Abstract
Objectives
We examined the molecular basis of the emergence of mupirocin resistance in a methicillin-resistant Staphylococcus aureus (MRSA) strain colonizing a nursing home resident undergoing mupirocin prophylaxis.
Patient and methods
A persistent carrier of mupirocin-susceptible MRSA participated in a trial of mupirocin for nasal decolonization among nursing home residents. During prophylaxis a high-level mupirocin-resistant MRSA emerged in the nasal isolates from this patient. S. aureus and coagulase-negative staphylococci were isolated prior to, during and after 14 days of mupirocin treatment. The staphylococcal isolates and their plasmids were examined by molecular genetic methods.
Results
Allmupirocin-susceptible and-resistant MRSA isolates possessed the same genotype. The patient was also colonized by a single mupirocin-resistant Staphylococcus epidermidis strain. The mupirocin-resistant MRSA and S. epidermidis strains harboured identical plasmids that carried the mupA determinant and genes for conjugative DNA transfer in staphylococci. These plasmids could be transferred in vitro from both clinical isolates to S. aureus RN2677.
Conclusions
The MRSA strain contained a conjugative plasmid expressing mupA that was identical with that found in the S. epidermidis strain which colonized the patient. These findings suggest that transfer of mupA from S. epidermidis to MRSA probably occurred during mupirocin prophylaxis.
Keywords: MRSA, nasal decolonization, coagulase-negative, staphylococci
Introduction
Mupirocin prevents bacterial protein synthesis by inhibiting isoleucyl-tRNA synthetase (IleRS). Application of mupirocin to the anterior nares of patients colonized with Staphylococcus aureus may eliminate the carriage of these organisms and is important in preventing the spread and development of staphylococcal infections.1 However, mupirocin resistance has emerged in staphylococci, including methicillin-resistant S. aureus (MRSA). Low-level resistance (mupirocin MIC = 4–256 mg/L) results from mutational change within the native IleRS,2 whereas high-level, transferable, resistance (mupirocin MIC ≥512 mg/L) is mediated by the mupA determinant that encodes an alternate IleRS.3
Widespread use of mupirocin has resulted in increased prevalence of high-level mupirocin resistance, particularly among coagulase-negative staphylococci (CoNS).4 Accordingly, it has been suggested that mupirocin-resistant CoNS might be an important source of the mupA determinant in MRSA, but evidence for transfer from CoNS during mupirocin prophylaxis has not been obtained.3
During a study to evaluate the efficacy of mupirocin in eliminating nasal carriage of S. aureus among nursing home residents, high-level resistance to mupirocin emerged in the MRSA that persistently colonized a participant in the study.1 Since the patient was also colonized by mupirocin-resistant CoNS, we investigated whether mupirocin resistance in MRSA originated from the mupirocin-resistant CoNS.
Patient and methods
The patient was a 73-year-old male who had a chronic tracheostomy and a 5 year history of recurrent MRSA infections prior to enrolment in an Institutional Review Board-approved study. He was persistently colonized in his nares with mupirocin-susceptible MRSA and received mupirocin ointment (2% in polyethylene glycol base) in the nares twice daily for 14 days. Further nasal samples were obtained during the course of mupirocin prophylaxis, the day after completion (day 15 of study) and a week later (day 22). The susceptibilities of isolates identified as S. aureus or CoNS to methicillin and mupirocin were determined by standard procedures.1,2
Staphylococcal nasal isolates recovered from the patient were also typed by PFGE1 and MRSA isolates were further characterized by spa typing.5 PCR amplification and sequencing of the native ileS genes of MRSA isolates were performed to detect mutations which may confer resistance to mupirocin.2 Filter matings were performed using S. aureus strains RN2677 (novobiocin- and rifampicin-resistant) as the recipient and RN4220 (pGO1) as a control donor.6 Plasmid DNA isolated from staphylococci using the Qiagen Midi Plasmid Extraction Kit (Qiagen, Crawley, UK) was analysed by Southern hybridization with the AlkPhos Direct kit (Amersham Biosciences, Buckinghamshire, UK).
Results and discussion
Within 4 days of administering mupirocin, multiple samples taken from the nares of the patient were negative for MRSA. However, a high-level mupirocin-resistant MRSA isolate was cultured from a nasal sample obtained the day after completion of the 14 day mupirocin regimen. High-level mupirocin-resistant MRSA isolates were again recovered a week later. PFGE typing (data not shown) revealed that all MRSA isolates were genetically identical, a finding confirmed by spa typing5 (data not shown). The isolation of identical PFGE and spa types suggested that mupirocin resistance might have been acquired by the patient’s pre-therapy MRSA strain.
