Abstract
To provide multiple conjugating sites on cyclic peptides for their increasing biomedical applications, a tailed cyclic RGD peptide, c[RGDfE(GGGKK-NH2)] was designed with c(RGDfE) linked through Glu to a tail consisting of a spacer of three Gly residues and a linker of two Lys residues. The spacer is used to increase the mobility and binding ability of the c(RGDfE) ligand, and the linker is used to proved multiple active sites for conjugating other molecules or biomaterials. We found that the sequence of Glu(Gly)-OAll leads to glutarimide formation, which disrupts the formation of cyclic RGD peptides. However, our results show that glutarimide formation is sequence dependent and can be inhibited by incorporating an amino acid like Lys(Boc) with steric hindrance from the protecting group. To prevent glutarimide formation, Ser(tBu) was used to replace the glycine in the GGG spacer adjacent to the residue of Glu, and a tailed cyclic RGD peptide, c[RGDfE(SGGKK-NH2)] was successfully obtained.
Keywords: Glutarimide formation, Cyclic RGD peptide, Solid-phase peptide synthesis, Glutamic acid, Allyl protecting group
INTRODUCTION
Cyclic peptides are important synthetic targets because of their importance in biomedical uses as cell-specific adhesive ligands, protein mimics, antibiotics and other therapeutic agents [1–3]. The constraint imposed by cyclization results in increased stability in solution and resistance to proteolysis, as well as higher specificity and enhanced biological activity, in comparison with linear analogues [4,5]. Cyclic peptides can be synthesized by cyclizing linear peptides either in solution or on resin. The on-resin method has been more efficient to prepare cyclic peptides than the solution-phase approach. The principle of on-resin cyclization is based on anchoring the peptide to the resin through a side chain and applying orthogonal protecting groups for ring closure. It possesses advantages, such as time-saving, less side reactions and easy purification.
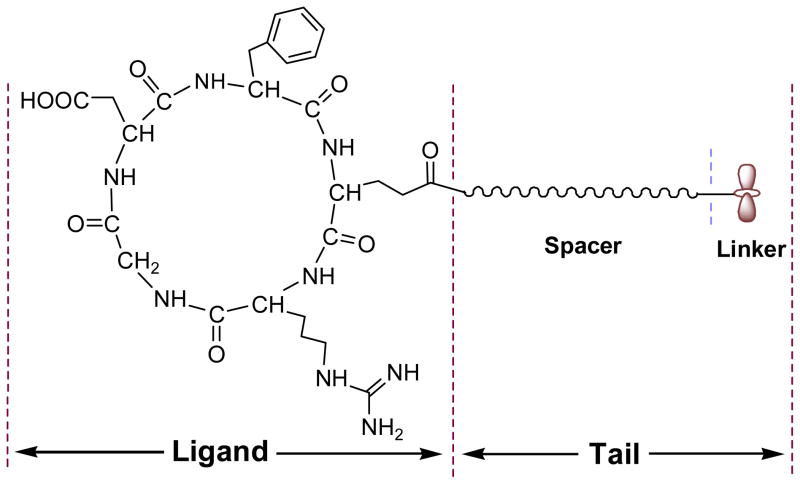
Cyclic RGD peptides, like cyclo(Arg-Gly-Asp-D-Phe-Val) [c(RGDfV)], efficiently compete with native adhesion proteins like vitronectin and fibronectin for specific binding to integrins [6]. To provide conjugating sites for their increasing applications in drug delivery, tumor imaging, surface modification and tissue engineering [7–10], a tailed cyclic peptide of c[RGDfE(tail)] was designed in our laboratory with c(RGDfE) linked to a tail consisting of a spacer and a linker, as shown in Figure 1. The spacer is used to increase the mobility of the c(RGDfE) ligand, which may lead to enhanced binding ability. The linker may consist of multiple active sites for conjugating with other molecules or biomaterials. For example, a tailed cyclic peptide with a linker consisting of multiple Lys residues, has the potential use for coupling with acryloyl-PEG-NHS to make bioactive PEG diacrylate (PEGDA) [11]. Compared with c(RGDfK) with limited spacer length and only one free amino group on Lys for conjugation, the design of tailed cyclic RGD peptides has the advantage to control the flexibility through the spacer and provide multiple conjugating sites through the linker.
