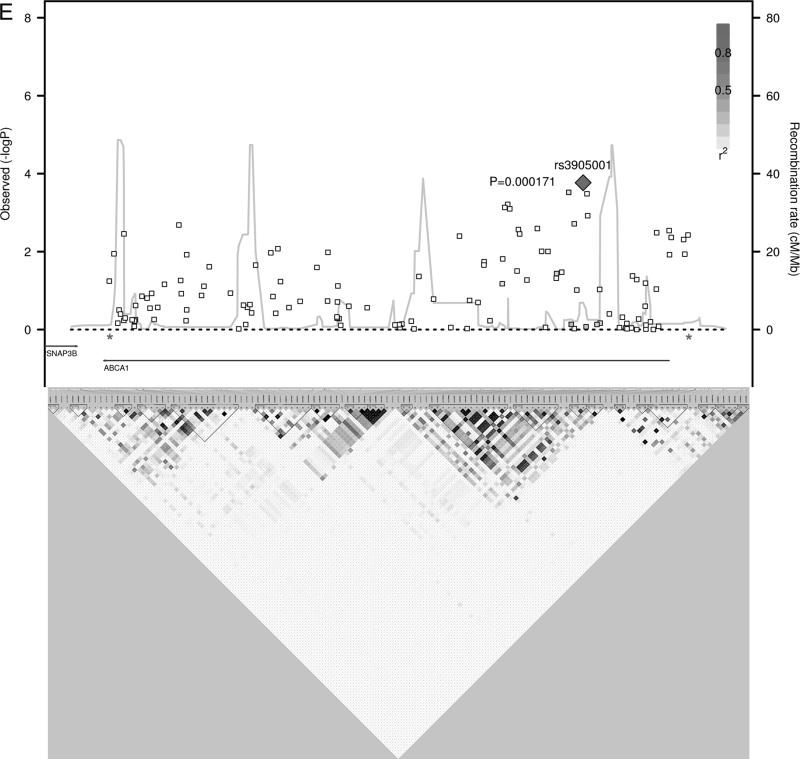
Figure 1.
HDL-C Regional Association plots. P-values from SNPs in loci with multiple, independent association signals (A) CETP, (B) LPL, (C) LIPG, (D) APOA1/A4/A5/C3, (E) ABCA1 are plotted with linkage disequilibrium plots, which have been enlarged to show the region between the asterisks.
Discussion
This study represents a comprehensive candidate gene approach to identifying loci affecting HDL-C in humans, genotyping genes selected based on known biology of HDL metabolism, data from mouse studies, published genetic association studies, and available first-generation GWAS data (Supplemental Table 1). An extreme HDL phenotype case-control cohort was utilized for discovery purposes with extensive replication in populations unselected for lipids. We also utilized these studies to perform a meta-analysis of over 7,800 individuals. The extreme case-control discovery cohort design for the quantitative trait HDL-C appeared to efficiently identify associated SNPs. All of 31 SNPs reaching the significance threshold in the discovery cohort replicated at a significance threshold of P < 0.05 and all but one were significantly associated in the meta-analysis. All of the 40 SNPs significantly associated with HDL-C in the combined QTL analysis utilizing studies unselected for lipids (in this paper used to replicate the discovery cohort results) were at least suggestively associated in the discovery cohort. Similarly, only one of 72 total SNPs reaching the significance threshold after meta-analysis failed to be even nominally associated in the discovery cohort. Additionally, cohorts composed of extreme individuals may be ideally suited for identifying low-frequency, causative alleles of large effect in candidate loci when such alleles are adequately represented. The IBC array allowed higher density genotyping than commercially available genome wide platforms, with targeted coverage of candidate genes and an emphasis on potentially functional coding variants.
Several of the HDL-C GWAS loci reported to date are replicated here and suggestive evidence for an association with HDL-C for several additional candidate genes is provided, many of which have not been reported in GWAS studies to date and may be poorly covered on commercially available platforms11. Prior candidate gene studies have explored many of these loci with variable results, likely due to small sample sizes and limited genotyping9, 10. Interest in these loci persists due to compelling biological and animal data such as for ABCG1, GPR109A, and PCSK5. ABCG1 knock-out mice exhibit decreased HDL-C on a high-fat diet and has been shown to mediate cholesterol efflux to HDL17, 18. GPR109A has been shown to be the receptor for nicotinic acid19, which is used clinically to raise HDL-C levels. PCSK5 is a proprotein convertase that has been shown to modulate HDL-C levels through inactivation of endothelial lipase20, 21. A genetic variant in PCSK5 was also recently shown to segregate with low HDL-C in a human pedigree22.
