Abstract
Modeling and analysis of MR images of the early developing human brain is a challenge because of the transient nature of different tissue classes during brain growth. To address this issue, a statistical model that can capture the spatial variation of structures over time is needed. Here, we present an approach to building a spatio-temporal model of tissue distribution in the developing brain which can incorporate both developed tissues as well as transient tissue classes such as the germinal matrix by using constrained higher order polynomial models. This spatio-temporal model is created from a set of manual segmentations through groupwise registration and voxelwise non-linear modeling of tissue class membership, that allows us to represent the appearance as well as disappearance of the transient brain structures over time. Applying this model to atlas-based segmentation, we generate age-specific tissue probability maps and use them to initialize an EM segmentation of the fetal brain tissues. The approach is evaluated using clinical MR images of young fetuses with gestational ages ranging from 20.57 to 24.71 weeks. Results indicate improvement in performance of atlas-based EM segmentation provided by higher order temporal models that capture the variation of tissue occurrence over time.
1 Introduction
This work is motivated by the study of early human brain development in utero from fetal magnetic resonance (MR) imaging. Automated labeling of tissues is a key initial step in the accurate quantification of brain development. This allows us to model the normal development process and to extract rules to detect growth patterns that may be related to abnormal outcomes. In order to meaningfully label tissues present within a given brain image, it is necessary to interpret the anatomy in relation to its developmental stage. As identified by previous studies of neonates [1, 2], infants [3], young children [4] and adults [5], the use of an age-specific atlas can significantly improve the results of automated analysis of brain image data.
The key challenges in segmentation of the early developing brain, as opposed to the developed adult brain, are the dramatic changes occurring over very short timescales [6]. These impose not simply shape changes but underlying changes in morphology whereby transient tissue classes appear and then disappear from different brain regions over time. Such cases would require the construction of many distinct atlases for different weeks of brain development.
Instead, here we describe an approach to modeling a complete 4-dimensional atlas of tissue distribution within the human fetal brain which can describe not only the variation in the presence of tissues, but also the appearance and complete disappearance of tissues classes over time through a temporally parametrized probability distribution for each voxel in the brain. Specifically, the approach makes use of a polynomial temporal models of tissue probabilities that can capture peaks in the occurrence of given tissue types over time.
In order to construct such a model, repeated serial imaging of the same anatomy is not a viable route. It is not feasible to repeatedly image pregnant women with a short enough time interval to capture subtle changes in morphology and morphometry. As a result, we have to use a large deformation groupwise diffeomorphic registration to bring collections of images of different fetuses with different ages into correspondence and construct models of common variation of tissue types over time.
2 Methods
2.1 MR Image Acquisition and Processing
Fetal MR imaging is performed in our institution on a 1.5T scanner without sedation or contrast agent administration. For each subject, multiple stacks of single-shot fast spin-echo (SSFSE) T2-weighted slice images (in plane resolution 0.469mm × 0.469mm, slice thickness ≈ 3mm) are acquired in the approximately axial, sagittal and coronal plane with respect to the fetal brain. The MR sequence parameters (TR = 4000–8000ms, TE = 91ms) were originally designed for clinical scans and cannot be adjusted for image analysis purposes. After acquisition, image stacks are registered using an intersection-based technique [7] to account for spontaneous fetal movement during scanning and reconstructed into 3D volumes with isotropic resolution 0.469mm × 0.469mm × 0.469mm. Reconstructed volumes are manually segmented into regions of cortical grey matter (GM), white matter (WM), the germinal matrix (GMAT) and ventricles (VENT).
2.2 Groupwise Registration of Segmentations
In order to relate data from multiple fetal subjects with different anatomies and tissue morphologies, a groupwise registration is performed between the tissue segmentations of the training dataset. Each base WM map is first globally aligned to the WM map of one of the subjects using an affine transformation of each anatomy driven by maximization of the normalized mutual information [8]. This provides a starting estimate for further collective alignment of the subjects into a common average space. In order to avoid extreme warping between subjects with and without the germinal matrix in different brain regions, only two basic tissue classes are used for this registration – grey matter (GM), and white matter/germinal matrix combined (WM+GMAT). Each binary tissue region in each subject s is blurred and used to form a smooth map of tissue probability ps(x) = [pGM,s(x) pWM+GMAT,s(x) ] with components varying between 0 and 1 and representing the presence or absence of GM and WM+GMAT, respectively.
A combined registration metric
| (1) |
describing the similarity between each set of subject tissue maps ps(x) and the current group-averaged tissue map p̄(x) is formed from the sum of squared differences in probability of the two tissue classes in each pair of subjects. The derivative of (1) with respect to the spatial mapping us(x) between subject maps is used as a force to drive a symmetric groupwise multiple-elastic registration algorithm. The tissue content difference force is balanced by a linear elastic energy
| (2) |
where μ and λ are constants determining the strength of the regularization. The resulting set of diffemorphic maps provides mapping between the average shape of the group and each of the training anatomies.