Figure 1.
Tailed cyclic peptide of c[RGDfE(tail)] with c(RGDfE) linked to a tail consisting of a spacer and a linker.
Allylic groups have been used as the common orthogonal protecting groups for on-resin cyclization with Fmoc strategy [12,13]. For example, allyl (All) and allyloxycarbonyl (Aloc) groups, for protecting carboxylic and amino groups, respectively, enable the construction of a lactam bridge selectively between carboxylic and amino groups while the peptide is still attached to the resin. One major problem with using aspartic acid for orthogonal peptide synthesis is the formation of aspartimide [14–17]. Glutamic acid has a similar structure to aspartic acid. However, to the best of our knowledge, there is little research on studying glutarimide formation[18,19].
Here, we report on unexpected glutarimide formation in the orthogonal solid-phase synthesis of tailed cyclic peptides using Fmoc-protected glutamic acid α-allyl ester (Fmoc-Glu-OAll). Initially, c[RGDfE(GGGKK-NH2)] was designed with c(RGDfE) linked through Glu to a tail consisting of a spacer of three Gly residues (GGG) and a linker of two Lys residues (KK). We found that the sequence of Glu(Gly)-OAll leads to glutarimide formation, which disrupts the formation of cyclic RGD peptides. MALDI-TOF MS and 1H NMR were used to confirm glutarimide formation. The prevention of glutarimide formation was studied by incorporating amino acids with steric hindrance next to Glu.
MATERIALS AND METHODS
Fmoc-protected amino acids were purchased from AnaSpec Inc. (San Jose, CA). PAL resin with a loading of 0.36 mmol/g was purchased from PE Biosystems (Foster City, CA). HATU was purchased from GenScript Corp. (Piscataway, NJ). The cleavage cocktail of Reagent K was prepared prior to use according to TFA/phenol/H2O/EDT/TIPS (85/5/5/2.5/2.5). Mass spectrometry was performed on a Bruker Biflex III MALDI-TOF mass spectrometer by dissolving the sample and the matrix of 2,5-dihydroxybenzoic acid in 1:1 (v/v) ethanol and water, and mass spectra were typically accumulated from 200 laser shots. 1H NMR spectra were recorded on a Bruker 400 MHz spectrometer.
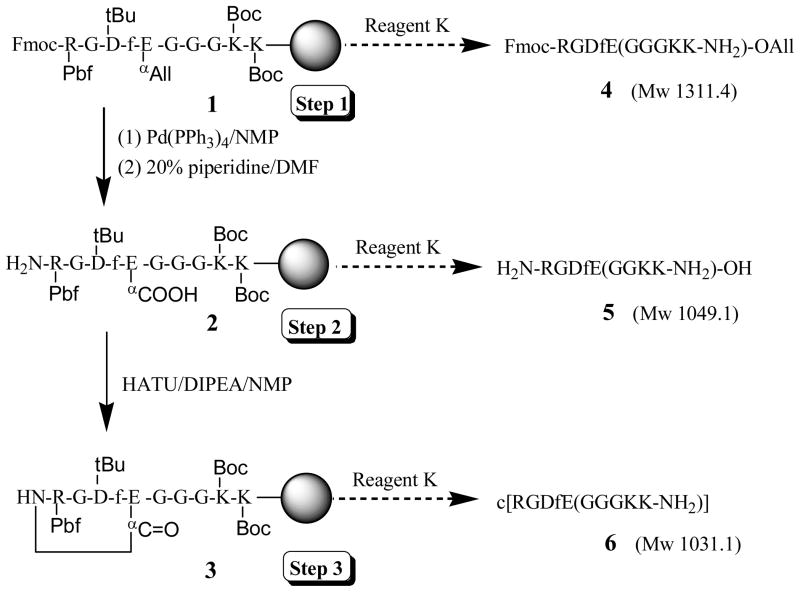
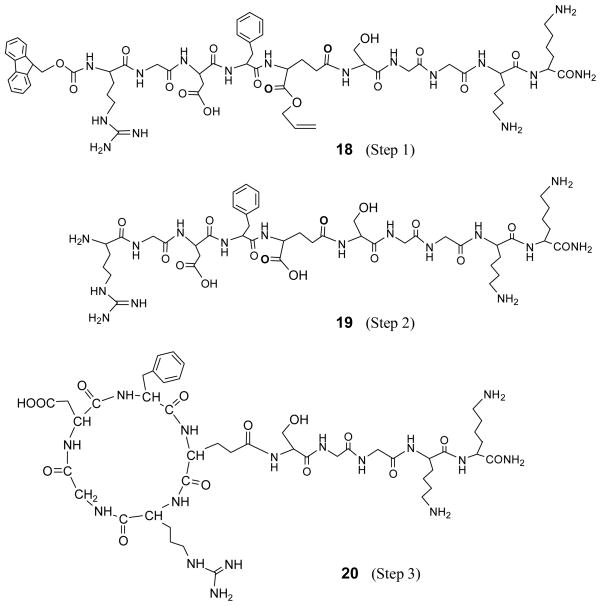