Evidence is also provided for multiple independent associations with HDL-C within five loci (CETP, LPL, LIPG, APOA1/A4/A5/C3, ABCA1), consistent with the findings published by Talmud et al using the same IBC array. They identified multiple independent associations with HDL-C in CETP, LPL, LIPC, and APOA523. A recent meta-analysis of 46 lipid GWASs also identified 6 HDL-C loci with at least a second independent association with HDL-C including LPL, ABCA1, APOA1/A4/A5/C3, ZNF664, LIPC, and CETP6. We note that the minor alleles of the SNPs that are independently associated may have effects in opposite directions and suggest that genes harbor multiple causative variants across the frequency spectrum from common to rare variants. As an example, we replicated the previously published finding in the endothelial lipase gene, LIPG, showing that common SNPs 3’ to the gene and the low frequency nonsynonymous variant, N396S (rs77960347), independently associate with HDL-C8. The 3’ SNPs may affect transcript levels while the N396S variant has been shown to have reduced phospholipase activity in vitro and in vivo8.
It is also possible that a single locus may harbor more than one causative gene. This may be the case at the APOA1/A4/A5/C3 locus. We identified two independent signals in the region, one the APOA5 nonsynonymous variant I44M (rs12287066) and one an APOA1 intronic SNP (rs12099358, Figure 1D). Interestingly, meta-analysis of 46 GWASs also identified two independent SNPs in the locus with the first SNP near APOA5 and the second SNP near APOA16. Furthermore, Chasman et al showed that triglyceride phenotype associations accumulate near APOA5 but HDL-related phenotypic associations predominated near APOA124, suggesting two independent effects at the locus, possibly from two different genes.
Our analysis identified nonsynonymous variants as the most highly associated variants with HDL-C for APOA5, APOB, PON2, and LRP2 (Table 2). We identified several other non-synonymous variants that, while not the SNP with the strongest evidence for association with HDL-C for the gene, may nonetheless have real and important effects on HDL-C levels. Specifically, two of our candidate loci (CETP and LIPG) harbor nonsynonymous variants significantly associated with HDL-C independent of the most significantly associated SNPs in the gene. It is possible that some of the other nonsynonymous SNPs identified in our study are also independently associated with HDL-C, but our study continues to be underpowered to detect the independent association. GWAS platforms emphasizing common variants (MAF > 0.05) are likely to miss low frequency nonsynonymous variants, which are difficult to impute due to their low frequency. The higher power (and larger numbers) needed to identify low-frequency causative variants suggest that even larger sample sizes analyzed on the IBC array may be able to identify rare nonsynonymous causative variants.
It is important to note that association, even of nonsynonymous variants, is not necessarily indicative of causality. The APOB SNP with the most significant HDL-C association in a meta-analysis of 46 GWASs was the nonsynonymous variant S4338N (rs1042034, MAF=0.22)6 which is in strong LD (r2=0.98) with another nonsynonymous APOB variant P2739R (rs676210, MAF=0.22) also significantly associated with HDL-C in the meta-analysis and both variants are associated with HDL-C in our study. The most significant APOB SNP in our study (A4481T, rs1801695, MAF=0.03) was unavailable in a majority of the 46 GWASs in the meta-analysis, even after imputation. Additional studies with these nonsynonymous variants will be necessary to determine which, if any, of these variants affect protein function and influence HDL-C.
In conclusion, this HDL-C candidate gene association study in over 7,800 individuals using a dense genotyping array provides evidence that a single locus associated with HDL-C may harbor multiple causative variants, sometimes with effects in opposite directions, and with various putative mechanisms of effect. This study replicates many previously reported GWAS loci and provides suggestive evidence of association with HDL-C for several additional candidate genes. Furthermore, this study provides evidence that the extreme case-control discovery cohort design for studying quantitative traits can be efficiently used to identify associated SNPs.
Supplementary Material
Acknowledgments
We would like to thank the individuals and families from all of these studies for their participation as well as the referring physicians both in and outside the University of Pennsylvania Health System for their help with recruitment of the HHDL study. We also thank the research nurses and other staff who undertook the recruitment and phenotyping.
Funding Sources: This work was supported by a National Heart, Lung and Blood Institute Ruth L. Kirschstein National Research Service Award (F30, to A.C.E.), an NIH R01 HL089309 (to D.J.R.) and a Doris Duke Charitable Foundation Distinguished Clinical Scientist Award (to D.J.R.). The MONICA/KORA Augsburg studies were financed by the Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany and supported by grants from the German Federal Ministry of Education and Research (BMBF). Part of this work was financed by the German National Genome Research Network (NGFNPlus, project number 01GS0834) and through additional funds from the University of Ulm. Furthermore, the research was supported within the Munich Center of Health Sciences (MC Health) as part of LMU innovative. Recruitment and genotyping of the GRAPHIC cohort was funded by the British Heart Foundation. N.J.S. holds a British Heart Foundation Chair of Cardiology and M.D.T. holds a Medical Research Council Clinician Scientist Fellowship (G0501942). N.J.S and M.T are supported by the Leicester NIHR Biomedical Research Unit in Cardiovascular Disease. This study is part of the research portfolio supported by the Leicester NIHR Biomedical Research Unit in Cardiovascular Disease.