All pre-therapy mupirocin-susceptible CoNS strains were eradicated by intranasal application of mupirocin within 3 days of starting treatment. These susceptible organisms were supplanted by a high-level mupirocin-resistant CoNS that persisted in all subsequent nasal samples obtained from the patient for the remainder of the study. PFGE analysis of CoNS isolates indicated that while mupirocin-susceptible isolates were distinct strains, all mupirocin-resistant CoNS isolates represented the same strain (data not shown). Therefore, the patient probably became colonized by a single mupirocin-resistant CoNS clone, which was identified as Staphylococcus epidermidis.
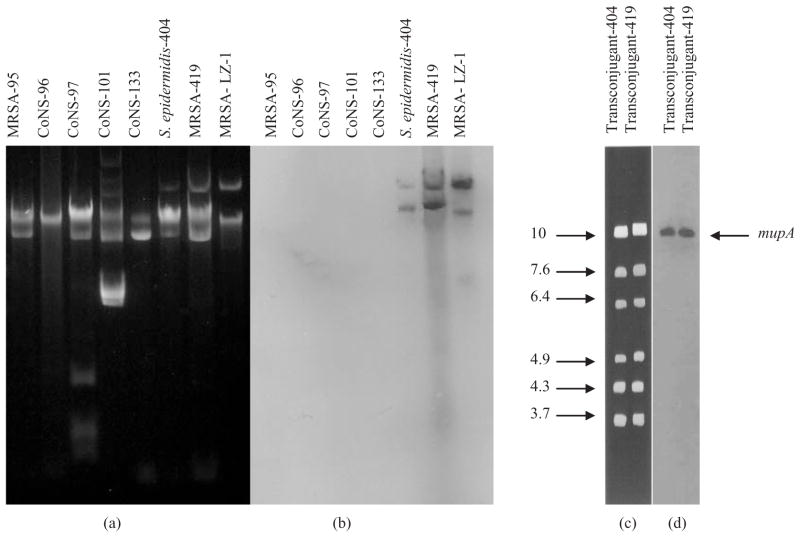
Mutations within ileS may confer resistance to mupirocin in S. aureus.2 However, no mutations were detected within PCR amplicons of the ileS from the mupirocin-resistant MRSA strain. The genetic basis of mupirocin resistance among staphylococcal nasal isolates (CoNS and MRSA) was examined by Southern hybridization using a mupA probe prepared by PCR amplification from the mupA-positive high-level mupirocin-resistant S. aureus strain, LZ-1.7,8 Only isolates that expressed high-level resistance to mupirocin contained plasmids that hybridized to the mupA probe (Figure 1).
Figure 1.
Agarose gel electrophoresis of plasmid DNA extracted from representative mupirocin-susceptible and mupirocin-resistant MRSA and CoNS and Southern hybridization with the mupA probe. (a) Total plasmid DNA from representative staphylococcal nasal isolates; and (b) corresponding hybridization with the mupA probe. MRSA-95, pre-mupirocin prophylaxis mupirocin-susceptible MRSA isolate; CoNS-96, CoNS-97 and CoNS-101, pre-mupirocin prophylaxis mupirocin-susceptible CoNS isolates; CoNS-133, mupirocin-susceptible CoNS isolated on the first day of prophylaxis; S. epidermidis-404, post-mupirocin prophylaxis mupirocin-resistant CoNS isolate; MRSA-419, post-mupirocin prophylaxis mupirocin-resistant MRSA isolate; MRSA-LZ-1, mupA-positive MRSA control. (c) EcoRI restriction digests of plasmid DNA from mupirocin-resistant transconjugant-404 and transconjugant-419 obtained from matings of mupirocin-resistant S. epidermidis-404 and MRSA-419, respectively, with RN2677; and (d) corresponding hybridization with the mupA probe. Approximate sizes of DNA fragments are shown in kilobases (kb).
Transfer of the mupA determinant from clinical isolates was investigated using the mupirocin-resistant S. epidermidis and MRSA strains as donors and S. aureus RN2677 as the recipient. The mupA gene was transferred by conjugation with a frequency of transfer of ~10−6 per donor for MRSA and 10−9 per donor for S. epidermidis. Southern analysis confirmed that mupA was borne on a conjugative replicon, since plasmid DNA obtained from mupirocin-resistant transconjugants resulting from matings of mupirocin-resistant MRSA/S. epidermidis with S. aureus RN2677 hybridized with both a 6.3 kb DNA probe prepared from the conjugal transfer region of plasmid pGO19 and the mupA probe (Figure 1). EcoRI restriction of the plasmids from MRSA and S. epidermidis revealed that they were indistinguishable and ~37 kb in size (Figure 1).