Peptides were synthesized by a three-step synthesis method in a 0.25 mmol scale with PAL resin to produce an amide C-terminus on a 433A automatic peptide synthesizer (Applied Biosystems). An example for synthesizing designed c[RGDfE(GGGKK-NH2)] was shown in Scheme 1. Standard Fmoc strategy was used in Step 1. Removal of the allyl group in Step 2 was performed manually on Peptide Synthesizer by three equivalents of Pd(PPh3)4 in a mixture solvent of acetic acid/NMM/DCM (1.6/0/8/30) for 2 h, followed by washing sequentially by 0.5% DEDTC in DIPEA/NMP, DCM and NMP. Then, 20% piperidine in DMF was transferred to the reaction vessel to remove Fmoc. The resulting linear peptides were cyclized in Step 3 using four equivalents of HATU in DIPEA/NMP (1:9). Peptides were cleaved by Reagent K at room temperature for 2–3 h, followed by filtration, precipitation in cold ether, centrifugation and drying under vacuum. Crude peptides were further purified by reverse phase-HPLC on a Waters 2690 Alliance system equipped with a Waters 2487 UV detector and an Alltech Hyperprep PEP 100A C18 column (250 mm × 22 mm, 8 μm), using a 30–50% acetonitrile gradient in water with 0.01% TFA (v/v).
Scheme 1.
Three-step synthesis of designed c(RGDfE) linked with a tail of GGGKK
RESULTS AND DISCUSSION
In the initial experiment, a spacer of three Gly residues (GGG) was used to extend the linker out, and a linker of two Lys residues (KK) was used to provide two free amine groups as active sites for further conjugation with other biomaterials. The solid-phase synthesis of tailed cyclic peptides included three major steps, as shown in Scheme 1. First, a protected linear peptide on PAL resin (1) was synthesized. Then, allyl and Fmoc groups were removed sequentially by Pd(PPh3) and piperidine to produce a deprotected linear peptide on the resin (2). Finally, cyclization of 2 was carried out using HATU for the expected formation of the cyclic peptide of c[RGDfE(GGGKK-NH2)] on the resin (3). Theoretically, three free peptides, including an Fmoc/allyl protected linear peptide (4, Mw 1311.4), a deprotected linear peptide (5, Mw 1049.1) and a cyclic peptide (6, Mw 1031.1) would be obtained from their cleavage from the peptide-bound resins 1–3, respectively, by using Reagent K containing 85% TFA.