2.3 Voxelwise Statistical Modeling
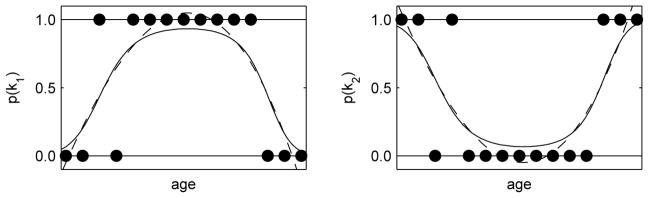
After spatial normalization, we perform statistical modeling to create a probabilistic spatio-temporal model of tissue distribution in the fetal brain. Based on a set of s segmented images corresponding to gestational ages ts, the model uses the age of the fetus as an independent variable and tissue class probabilities pks(x) derived from manual segmentations as target values for each voxel x in the average shape space. As statistical modeling directly in probability space may result in values that are not valid probabilities as shown in Fig. 1, we use an alternative LogOdds representation of probabilities [9].
Fig. 1.
An example of fitting a 3rd degree polynomial to probabilities of two mutually exclusive classes k1 and k2 (dots) directly in probability space (dashed line, note invalid probability values p < 0 and p > 1) and in the space of LogOdds (solid line).
The LogOdds
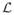 (pks(x)) of a probability pks(x) is the logarithm of the odds between the probability pks(x) and its complement p̄ks(x) = 1 − pks(x).
(pks(x)) of a probability pks(x) is the logarithm of the odds between the probability pks(x) and its complement p̄ks(x) = 1 − pks(x).
| (3) |
In the context of brain segmentation, pks(x) is the probability that voxel x in subject s is assigned to tissue class k whereas its complement p̄ks(x) is the probability of the assignment to any other tissue type. The inverse of the LogOdds function
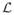 (·) is the standard logistic function
(·) is the standard logistic function
| (4) |
A spatio-temporal atlas is created by temporal modeling of changes in tissue probabilities pks(x) at each point x of the average shape space. For each x, tissue class probabilities pks(x) (k ∈ {1, 2, …, K}) are transfered first to the space of LogOdds using (3). Then, a temporal model is obtained by interpolation between values of lks(x) corresponding to gestational ages ts of the s subjects. Time-related changes in probabilities pk(x) are modeled using a D-th degree polynomial in the space of LogOdds
| (5) |
with coefficients akd(x) found through least squares fitting. To obtain a full probabilistic spatio-temporal model , all need to be transfered back to probability space using (4) and normalized across K tissue classes to assure that are legitimate probabilities.
| (6) |
The spatio-temporal model p̂(x, t) can be used for generation of age-specific probabilistic atlases p̂(x) for any value of gestational age t. Such tissue probability maps may serve as sources of spatially varying priors for atlas-based segmentation of the fetal brain.
3 Results
3.1 Population
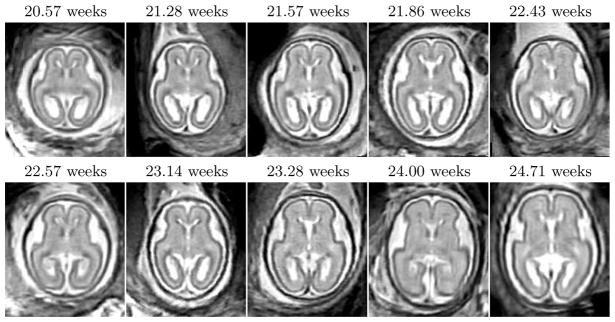
Clinical MR imaging was performed on a 1.5T scanner for a group of 10 fetal subjects with normal brain development. The gestational ages of the fetuses were almost uniformly distributed between 20.57 and 24.71 weeks (Fig. 2). For each subject, a high-resolution volume was reconstructed and manually segmented into regions of grey matter (GM), white matter (WM), the germinal matrix (GMAT) and ventricles (VENT). The resulting tissue label maps were verified by pediatric neuroradiologists with experience in fetal imaging and served as reference for further experiments. The reconstructed volumes were spatially normalized by groupwise registration of their respective manual segmentations as described in Section 2.2. After registration, the tissue label maps were transformed to the average space using nearest-neighbor interpolation.
Fig. 2.
Axial views of rigidly aligned reconstructed MR images of 10 fetal subjects (20.57 – 24.71 weeks GA) demonstrating variability in brain size and shape.