Footnotes
Conflict of Interest Disclosures: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The framingham study. Am J Med. 1977;62:707–714. doi: 10.1016/0002-9343(77)90874-9. [DOI] [PubMed] [Google Scholar]
- 2.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, Byun J, Vuletic S, Kassim S, Singh P, Chea H, Knopp RH, Brunzell J, Geary R, Chait A, Zhao XQ, Elkon K, Marcovina S, Ridker P, Oram JF, Heinecke JW. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of hdl. J Clin Invest. 2007;117:746–756. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuchel M, Rader DJ. Macrophage reverse cholesterol transport: Key to the regression of atherosclerosis? Circulation. 2006;113:2548–2555. doi: 10.1161/CIRCULATIONAHA.104.475715. [DOI] [PubMed] [Google Scholar]
- 4.Rader DJ. Molecular regulation of hdl metabolism and function: Implications for novel therapies. J Clin Invest. 2006;116:3090–3100. doi: 10.1172/JCI30163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–1156. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- 6.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Cecile JWJA, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr., Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romeo S, Pennacchio LA, Fu Y, Boerwinkle E, Tybjaerg-Hansen A, Hobbs HH, Cohen JC. Population-based resequencing of angptl4 uncovers variations that reduce triglycerides and increase hdl. Nat Genet. 2007;39:513–516. doi: 10.1038/ng1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmondson AC, Brown RJ, Kathiresan S, Cupples LA, Demissie S, Manning AK, Jensen MK, Rimm EB, Wang J, Rodrigues A, Bamba V, Khetarpal SA, Wolfe ML, Derohannessian S, Li M, Reilly MP, Aberle J, Evans D, Hegele RA, Rader DJ. Loss-of-function variants in endothelial lipase are a cause of elevated hdl cholesterol in humans. J Clin Invest. 2009;119:1042–1050. doi: 10.1172/JCI37176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boes E, Coassin S, Kollerits B, Heid IM, Kronenberg F. Genetic-epidemiological evidence on genes associated with hdl cholesterol levels: A systematic in-depth review. Exp Gerontol. 2009;44:136–160. doi: 10.1016/j.exger.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissglas-Volkov D, Pajukanta P. Genetic causes of high and low serum hdl-cholesterol. J Lipid Res. 2010;51:2032–2057. doi: 10.1194/jlr.R004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k snp array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable snp genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 13.Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat Meth. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 14.Lehrke M, Millington SC, Lefterova M, Cumaranatunge RG, Szapary P, Wilensky R, Rader DJ, Lazar MA, Reilly MP. Cxcl16 is a marker of inflammation, atherosclerosis, and acute coronary syndromes in humans. J Am Coll Cardiol. 2007;49:442–449. doi: 10.1016/j.jacc.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiersma H, Nijstad N, de Boer JF, Out R, Hogewerf W, Van Berkel TJ, Kuipers F, Tietge UJ. Lack of abcg1 results in decreased plasma hdl cholesterol levels and increased biliary cholesterol secretion in mice fed a high cholesterol diet. Atherosclerosis. 2009 doi: 10.1016/j.atherosclerosis.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy MA, Barrera GC, Nakamura K, Baldan A, Tarr P, Fishbein MC, Frank J, Francone OL, Edwards PA. Abcg1 has a critical role in mediating cholesterol efflux to hdl and preventing cellular lipid accumulation. Cell Metab. 2005;1:121–131. doi: 10.1016/j.cmet.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, Offermanns S. Puma-g and hm74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med. 2003;9:352–355. doi: 10.1038/nm824. [DOI] [PubMed] [Google Scholar]
- 20.Seidah NG, Khatib AM, Prat A. The proprotein convertases and their implication in sterol and/or lipid metabolism. Biol Chem. 2006;387:871–877. doi: 10.1515/BC.2006.110. [DOI] [PubMed] [Google Scholar]
- 21.Jin W, Wang X, Millar JS, Quertermous T, Rothblat GH, Glick JM, Rader DJ. Hepatic proprotein convertases modulate hdl metabolism. Cell Metab. 2007;6:129–136. doi: 10.1016/j.cmet.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iatan I, Dastani Z, Do R, Weissglas-Volkov D, Ruel I, Lee JC, Huertas-Vazquez A, Taskinen MR, Prat A, Seidah NG, Pajukanta P, Engert JC, Genest J. Genetic variation at the proprotein convertase subtilisin/kexin type 5 gene modulates high-density lipoprotein cholesterol levels. Circ Cardiovasc Genet. 2009;2:467–475. doi: 10.1161/CIRCGENETICS.109.877811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, Casas JP, Sofat R, Kumari M, Rodriguez S, Johnson T, Newhouse SJ, Dominiczak A, Samani NJ, Caulfield M, Sever P, Stanton A, Shields DC, Padmanabhan S, Melander O, Hastie C, Delles C, Ebrahim S, Marmot MG, Smith GD, Lawlor DA, Munroe PB, Day IN, Kivimaki M, Whittaker J, Humphries SE, Hingorani AD. Gene-centric association signals for lipids and apolipoproteins identified via the humancvd beadchip. Am J Hum Genet. 2009;85:628–642. doi: 10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, Cupples LA, Hamsten A, Kathiresan S, Malarstig A, Ordovas JM, Ripatti S, Parker AN, Miletich JP, Ridker PM. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS Genet. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.