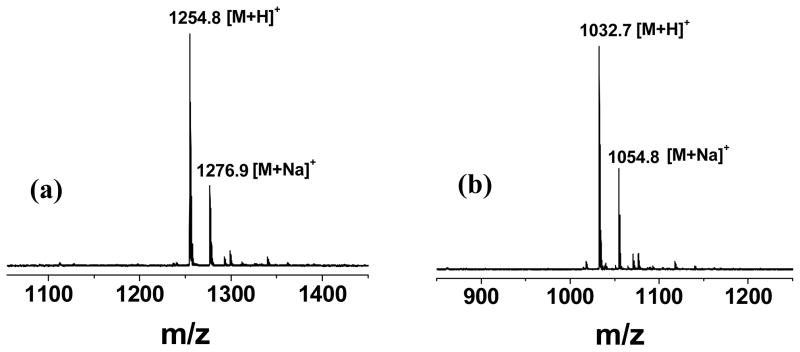
MALDI-TOF MS analysis was performed to verify the peptides obtained in each step in Scheme 1. Unexpectedly, as shown in Figure 2, the Mw of the peptides obtained in Steps 1 and 2 were 1253.8 and 1031.3, respectively. They didn’t match the desirable structures of peptides 4 and 5. Further analysis shows that the Mw of the obtained peptide in Step 1 is 58 less than the expected structure of 4, which is equal to the Mw of allyl alcohol. This indicates the allyl group may be eliminated during Step 1. The peptides obtained in Step 3 show two major products with Mw of 1083.8 and 1115.9, with ΔMw23 of 52.1 and 84.5, respectively. We did combinational calculations including adducts with the possible hydrolytic products from HATU, but still didn’t find a satisfied explanation due to the complexity of the hydrolysis of HATU [20,21].
Figure 2.
MALDI-TOF MS spectra of peptides obtained in Step 1 (a) and Step 2 (b) in the three-step synthesis of peptides with a GGG spacer.
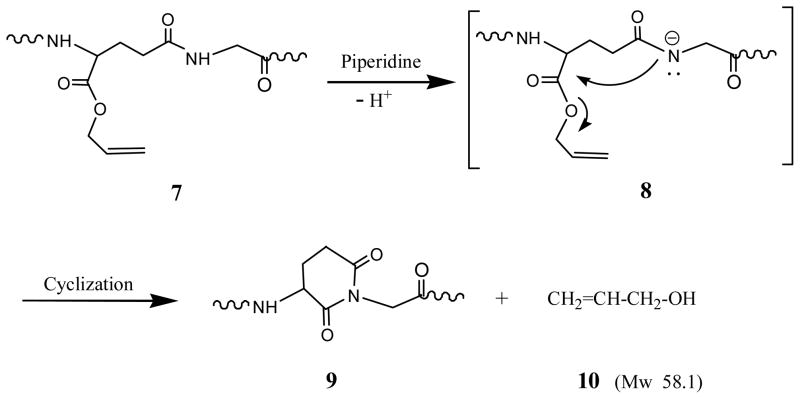
To confirm the elimination of the allyl group, 1H NMR analysis was carried out. Fmoc-Glu-OAll in CDCl3 shows two characteristic protons peaks for the double bond of the ally group at 5.25 ppm (2H, -CH=CH2) and 5.95 ppm (1H, -CH=CH2), which disappeared in the NMR spectrum of the peptide in D2O obtained in Step 1 in Scheme 1. No allyl proton signals were observed from the peptide obtained in Step 1 in Scheme 1, suggesting the elimination of the allyl group in this step. This is also consistent with ΔMw12 of 222 (Table 1), which equals the molecular weight of one Fmoc leaving group. As we know, aspartimide is easy to form when using the sequence of Asp-Gly in peptide synthesis. Our results indicate that the sequence of Glu(Gly)-OAll (7), as shown in Scheme 2, follows the same trend to form a cyclic imide, glutarimide (9). In the transition state (8), the attack of the NH group of Gly on the O-allyl protected α-carboxyl group of Glu is catalyzed by bases like piperidine. The leaving of the allyl group and the closure of a six-member ring proceed simultaneously to form glutarimide with a structure of 3-amino piperidine 2,5-dione (Apd). The Mw of the resulting Apd-Gly (9) is 58 less than that of Glu(Gly)-OAll due to the elimination of one molecule of allyl alcohol (10), which is consistent with the Mw difference observed in Step 1.
Table 1.