3.2 Model Construction and Generation of Age-specific Atlases
Based on the values of pks(x) and the corresponding gestational ages ts of the 10 fetal subjects, we created a spatio-temporal model p̂(x, t) with D = 0. This is equivalent to simple averaging of spatially normalized tissue label maps as is conventionally performed in atlas construction. The spatially varying tissue probability maps generated from such a model are not time-dependent and are shown in Fig. 3. To model dynamic changes in tissue presence at different gestational ages, we also created spatio-temporal models p̂(x, t) with D = 1 and D = 2 (higher order models with D > 2 were unstable for the available number of training subjects). From these models, we generated sets of age-specific probabilistic atlases p̂(x), examples of which are shown in Fig. 4. Visual inspection of these tissue probability maps confirms that higher order spatio-temporal models correctly capture age-related changes in the morphology of the fetal brain such as the rapidly decreasing volume of the germinal matrix (especially in the areas of the occipital lobes) and shrinking of ventricles with respect to the overall brain volume.
Fig. 3.
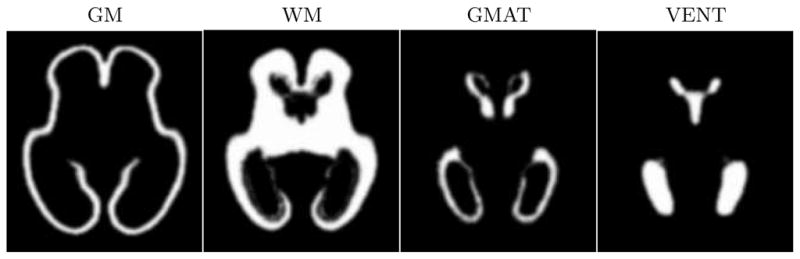
Tissue probability maps generated from a spatio-temporal model with D = 0. This is equivalent to simple averaging of spatially normalized tissue label maps.
Fig. 4.
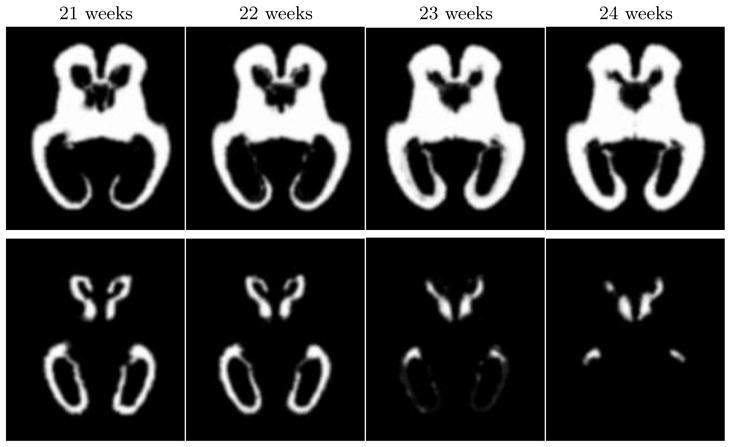
Age-specific tissue probability maps for white matter (top row) and the germinal matrix (bottom row) generated from a spatio-temporal model with D = 2.
3.3 Application to Atlas-based Tissue Segmentation
Based on previously reported results for infants [3] and young children [4], we expected that the use of age-specific atlases may also improve the performance of atlas-based segmentation of the fetal brain. To verify this hypothesis, we transformed MR images of the 10 fetal subjects to the average space and automatically segmented them using conventional EM algorithm [10, 11]. For each subject, we generated age-matched probabilistic atlases p̂(x) from spatio-temporal models p̂(x, t) with D = 0, D = 1 and D = 2 and used them as sources of spatially-varying tissue priors to drive the segmentation process.
For quantitative evaluation, we calculated the values of the Dice similarity coefficient (DSC) [12] between the results of automatic segmentation and respective manual segmentations of each subject. Table 1 presents the average DSC performance of automatic EM segmentation with probabilistic atlases generated from spatio-temporal models with D = 0, D = 1 and D = 2 for four different types of fetal brain structures. For D = 1 and D = 2, the significance of DSC improvements with respect to D = 0 was determined by a paired t-test.
Table 1.