The increasing prevalence of transferable mupirocin resistance among CoNS species could be an important threat to the future use of mupirocin against MRSA.3,4 In this study we obtained direct evidence for the transfer of mupirocin resistance from S. epidermidis to S. aureus in a clinical situation involving the use of mupirocin. Indeed, it appears that after mupirocin prophylaxis had commenced, the patient became colonized with a single mupirocin-resistant S. epidermidis strain that transferred plasmid-borne mupA to mupirocin-susceptible MRSA resulting in mupirocin treatment failure. It is likely that mupirocin-resistant CoNS was a minor endogenous strain that was not detected upon initial screening and emerged under the selective pressure of mupirocin. In a prior study, 90% of our laboratory staff and patients had high-level mupirocin-resistant CoNS emerge during therapy that persisted following treatment.10
Studies of S. aureus colonization in our nursing home residents have shown that 82% of persistent carriers harbour the same strain for many months.11 Therefore, it is unlikely that the patient described here became recolonized with a different MRSA strain following treatment with mupirocin. It is also unlikely that he acquired a high-level mupirocin-resistant strain from other patients enrolled in the study, since mupirocin-resistant MRSA was not isolated from these participants.
In conclusion, it appears that high-level mupirocin resistance was acquired by the patient’s pre-therapy MRSA strain through conjugative transfer of mupirocin resistance from S. epidermidis during nasal decolonization prophylaxis with mupirocin.
Acknowledgments
J. G. H. acknowledges PhD funding from the Association of Commonwealth Universities, UK. S. F. B. and L. M. acknowledge funding from the National Institute on Aging (NIA) and Claude D. Pepper Older Americans Independence Centers (AG 08808) and advice from Dr Carol Kauffman. Career development funding to L. M. is provided by NIA (K23 AG022463). Mupirocin was a gift from GlaxoSmithKline Pharmaceuticals (Harlow, UK)
Footnotes
Transparency declarations
The authors have no professional or personal financial conflicts to declare that would have unduly influenced the results and conclusions of this research.
References
- 1.Mody L, Kauffman CA, McNeil SA, et al. Mupirocin-based decolonization of Staphylococcus aureus carriers in residents of 2 long-term care facilities: a randomized, double-blind, placebo-controlled trial. Clin Infect Dis. 2003;37:1467–74. doi: 10.1086/379325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurdle JG, O’Neill AJ, Chopra I. The isoleucyl-tRNA synthetase mutation V588F conferring mupirocin resistance in glycopeptide-intermediate Staphylococcus aureus is not associated with a significant fitness burden. J Antimicrob Chemother. 2004;53:102–4. doi: 10.1093/jac/dkh020. [DOI] [PubMed] [Google Scholar]
- 3.Cookson BD. The emergence of mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J Antimicrob Chemother. 1998;41:11–8. doi: 10.1093/jac/41.1.11. [DOI] [PubMed] [Google Scholar]
- 4.Deshpande LM, Fix AM, Pfaller MA, et al. Emerging elevated mupirocin resistance rates among staphylococcal isolates in the SENTRY Antimicrobial Surveillance Program (2000): correlations of results from disk diffusion, E test and reference dilution methods. Diagn Microbiol Infect Dis. 2002;42:283–90. doi: 10.1016/s0732-8893(01)00328-5. [DOI] [PubMed] [Google Scholar]
- 5.Harmsen DH, Claus WW, Rothganger J, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003;41:5442–8. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Archer GL, Johnston JL. Self-transmissible plasmids in staphylococci that encode resistance to aminoglycosides. Antimicrob Agents Chemother. 1983;24:70–7. doi: 10.1128/aac.24.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kauffman CA, Terpenning MS, He X, et al. Attempts to eradicate methicillin-resistant Staphylococcus aureus from a long-term care facility with the use of mupirocin ointment. Am J Med. 1993;94:371–8. doi: 10.1016/0002-9343(93)90147-h. [DOI] [PubMed] [Google Scholar]
- 8.Janssen DA, Zarins LT, Schaberg DR, et al. Detection and characterization of mupirocin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1993;37:2003–6. doi: 10.1128/aac.37.9.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas WD, Archer GL. Identification and cloning of the conjugative transfer region of Staphylococcus aureus plasmid pGO1. J Bacteriol. 1989;171:684–91. doi: 10.1128/jb.171.2.684-691.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley S, Cinti S, Ramsey M, et al. Mupirocin resistance (MUP-R) in Staphylococcus aureus and coagulase negative staphylococci (CNS). Abstracts of the Ninety-fifth Annual Meeting of the American Society for Microbiology; Washington, DC. Washington, DC, USA: American Society for Microbiology; 1995. p. Abstract A-29.p. 148. [Google Scholar]
- 11.Bradley SF, Terpenning MS, Ramsey MA, et al. Methicillin-resistant Staphylococcus aureus: colonization and infection in a long-term facility. Ann Intern Med. 1991;115:417–22. doi: 10.7326/0003-4819-115-6-417. [DOI] [PubMed] [Google Scholar]