Mw of peptides obtained in the three-step synthesis of tailed cyclic RGD peptides of c[RGDfE(tail)].
| Tail | Mw from MALDI-TOF MS | ΔMw12a | ΔMw23b | ||
|---|---|---|---|---|---|
| Step 1 | Step 2 | Step 3 | |||
| GGGKK | 1253.8 | 1031.7 | N/Ac | 222.1 | N/Ac |
| KK | 1139.8 | 878.0 | 860.1 | 261.8 | 17.9 |
| SGGKK | 1340.8 | 1078.8 | 1060.7 | 262.0 | 18.1 |
Mw difference between the peptides obtained in Steps 1 and 2.
Mw difference between the peptides obtained in Steps 2 and 3.
Not applicable.
Scheme 2.
Mechanism of glutarimide formation in Glu(Gly)-OAll
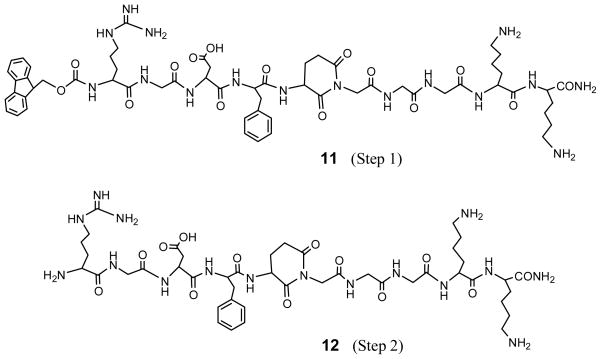
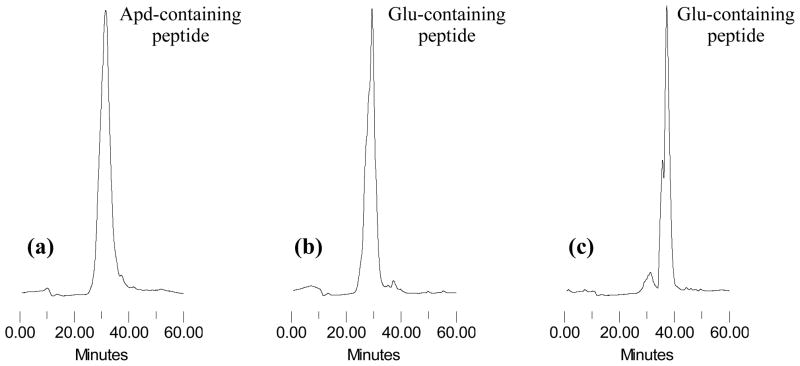
Based on the above analysis, the actual structures of the peptides formed in Steps 1 and 2 in Scheme 1 are 11 and 12, as shown in Figure 3. They have an Apd unit instead of a Glu residue. HPLC analysis, as shown in Figure 4a demonstrates that Fmoc-RGDfE(GGGKK)-OAll resulted in a quantitative Apd formation (100%) in Step 1, and the yield results were listed in Table 2. Mass spectra (Figure 2) shows that 12 is the major products in Step 2, and there were no further addition products with piperidine. Thus, glutarimide is stable in basic conditions like piperidine due to its six-member ring structure, but 12 may react with byproducts from HATU hydrolysis to form peptide adducts in Step 3 [19]. Glutarimide formation from the sequence of Glu(Gly)-OAll in Step 1 results in no carboxylic group available from Glu in 12, and thereafter disrupts the formation of cyclic RGD peptides between Arg and Glu in Step 3. This analysis provides a reasonable explanation on what happened in the orthogonal solid-phase synthesis of designed cyclic RGDfE peptides with a GGGKK tail through Glu.
Figure 3.
Peptide structures obtained in Step 1 (11) and Step 2 (12) in the three-step synthesis of designed c(RGDfE) with a GGG spacer.
Figure 4.
HPLC profiles of crude products obtained in Step 1 in synthesis of cyclic RGDfE peptides with tails of GGGKK (a), KK (b), and SGGKK (c).
Table 2.