Average DSC performance of atlas-based EM segmentation of 10 fetal subjects with probabilistic atlases generated from spatio-temporal models of degree D. The p values represent the significance of improvement vs. simple averaging (D = 0).
| Structure | D = 0 | D = 1 | D = 2 | ||
|---|---|---|---|---|---|
| DSC | DSC | p | DSC | p | |
| GM | 0.834 ± 0.014 | 0.835 ± 0.013 | 0.168 | 0.836 ± 0.013 | 0.180 |
| GMAT | 0.642 ± 0.131 | 0.752 ± 0.069 | 0.012 | 0.761 ± 0.081 | 0.011 |
| WM | 0.905 ± 0.012 | 0.917 ± 0.014 | 0.038 | 0.918 ± 0.014 | 0.034 |
| VENT | 0.857 ± 0.040 | 0.862 ± 0.029 | 0.411 | 0.867 ± 0.029 | 0.099 |
For all degrees of the spatio-temporal model, automatic atlas-based EM segmentation yields acceptable performance (DSC > 0.8) for grey matter, white matter and ventricular CSF. Similar DSC values were previously reported for automatic brain segmentation in neonates [1, 2] and infants [3]. The inferior average performance for the germinal matrix (DSC = 0.642) for D = 0 arises mainly from oversegmenting of this tissue type for subjects older than 23 weeks. For these subjects, up to 8% of voxels actually representing white matter is incorrectly assigned to GMAT, bringing down the average performance for this subgroup (DSC = 0.475).
The use of age-specific atlases generated from spatio-temporal models with D = 1 and D = 2 improves the DSC performance for all four brain structures and statistically significant changes can be observed for the germinal matrix and white matter. Moreover, the standard error for GMAT is reduced almost by half, from 0.131 for D = 0 to 0.069 for D = 1, thanks to more consistent performance of GMAT segmentation across all gestational ages.
4 Conclusions
We presented an approach to building a spatio-temporal model of tissue distributions in the developing fetal brain which can incorporate both developed tissues as well as transient tissue classes such as the germinal matrix. The model is created from a set of manual segmentations through groupwise registration and voxelwise non-linear modeling of tissue class membership, that allows us to represent the appearance as well as disappearance of brain structures over time.
Based on the spatio-temporal model, we generated a set of age-specific tissue probability maps and used them as sources of tissue priors for atlas-based EM segmentation. Experiments performed using clinical MR scans of 10 fetal subjects demonstrated that the use of age-specific atlases improves the performance of automatic brain segmentation as measured by the average DSC values.
The presented study will be extended by spatio-temporal modeling of tissue contrast changes in fetal MR images. This will allow us to create age-specific synthetic MR images that can be used as targets for registration of new subjects to the average shape space before they can be segmented with age-specific probabilistic atlases generated from the spatio-temporal model.
Acknowledgments
This work was primarily funded by NIH Grant R01 NS 55064. The work of Orit A. Glenn and imaging studies were funded by NIH Grant K23 NS 52506.
Contributor Information
Piotr A. Habas, Email: piotr.habas@ucsf.edu.
Kio Kim, Email: kio.kim@ucsf.edu.
Colin Studholme, Email: colin.studholme@ucsf.edu.
References
- 1.Prastawa M, Gilmore JH, Lin W, Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med Image Anal. 2005;9(5):457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Xue H, Srinivasan L, Jiang S, Rutherford M, Edwards AD, Rueckert D, Hajnal JV. Automatic segmentation and reconstruction of the cortex from neonatal MRI. Neuroimage. 2007;38(3):461–477. doi: 10.1016/j.neuroimage.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 3.Murgasova M, Dyet L, Edwards D, Rutherford M, Hajnal J, Rueckert D. Segmentation of brain MRI in young children. Acad Radiol. 2007;14(11):1350–1366. doi: 10.1016/j.acra.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Yoon U, Fonov VS, Perusse D, Evans AC. The effect of template choice on morphometric analysis of pediatric brain data. Neuroimage. 2009;45(3):769–777. doi: 10.1016/j.neuroimage.2008.12.046. [DOI] [PubMed] [Google Scholar]
- 5.Studholme C, Cardenas VA, Weiner MW. Multiscale image and multiscale deformation of brain anatomy for building average brain atlases. Medical Imaging: Image Processing Proc SPIE. 2001;4322 [Google Scholar]
- 6.Prayer D, Kasprian G, Krampl E, Ulm B, Witzani L, Prayer L, Brugger PC. MRI of normal fetal brain development. Eur J Radiol. 2006;57(2):199–216. doi: 10.1016/j.ejrad.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Kim K, Hansen MF, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Intersection-based registration of slice stacks to form 3D images of the human fetal brain. Proc. 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro; 2008. pp. 1167–1170. [Google Scholar]
- 8.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognit. 1999;32(1):71–86. [Google Scholar]
- 9.Pohl KM, Fisher J, Shenton M, McCarley RW, Grimson WEL, Kikinis R, Wells WM. Medical Image Computing and Computer-Assisted Intervention. Vol. 4191. LNCS; 2006. Logarithm odds maps for shape representation; pp. 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based bias field correction of MR images of the brain. IEEE Trans Med Imaging. 1999;18(10):885–896. doi: 10.1109/42.811268. [DOI] [PubMed] [Google Scholar]
- 11.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of MR images of the brain. IEEE Trans Med Imaging. 1999;18(10):897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- 12.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]