Peptide yields in the first step synthesis of tailed cyclic RGD peptides determined by HPLC.
| Peptide | Expected | % Glu-containing | % Apd-containing | Found |
|---|---|---|---|---|
| Mw | peptide | peptide | Mw | |
| Fmoc-RGDfE(GGGKK)-OAll | 1311.4 | 0 | 100a | 1253.5 |
| Fmoc-RGDfE(KK)-OAll | 1140.0 | 99.1 | 0 | 1139.8 |
| Fmoc-RGDfE(SGGKK)-OAll | 1341.4 | 93.1 | 0 | 1341.3 |
Mw of Apd-containing peptide is 1253.4
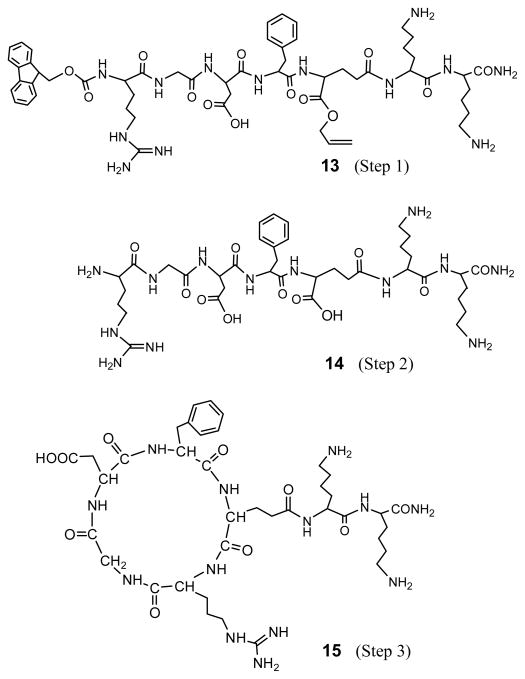
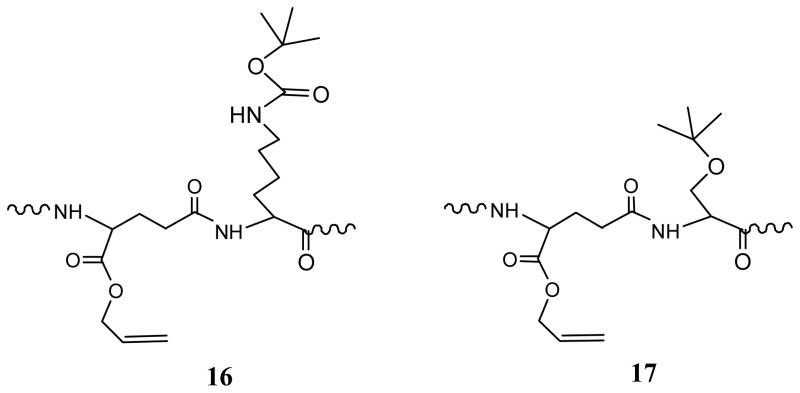
A control peptide, c[RGDfE(KK-NH2)] without the GGG spacer, was synthesized in the similar three-step synthesis as shown in Scheme 1, in order to elucidate the sequence effect of glycine on glutarimide formation. Mass analysis shows that all three peptides had the Mw as expected. The Mw difference between 13 and 14 (ΔMw12) is 262, equal to one Fmoc group plus one allyl group, while the Mw difference between 13 and 15 (ΔMw23) is 18, equal to one molecule of water (Table 1). These results show that both Fmoc and allyl groups remained in Step 1 and were removed in Step 2, and formed a ring structure in Step 3. Their structures are shown in Figure 5. HPLC analysis of the peptides obtained in Step 1 also shows no glutarimide formation, as shown in Figure 4b. It demonstrates that the hindrance from the Boc protecting group, as shown in Figure 6 (16), can prevent the NH of lysine to attack the O-allyl protected carboxyl of Glu, leading to no glutarimide formation in Step 1 (Table 2). Therefore, glutarimide formation is sequence dependent and can be inhibited by an amino acid with steric hindrance.
Figure 5.
Peptide structures obtained in the three-step synthesis of designed c[RGDfE(KK-NH2)] without the GGG spacer.
Figure 6.
Sequence structures of Glu[Lys(Boc)]-OAll (16) and Glu[Ser(tBu)]-OAll (17).
To prevent glutarimide formation in the sequence of Glu(Gly)-OAll while still preserving a spacer between the ligand and the linker, c[RGDfE(SGGKK-NH2)] was designed by using Ser(tBu) to replace the glycine in the GGG spacer adjacent to the residue of Glu. The sequence of Glu[Ser(tBu)]-OAll (17) (Figure 6) is expected to provide the similar steric hindrance to prevent glutarimide formation as Glu[Lys(Boc)]-OAll (16). HPLC analysis of the peptides obtained in Step 1 also shows no glutarimide formation, as shown in Figure 4c. The results from MALDI-TOF mass analysis (Tables 1 and 2) show that the Fmoc and allyl groups were still remained in the Step 1 peptide, and HPLC analyssi without glutarimide formation, as shown in Figure 7. The peptides 18–20 obtained in the three-step synthesis had the expected Mw with ΔMw12 of 262 and ΔMw23 of 18. It indicates that the tBu protective group of serine, like the Boc protective group of lysine, can provide steric hindrance to prevent glutarimide formation. As a result, cyclic RGD peptide 20 with a SGGKK tail was formed in Step 3 from the cyclization of 19 between Arg and Glu.
Figure 7.
Peptide structures obtained in the three-step synthesis of c[RGDfE(SGGKK-NH2)].
CONCLUSIONS
We present here the occurrence of glutarimide formation in orthogonal solid-phase synthesis of cyclic RGD peptides linked to a GGGKK tail using Fmoc-Glu-OAll. The Glu(Gly)-OAll sequence leads to glutarimide formation, which disrupts the formation of cyclic RGD peptides. The results show that glutarimide formation is sequence dependent and can be prevented by incorporating a sterically hindered amino acid like Lys(Boc) adjacent to the residue of Glu. To prevent glutarimide formation while still preserving a spacer between the c(RGDfE) ligand and the linker of KK, Ser(tBu) was used to replace the glycine in the GGG spacer adjacent to the residue of Glu, and a tailed cyclic RGD peptide, c[RGDfE(SGGKK-NH2)] was successfully obtained.
Acknowledgments
This project was supported by the National Institutes of Health (Grant EB002067). We gratefully thank Faina Kligman and Eric H. Anderson for some valuable discussion and the facilities provided by Center for Cardiovascular Biomaterials
Abbreviations
As recommended in J. Pep. Sci. 2006; 12: 1-12, with the following additions and variations:
- All
allyl
- Aloc
allyloxycarbonyl
- Apd
3-amino piperidine 2,5-dione
- c(RGDfE)
cyclo(Arg-Gly-Asp-D-Phe-Glu)
- DEDTC
sodium diethyldiethio-carbamate trihydrate
- DIPEA
diisopropylethylamine
- EDT
1,2-ethanedithiol
- NHS
N-hydroxyl succinimide
- NMP
1-methyl-2-pyrrolidinone
- Pd(PPh3)4
tetrakis-(triphenyl phosphine) palladium (0)
- RGD
Arg-Gly-Asp
- tBu
t-butyl ether
- TIPS
triisopropylsilane
References
- 1.Lambert JN, Mitchell JP, Roberts KD. The synthesis of cyclic peptides. J Chem Soc, Perkin Trans. 2001;1:471–484. [Google Scholar]
- 2.Davies JS. The cyclization of peptides and depsipeptides. J Pep Sci. 2003;9:471–501. doi: 10.1002/psc.491. [DOI] [PubMed] [Google Scholar]
- 3.Schaffner P, Dard MM. Structure and function of RGD peptides involved in bone biology. Cell Mol Life Sci. 2003;60:119–132. doi: 10.1007/s000180300008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizo J, Gierasch LM. Constrainted peptides: models of bioactive peptides and protein substructures. Annu Rev Biochem. 1992;61:387–418. doi: 10.1146/annurev.bi.61.070192.002131. [DOI] [PubMed] [Google Scholar]
- 5.Hruby VJ, Al-Obeidi F, Kazmierski W. Emerging approaches in the molecular design of receptor-selective peptide ligands: conformational, topographical and dynamic consideration. Biochem J. 1990;268:249–262. doi: 10.1042/bj2680249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thumshirn G, Hersel U, Goodman SL, Kessler H. Multimeric cyclic RGD peptides as poteintial tools for tumor targeting: solid-phase peptide synthesis and chemoselective oxime ligation. Chem Eur J. 2003;9:2717–2725. doi: 10.1002/chem.200204304. [DOI] [PubMed] [Google Scholar]
- 7.Nasongkla N, Shuai X, Ai H, Weinberg BD, Pink J, Boothman DA, Gao J. cRGD-functionalized polymer micelles for targeted doxorubicin delivery. Angew Chem Int Ed. 2004;43:6323–6327. doi: 10.1002/anie.200460800. [DOI] [PubMed] [Google Scholar]
- 8.Liu S. Radiolabeled multimeric cyclic RGD peptides as integrin αvβ3 targeted radiotracers for tumor imaging. Mol Pharm. 2006;3:472–487. doi: 10.1021/mp060049x. [DOI] [PubMed] [Google Scholar]
- 9.Auernheimer J, Dahmen C, Hersel U, Bausch A, Kessler H. Photoswithched cell adhesion on surfaces with RGD peptides. J Am Chem Soc. 2005;127:16107–16110. doi: 10.1021/ja053648q. [DOI] [PubMed] [Google Scholar]
- 10.Jeschke B, Meyer J, Jonczyk A, et al. RGD-peptides for tissue engineering of articular cartilage. Biomaterials. 2002;23:3455–3463. doi: 10.1016/s0142-9612(02)00052-2. [DOI] [PubMed] [Google Scholar]
- 11.Zhu J, Beamish JA, Tang C, Kottke-Marchant K, Marchant RE. Extracellular matrix-like cell-adhesive hydrogels from RGD-containing poly(ethylene glycol) diacrylate. Macromolecules. 2006;39:1305–1307. [Google Scholar]
- 12.Brase S, Kirchhoff JH, Kobberling J. Palladium-catalysed reactions in solid phase organic synthesis. Tetrahedron. 2003;59:885–939. [Google Scholar]
- 13.Kates SA, Daniels SB, Alberticio F. Automated allyl cleavage for continuous-flow synthesis of cyclic and branched peptides. Anal Biochem. 1993;212:303–310. doi: 10.1006/abio.1993.1334. [DOI] [PubMed] [Google Scholar]
- 14.Tam JP, Riemen MW, Merrifield RB. Mechanisms of aspartimide formation: the effects of protecting groups, acid, base, temperature and time. Pep Res. 1988;1:6–18. [PubMed] [Google Scholar]
- 15.Mergler M, Sax DB, Weiler P, Vorherr T. The aspartimide problem in Fmoc-based SPPS. Part I. J Pep Sci. 2003;9:36–46. doi: 10.1002/psc.430. [DOI] [PubMed] [Google Scholar]
- 16.Flora D, Mo H, Mayer JP, Khan MA, Yan LZ. Detection and control of aspartimide formation in the synthesis of cyclic peptides. Bioorg Med Chem Lett. 2005;15:1065–1068. doi: 10.1016/j.bmcl.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 17.Stathopoulos P, Papas S, Kostidis S, Tsikaris V. α- and β-Aspartyl peptide ester formation via aspartimide ring opening. J Pep Sci. 2005;11:658–664. doi: 10.1002/psc.675. [DOI] [PubMed] [Google Scholar]
- 18.Battersby AR, Robinson JC. Studies on specific chemical fission of peptide links. Part I. The rearrangement of aspartyl and glutamyl peptides. J Chem Soc. 1955;77:259–269. [Google Scholar]
- 19.Feinberg RS, Merrifield RB. Modification of peptides containing glutamic acid by hydrogen fluoride-anisole mixtures. γ-Acylation of anisole or the glutamyl nitrogen. J Am Chem Soc. 1975;97:3485–3757. doi: 10.1021/ja00845a034. [DOI] [PubMed] [Google Scholar]
- 20.Albericio F, Bofill JM, El-Faham A, Kates SA. Use of onium salt-based coupling reagents in peptide synthesis. J Org Chem. 1998;63:9678–9683. [Google Scholar]
- 21.Alsina J, Barany G, Albericio F, Kates SA. Pyrrolidide formation as a side reaction during activation of carboxylic acids by phosphonium salt coupling reagents. Lett Pep Sci. 1999;6:243–245. [Google Scholar]